Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
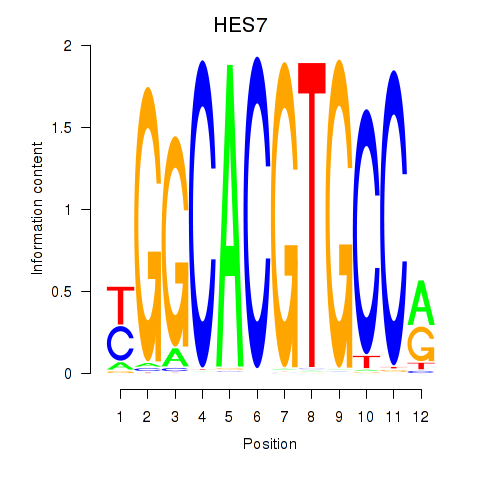
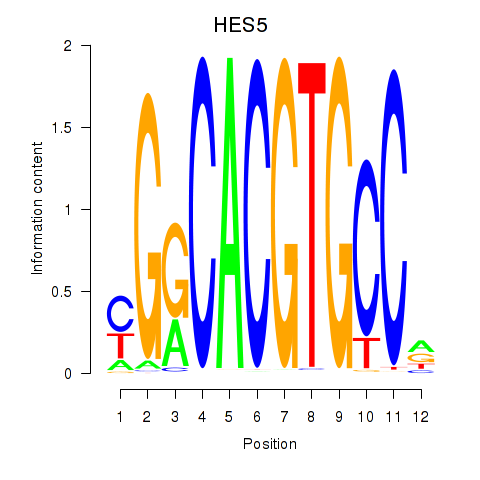
Results for HES7_HES5
Z-value: 0.55
Transcription factors associated with HES7_HES5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HES7
|
ENSG00000179111.4 | hes family bHLH transcription factor 7 |
|
HES5
|
ENSG00000197921.5 | hes family bHLH transcription factor 5 |
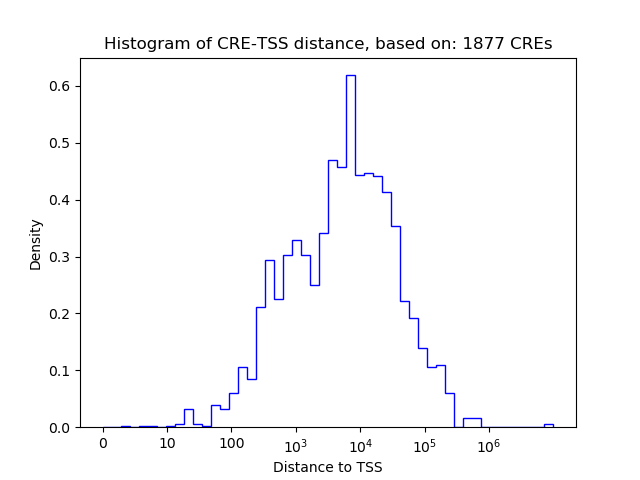
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr1_2473159_2473310 | HES5 | 11550 | 0.089609 | -0.35 | 3.5e-01 | Click! |
| chr1_2472569_2472730 | HES5 | 10965 | 0.090434 | 0.21 | 5.9e-01 | Click! |
| chr1_2466100_2466356 | HES5 | 4544 | 0.109246 | -0.21 | 5.9e-01 | Click! |
| chr1_2471790_2472028 | HES5 | 10225 | 0.091524 | -0.16 | 6.8e-01 | Click! |
| chr1_2472403_2472554 | HES5 | 10794 | 0.090680 | 0.14 | 7.1e-01 | Click! |
| chr17_8030164_8030365 | HES7 | 2854 | 0.116782 | 0.37 | 3.3e-01 | Click! |
| chr17_8025338_8025489 | HES7 | 1989 | 0.150994 | 0.35 | 3.6e-01 | Click! |
| chr17_8026134_8026345 | HES7 | 1163 | 0.265348 | 0.29 | 4.6e-01 | Click! |
| chr17_8026960_8027182 | HES7 | 331 | 0.748773 | 0.25 | 5.2e-01 | Click! |
| chr17_8025866_8026098 | HES7 | 1420 | 0.212818 | 0.22 | 5.8e-01 | Click! |
Activity of the HES7_HES5 motif across conditions
Conditions sorted by the z-value of the HES7_HES5 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
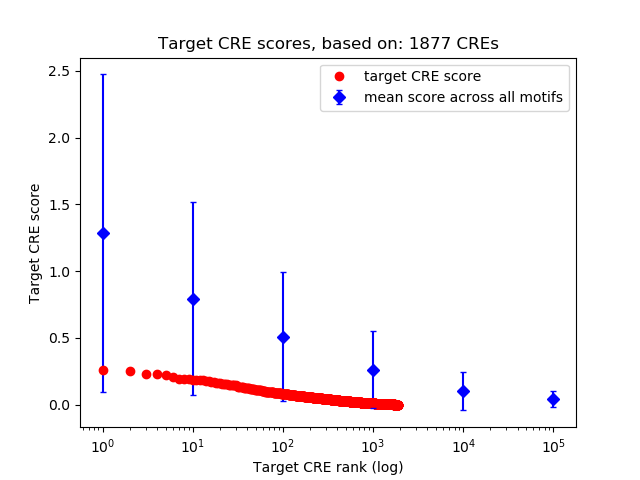
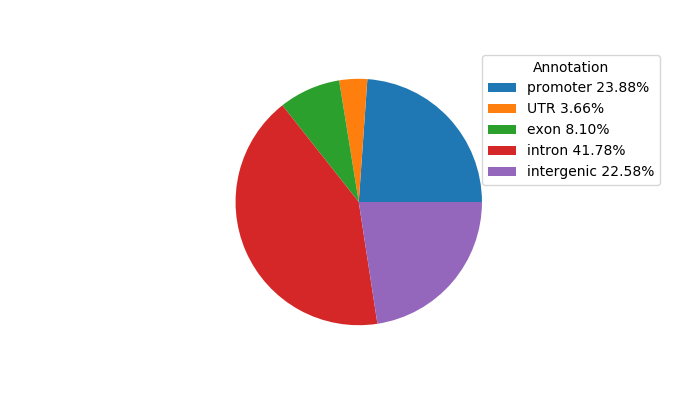
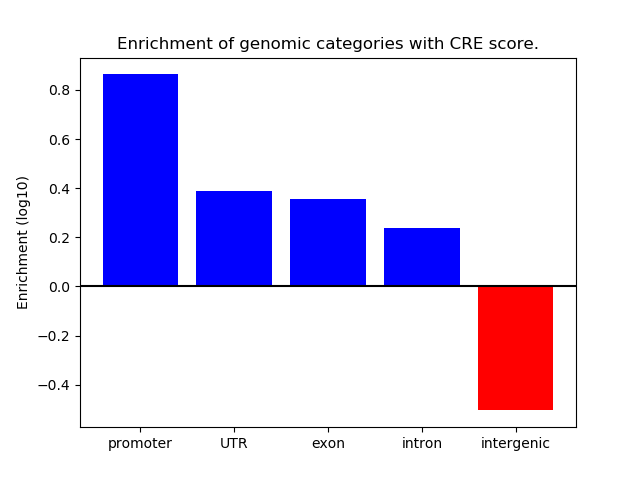
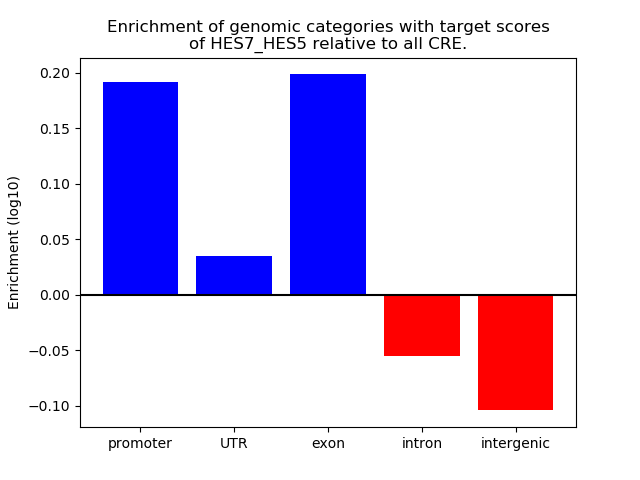
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr7_55604345_55604638 | 0.26 |
VOPP1 |
vesicular, overexpressed in cancer, prosurvival protein 1 |
896 |
0.67 |
| chr22_20785559_20785830 | 0.25 |
SCARF2 |
scavenger receptor class F, member 2 |
6418 |
0.12 |
| chr10_11892041_11892192 | 0.23 |
PROSER2 |
proline and serine-rich protein 2 |
18434 |
0.19 |
| chr5_77161267_77161418 | 0.23 |
TBCA |
tubulin folding cofactor A |
3262 |
0.38 |
| chr19_1418111_1418262 | 0.22 |
CTB-25B13.13 |
|
4286 |
0.08 |
| chr11_17519733_17519884 | 0.20 |
ABCC8 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 8 |
21359 |
0.17 |
| chr1_162342160_162342311 | 0.19 |
C1orf111 |
chromosome 1 open reading frame 111 |
4425 |
0.13 |
| chr17_71994614_71994765 | 0.19 |
RPL38 |
ribosomal protein L38 |
205032 |
0.02 |
| chr14_35586352_35586588 | 0.19 |
KIAA0391 |
KIAA0391 |
4582 |
0.22 |
| chr11_35640709_35641024 | 0.18 |
FJX1 |
four jointed box 1 (Drosophila) |
1131 |
0.58 |
| chr17_19213514_19213665 | 0.18 |
EPN2-AS1 |
EPN2 antisense RNA 1 |
4015 |
0.14 |
| chr6_85484508_85484659 | 0.18 |
TBX18 |
T-box 18 |
10346 |
0.3 |
| chr11_126336020_126336230 | 0.18 |
KIRREL3 |
kin of IRRE like 3 (Drosophila) |
25672 |
0.18 |
| chr1_220962364_220962515 | 0.18 |
MARC1 |
mitochondrial amidoxime reducing component 1 |
2338 |
0.3 |
| chr16_85675576_85675727 | 0.18 |
GSE1 |
Gse1 coiled-coil protein |
12387 |
0.16 |
| chr12_94242890_94243084 | 0.17 |
RP11-887P2.6 |
|
13044 |
0.2 |
| chr6_143771407_143771558 | 0.17 |
ADAT2 |
adenosine deaminase, tRNA-specific 2 |
328 |
0.47 |
| chr19_14544657_14544808 | 0.16 |
PKN1 |
protein kinase N1 |
427 |
0.74 |
| chr10_134496809_134496960 | 0.16 |
INPP5A |
inositol polyphosphate-5-phosphatase, 40kDa |
75454 |
0.1 |
| chr3_81783533_81783684 | 0.16 |
GBE1 |
glucan (1,4-alpha-), branching enzyme 1 |
9172 |
0.33 |
| chr7_74569134_74569285 | 0.16 |
GTF2IRD2B |
GTF2I repeat domain containing 2B |
12305 |
0.24 |
| chr6_3745494_3745645 | 0.16 |
RP11-420L9.5 |
|
5776 |
0.22 |
| chr9_92113326_92114071 | 0.15 |
SEMA4D |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D |
653 |
0.53 |
| chr7_99719791_99720047 | 0.15 |
TAF6 |
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
2455 |
0.09 |
| chrX_40132852_40133003 | 0.15 |
ENSG00000238920 |
. |
11422 |
0.29 |
| chr2_85734499_85734650 | 0.15 |
ENSG00000266577 |
. |
25175 |
0.08 |
| chr1_200983563_200983714 | 0.15 |
KIF21B |
kinesin family member 21B |
8898 |
0.19 |
| chr1_46154147_46154314 | 0.15 |
TMEM69 |
transmembrane protein 69 |
1344 |
0.32 |
| chr11_48029484_48029819 | 0.15 |
AC103828.1 |
|
7756 |
0.21 |
| chr2_240197215_240197647 | 0.15 |
ENSG00000265215 |
. |
29726 |
0.15 |
| chr9_140349393_140349643 | 0.14 |
NSMF |
NMDA receptor synaptonuclear signaling and neuronal migration factor |
357 |
0.79 |
| chr19_935004_935155 | 0.14 |
ARID3A |
AT rich interactive domain 3A (BRIGHT-like) |
2570 |
0.12 |
| chr10_6190085_6190387 | 0.14 |
ENSG00000201581 |
. |
1350 |
0.31 |
| chr5_156627118_156627269 | 0.13 |
ITK |
IL2-inducible T-cell kinase |
19356 |
0.12 |
| chr3_196015117_196015307 | 0.13 |
PCYT1A |
phosphate cytidylyltransferase 1, choline, alpha |
384 |
0.78 |
| chr1_23944860_23945102 | 0.13 |
MDS2 |
myelodysplastic syndrome 2 translocation associated |
8843 |
0.19 |
| chr17_58228988_58229139 | 0.13 |
CA4 |
carbonic anhydrase IV |
1760 |
0.34 |
| chrX_153248886_153249110 | 0.13 |
ENSG00000265176 |
. |
2450 |
0.14 |
| chr22_20120033_20120203 | 0.13 |
ZDHHC8 |
zinc finger, DHHC-type containing 8 |
647 |
0.5 |
| chr16_53489129_53489352 | 0.13 |
RBL2 |
retinoblastoma-like 2 (p130) |
5252 |
0.17 |
| chr5_171556780_171557174 | 0.13 |
ENSG00000266671 |
. |
48997 |
0.13 |
| chr11_63685536_63685687 | 0.12 |
RCOR2 |
REST corepressor 2 |
1295 |
0.32 |
| chr1_2326788_2326939 | 0.12 |
RER1 |
retention in endoplasmic reticulum sorting receptor 1 |
1419 |
0.24 |
| chr10_64843902_64844053 | 0.12 |
ENSG00000199446 |
. |
25922 |
0.19 |
| chr4_114679628_114679779 | 0.12 |
CAMK2D |
calcium/calmodulin-dependent protein kinase II delta |
2521 |
0.44 |
| chr12_47601769_47602062 | 0.12 |
PCED1B |
PC-esterase domain containing 1B |
8137 |
0.22 |
| chr19_1238693_1238844 | 0.12 |
C19orf26 |
chromosome 19 open reading frame 26 |
742 |
0.43 |
| chr1_223402859_223403010 | 0.11 |
TLR5 |
toll-like receptor 5 |
86310 |
0.09 |
| chrX_80065881_80066270 | 0.11 |
BRWD3 |
bromodomain and WD repeat domain containing 3 |
888 |
0.69 |
| chr3_25825314_25825465 | 0.11 |
NGLY1 |
N-glycanase 1 |
347 |
0.75 |
| chr1_9180295_9180446 | 0.11 |
GPR157 |
G protein-coupled receptor 157 |
8813 |
0.17 |
| chr5_14385266_14385417 | 0.11 |
TRIO |
trio Rho guanine nucleotide exchange factor |
94255 |
0.09 |
| chr11_33037755_33037906 | 0.11 |
DEPDC7 |
DEP domain containing 7 |
120 |
0.96 |
| chr10_105789345_105789496 | 0.11 |
COL17A1 |
collagen, type XVII, alpha 1 |
3306 |
0.22 |
| chr12_102165691_102165954 | 0.11 |
ENSG00000222255 |
. |
6535 |
0.11 |
| chr19_41922415_41922566 | 0.11 |
BCKDHA |
branched chain keto acid dehydrogenase E1, alpha polypeptide |
5863 |
0.09 |
| chr4_154380641_154380792 | 0.11 |
KIAA0922 |
KIAA0922 |
6782 |
0.26 |
| chr2_217542751_217542902 | 0.11 |
AC007563.5 |
|
16361 |
0.15 |
| chr16_29167036_29167187 | 0.11 |
CTB-134H23.3 |
|
48345 |
0.11 |
| chr20_55965760_55966107 | 0.10 |
RBM38 |
RNA binding motif protein 38 |
530 |
0.69 |
| chr16_89469756_89470099 | 0.10 |
RP1-168P16.2 |
|
9045 |
0.14 |
| chr5_139973795_139973946 | 0.10 |
ENSG00000200235 |
. |
14618 |
0.08 |
| chr13_22637652_22638073 | 0.10 |
ENSG00000244197 |
. |
273186 |
0.02 |
| chr1_27950347_27950704 | 0.10 |
FGR |
feline Gardner-Rasheed sarcoma viral oncogene homolog |
48 |
0.97 |
| chr5_1513019_1513170 | 0.10 |
LPCAT1 |
lysophosphatidylcholine acyltransferase 1 |
10998 |
0.22 |
| chr4_140476576_140476762 | 0.10 |
SETD7 |
SET domain containing (lysine methyltransferase) 7 |
693 |
0.59 |
| chr6_137144118_137144500 | 0.10 |
PEX7 |
peroxisomal biogenesis factor 7 |
586 |
0.76 |
| chr2_234150995_234151190 | 0.10 |
ATG16L1 |
autophagy related 16-like 1 (S. cerevisiae) |
9125 |
0.15 |
| chr19_42396817_42397275 | 0.10 |
ARHGEF1 |
Rho guanine nucleotide exchange factor (GEF) 1 |
1319 |
0.31 |
| chr4_103737282_103737795 | 0.10 |
UBE2D3 |
ubiquitin-conjugating enzyme E2D 3 |
9161 |
0.15 |
| chr2_232917540_232917691 | 0.10 |
AC105461.1 |
|
37185 |
0.16 |
| chr18_45755490_45755641 | 0.10 |
AC091150.1 |
HCG1818186; Uncharacterized protein |
23107 |
0.19 |
| chr14_104604037_104604479 | 0.10 |
CTD-2223O18.1 |
|
23 |
0.94 |
| chr10_42738180_42738331 | 0.10 |
ENSG00000264398 |
. |
7832 |
0.28 |
| chr6_111203655_111204086 | 0.10 |
AMD1 |
adenosylmethionine decarboxylase 1 |
7092 |
0.17 |
| chr19_54812861_54813012 | 0.09 |
LILRA3 |
leukocyte immunoglobulin-like receptor, subfamily A (without TM domain), member 3 |
2984 |
0.12 |
| chrX_70401426_70401720 | 0.09 |
RP5-1091N2.9 |
|
16452 |
0.12 |
| chr16_81129207_81129532 | 0.09 |
GCSH |
glycine cleavage system protein H (aminomethyl carrier) |
476 |
0.71 |
| chr1_204253068_204253219 | 0.09 |
RP11-203F10.5 |
|
7010 |
0.2 |
| chrX_39928105_39928256 | 0.09 |
BCOR |
BCL6 corepressor |
807 |
0.78 |
| chr7_65366081_65366232 | 0.09 |
VKORC1L1 |
vitamin K epoxide reductase complex, subunit 1-like 1 |
27800 |
0.16 |
| chr10_8306294_8306445 | 0.09 |
GATA3 |
GATA binding protein 3 |
209600 |
0.03 |
| chr14_105166825_105166976 | 0.09 |
INF2 |
inverted formin, FH2 and WH2 domain containing |
7301 |
0.13 |
| chr22_35730206_35730357 | 0.09 |
ENSG00000266320 |
. |
1352 |
0.4 |
| chr6_11332687_11332838 | 0.09 |
NEDD9 |
neural precursor cell expressed, developmentally down-regulated 9 |
49770 |
0.16 |
| chr13_76209699_76209926 | 0.09 |
RP11-173B14.5 |
|
318 |
0.71 |
| chrX_12937770_12937921 | 0.09 |
TLR8-AS1 |
TLR8 antisense RNA 1 |
11393 |
0.19 |
| chr15_22483712_22483863 | 0.09 |
IGHV1OR15-4 |
immunoglobulin heavy variable 1/OR15-4 (pseudogene) |
519 |
0.69 |
| chr17_80171257_80171663 | 0.09 |
RP13-516M14.2 |
|
643 |
0.41 |
| chrX_39179559_39179819 | 0.09 |
ENSG00000207122 |
. |
239151 |
0.02 |
| chr3_86416939_86417090 | 0.09 |
ENSG00000222934 |
. |
5732 |
0.34 |
| chr7_149485831_149485982 | 0.09 |
ZNF467 |
zinc finger protein 467 |
15338 |
0.19 |
| chr2_113925026_113925412 | 0.09 |
PSD4 |
pleckstrin and Sec7 domain containing 4 |
6329 |
0.15 |
| chr19_39892381_39892634 | 0.08 |
ZFP36 |
ZFP36 ring finger protein |
4946 |
0.08 |
| chr19_40225624_40225775 | 0.08 |
CLC |
Charcot-Leyden crystal galectin |
2969 |
0.19 |
| chr3_72158863_72159014 | 0.08 |
ENSG00000212070 |
. |
152641 |
0.04 |
| chr13_46365670_46365821 | 0.08 |
SIAH3 |
siah E3 ubiquitin protein ligase family member 3 |
60126 |
0.12 |
| chr11_44681020_44681182 | 0.08 |
CD82 |
CD82 molecule |
41281 |
0.15 |
| chr8_41734096_41734247 | 0.08 |
ANK1 |
ankyrin 1, erythrocytic |
20109 |
0.18 |
| chr11_9665537_9665688 | 0.08 |
SWAP70 |
SWAP switching B-cell complex 70kDa subunit |
20012 |
0.16 |
| chr13_114862694_114862923 | 0.08 |
RASA3-IT1 |
RASA3 intronic transcript 1 (non-protein coding) |
11487 |
0.22 |
| chr22_38214433_38214603 | 0.08 |
GALR3 |
galanin receptor 3 |
4871 |
0.1 |
| chr14_66308035_66308186 | 0.08 |
CTD-2014B16.3 |
Uncharacterized protein |
163131 |
0.04 |
| chr4_2819205_2819714 | 0.08 |
SH3BP2 |
SH3-domain binding protein 2 |
453 |
0.83 |
| chr20_57268529_57268804 | 0.08 |
NPEPL1 |
aminopeptidase-like 1 |
997 |
0.5 |
| chrX_130027668_130027939 | 0.08 |
ENOX2 |
ecto-NOX disulfide-thiol exchanger 2 |
9375 |
0.3 |
| chr15_55619953_55620104 | 0.08 |
PIGB |
phosphatidylinositol glycan anchor biosynthesis, class B |
8585 |
0.15 |
| chrX_9486353_9486504 | 0.08 |
TBL1X |
transducin (beta)-like 1X-linked |
16555 |
0.3 |
| chr2_75066333_75066484 | 0.08 |
HK2 |
hexokinase 2 |
4111 |
0.3 |
| chr22_50523315_50523466 | 0.08 |
MLC1 |
megalencephalic leukoencephalopathy with subcortical cysts 1 |
298 |
0.86 |
| chr10_82224653_82225144 | 0.08 |
TSPAN14 |
tetraspanin 14 |
5840 |
0.23 |
| chr22_42310292_42310562 | 0.08 |
SHISA8 |
shisa family member 8 |
143 |
0.92 |
| chr5_53831366_53831517 | 0.08 |
SNX18 |
sorting nexin 18 |
17848 |
0.26 |
| chr5_53838074_53838225 | 0.08 |
SNX18 |
sorting nexin 18 |
24556 |
0.25 |
| chr5_6718428_6718667 | 0.08 |
PAPD7 |
PAP associated domain containing 7 |
3802 |
0.32 |
| chr19_40440819_40441084 | 0.08 |
FCGBP |
Fc fragment of IgG binding protein |
418 |
0.82 |
| chr3_130542365_130542516 | 0.08 |
ENSG00000222783 |
. |
11501 |
0.2 |
| chr1_17019441_17019592 | 0.08 |
ENSG00000264742 |
. |
11766 |
0.12 |
| chr15_51476418_51476569 | 0.07 |
CYP19A1 |
cytochrome P450, family 19, subfamily A, polypeptide 1 |
30928 |
0.17 |
| chr9_139836823_139836974 | 0.07 |
FBXW5 |
F-box and WD repeat domain containing 5 |
280 |
0.75 |
| chr9_135092130_135092281 | 0.07 |
ENSG00000221105 |
. |
1943 |
0.41 |
| chr14_106967843_106968018 | 0.07 |
IGHV1-46 |
immunoglobulin heavy variable 1-46 |
142 |
0.82 |
| chr18_74280231_74280382 | 0.07 |
LINC00908 |
long intergenic non-protein coding RNA 908 |
39432 |
0.16 |
| chr15_31659811_31660031 | 0.07 |
KLF13 |
Kruppel-like factor 13 |
1564 |
0.55 |
| chr22_46997813_46997964 | 0.07 |
GRAMD4 |
GRAM domain containing 4 |
18411 |
0.21 |
| chr11_12398290_12398441 | 0.07 |
PARVA |
parvin, alpha |
367 |
0.89 |
| chr7_138435270_138435447 | 0.07 |
ATP6V0A4 |
ATPase, H+ transporting, lysosomal V0 subunit a4 |
23424 |
0.18 |
| chr17_42462131_42462433 | 0.07 |
ITGA2B |
integrin, alpha 2b (platelet glycoprotein IIb of IIb/IIIa complex, antigen CD41) |
406 |
0.76 |
| chr11_64064837_64065015 | 0.07 |
ENSG00000207024 |
. |
1417 |
0.16 |
| chrX_39963823_39964258 | 0.07 |
BCOR |
BCL6 corepressor |
7384 |
0.32 |
| chr7_157205057_157205285 | 0.07 |
DNAJB6 |
DnaJ (Hsp40) homolog, subfamily B, member 6 |
72656 |
0.1 |
| chr10_119303115_119303318 | 0.07 |
EMX2 |
empty spiracles homeobox 2 |
707 |
0.63 |
| chr2_240211896_240212145 | 0.07 |
ENSG00000265215 |
. |
15137 |
0.18 |
| chr6_159192572_159192723 | 0.07 |
ENSG00000265558 |
. |
6862 |
0.21 |
| chr4_113351506_113351657 | 0.07 |
RP11-402J6.1 |
|
84960 |
0.07 |
| chr10_73503024_73503387 | 0.07 |
C10orf105 |
chromosome 10 open reading frame 105 |
5624 |
0.22 |
| chr21_34395992_34396143 | 0.07 |
OLIG2 |
oligodendrocyte lineage transcription factor 2 |
2086 |
0.33 |
| chr11_2291133_2291433 | 0.07 |
ASCL2 |
achaete-scute family bHLH transcription factor 2 |
899 |
0.5 |
| chr8_141644530_141644748 | 0.07 |
AGO2 |
argonaute RISC catalytic component 2 |
1006 |
0.61 |
| chr12_124985454_124985605 | 0.07 |
NCOR2 |
nuclear receptor corepressor 2 |
5731 |
0.33 |
| chr6_167161616_167161767 | 0.07 |
RPS6KA2 |
ribosomal protein S6 kinase, 90kDa, polypeptide 2 |
9708 |
0.28 |
| chr16_4428001_4428209 | 0.07 |
VASN |
vasorin |
6256 |
0.13 |
| chr17_76310134_76310416 | 0.07 |
SOCS3 |
suppressor of cytokine signaling 3 |
45880 |
0.1 |
| chr13_114814477_114814986 | 0.07 |
RASA3 |
RAS p21 protein activator 3 |
28707 |
0.21 |
| chr16_85089467_85090023 | 0.07 |
KIAA0513 |
KIAA0513 |
7073 |
0.23 |
| chr4_94749842_94750092 | 0.07 |
ATOH1 |
atonal homolog 1 (Drosophila) |
75 |
0.98 |
| chr4_10551040_10551191 | 0.07 |
CLNK |
cytokine-dependent hematopoietic cell linker |
32026 |
0.23 |
| chr21_45564013_45564179 | 0.07 |
C21orf33 |
chromosome 21 open reading frame 33 |
8095 |
0.15 |
| chr6_151865308_151865459 | 0.07 |
CCDC170 |
coiled-coil domain containing 170 |
50208 |
0.13 |
| chr22_23527174_23527568 | 0.07 |
BCR |
breakpoint cluster region |
4819 |
0.16 |
| chrX_70763186_70763395 | 0.07 |
OGT |
O-linked N-acetylglucosamine (GlcNAc) transferase |
6426 |
0.2 |
| chr1_227133221_227133372 | 0.07 |
ADCK3 |
aarF domain containing kinase 3 |
5301 |
0.25 |
| chr4_2822258_2822409 | 0.06 |
SH3BP2 |
SH3-domain binding protein 2 |
1792 |
0.36 |
| chr12_1579383_1579534 | 0.06 |
WNT5B |
wingless-type MMTV integration site family, member 5B |
59599 |
0.11 |
| chr9_139303294_139303653 | 0.06 |
SDCCAG3 |
serologically defined colon cancer antigen 3 |
1247 |
0.26 |
| chr10_71905217_71905368 | 0.06 |
TYSND1 |
trypsin domain containing 1 |
1050 |
0.5 |
| chr17_5014893_5015044 | 0.06 |
ZNF232 |
zinc finger protein 232 |
168 |
0.53 |
| chr12_4928730_4928976 | 0.06 |
KCNA6 |
potassium voltage-gated channel, shaker-related subfamily, member 6 |
10511 |
0.2 |
| chr17_72304461_72304612 | 0.06 |
ENSG00000252785 |
. |
589 |
0.55 |
| chr14_97927674_97927825 | 0.06 |
ENSG00000240730 |
. |
68761 |
0.14 |
| chr14_105527301_105527518 | 0.06 |
GPR132 |
G protein-coupled receptor 132 |
3590 |
0.23 |
| chr5_131817472_131817699 | 0.06 |
IRF1 |
interferon regulatory factor 1 |
7752 |
0.13 |
| chr11_64108441_64109145 | 0.06 |
CCDC88B |
coiled-coil domain containing 88B |
1098 |
0.27 |
| chr2_10540752_10540903 | 0.06 |
HPCAL1 |
hippocalcin-like 1 |
19320 |
0.18 |
| chr19_5138285_5138436 | 0.06 |
CTC-482H14.5 |
|
39770 |
0.16 |
| chr21_45721457_45721805 | 0.06 |
PFKL |
phosphofructokinase, liver |
1697 |
0.21 |
| chr11_7561884_7562035 | 0.06 |
PPFIBP2 |
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
1550 |
0.38 |
| chr20_31096106_31096363 | 0.06 |
C20orf112 |
chromosome 20 open reading frame 112 |
2976 |
0.25 |
| chr17_42988559_42988831 | 0.06 |
GFAP |
glial fibrillary acidic protein |
676 |
0.51 |
| chr11_1767689_1767840 | 0.06 |
IFITM10 |
interferon induced transmembrane protein 10 |
4057 |
0.11 |
| chr9_96715477_96715717 | 0.06 |
BARX1 |
BARX homeobox 1 |
116 |
0.98 |
| chr7_149567890_149568136 | 0.06 |
ATP6V0E2 |
ATPase, H+ transporting V0 subunit e2 |
2044 |
0.26 |
| chr5_178054496_178054708 | 0.06 |
CLK4 |
CDC-like kinase 4 |
487 |
0.8 |
| chr8_142397145_142397301 | 0.06 |
PTP4A3 |
protein tyrosine phosphatase type IVA, member 3 |
4870 |
0.13 |
| chr6_139695930_139696081 | 0.06 |
CITED2 |
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 |
248 |
0.95 |
| chr10_102500685_102500836 | 0.06 |
PAX2 |
paired box 2 |
4708 |
0.28 |
| chr3_53157618_53157769 | 0.06 |
RFT1 |
RFT1 homolog (S. cerevisiae) |
6734 |
0.19 |
| chr14_99502237_99502388 | 0.06 |
AL162151.4 |
|
122441 |
0.06 |
| chr20_56232956_56233107 | 0.06 |
PMEPA1 |
prostate transmembrane protein, androgen induced 1 |
32649 |
0.17 |
| chr4_106227685_106227836 | 0.06 |
ENSG00000243383 |
. |
12945 |
0.26 |
| chr1_92196423_92196574 | 0.06 |
ENSG00000239794 |
. |
99133 |
0.08 |
| chr11_65380533_65380748 | 0.06 |
MAP3K11 |
mitogen-activated protein kinase kinase kinase 11 |
1073 |
0.26 |
| chr1_205247824_205248121 | 0.06 |
TMCC2 |
transmembrane and coiled-coil domain family 2 |
22643 |
0.15 |
| chr9_139790608_139790759 | 0.06 |
TRAF2 |
TNF receptor-associated factor 2 |
6696 |
0.08 |
| chr15_31325370_31325521 | 0.06 |
RP11-348B17.1 |
|
7036 |
0.18 |
| chr3_12438460_12438611 | 0.06 |
PPARG |
peroxisome proliferator-activated receptor gamma |
45564 |
0.15 |
| chr1_227917436_227917587 | 0.06 |
SNAP47 |
synaptosomal-associated protein, 47kDa |
1153 |
0.45 |
| chr7_74206755_74206906 | 0.06 |
GTF2IRD2 |
GTF2I repeat domain containing 2 |
14458 |
0.18 |
| chr12_46760698_46760932 | 0.06 |
SLC38A2 |
solute carrier family 38, member 2 |
1725 |
0.42 |
| chr3_196367115_196367900 | 0.06 |
PIGX |
phosphatidylinositol glycan anchor biosynthesis, class X |
861 |
0.37 |
| chr20_55012223_55012374 | 0.06 |
ENSG00000212084 |
. |
23115 |
0.11 |
| chr8_96192088_96192282 | 0.06 |
PLEKHF2 |
pleckstrin homology domain containing, family F (with FYVE domain) member 2 |
45881 |
0.15 |
| chr15_40453377_40453695 | 0.06 |
BUB1B |
BUB1 mitotic checkpoint serine/threonine kinase B |
268 |
0.9 |
| chr10_7299238_7299582 | 0.06 |
SFMBT2 |
Scm-like with four mbt domains 2 |
151297 |
0.04 |
| chrX_152518761_152518912 | 0.06 |
MAGEA1 |
melanoma antigen family A, 1 (directs expression of antigen MZ2-E) |
32721 |
0.15 |
| chr3_71413120_71413271 | 0.06 |
FOXP1 |
forkhead box P1 |
59284 |
0.14 |
| chr17_39932161_39932357 | 0.06 |
JUP |
junction plakoglobin |
3738 |
0.13 |
| chr9_31335901_31336264 | 0.06 |
ENSG00000211510 |
. |
561856 |
0.0 |
| chr2_37552318_37552512 | 0.06 |
PRKD3 |
protein kinase D3 |
464 |
0.79 |
| chr1_38142298_38142449 | 0.06 |
C1orf109 |
chromosome 1 open reading frame 109 |
13803 |
0.13 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.1 | GO:0042483 | negative regulation of odontogenesis(GO:0042483) |
| 0.0 | 0.1 | GO:0042668 | auditory receptor cell fate determination(GO:0042668) |
| 0.0 | 0.1 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.1 | GO:0045046 | protein import into peroxisome membrane(GO:0045046) |
| 0.0 | 0.1 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.0 | 0.0 | GO:0014874 | response to stimulus involved in regulation of muscle adaptation(GO:0014874) |
| 0.0 | 0.1 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 0.1 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 0.1 | GO:0008611 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.0 | GO:0021542 | dentate gyrus development(GO:0021542) |
| 0.0 | 0.0 | GO:0031558 | obsolete induction of apoptosis in response to chemical stimulus(GO:0031558) |
| 0.0 | 0.1 | GO:0072698 | protein localization to cytoskeleton(GO:0044380) protein localization to microtubule cytoskeleton(GO:0072698) |
| 0.0 | 0.1 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.1 | GO:0018027 | peptidyl-lysine dimethylation(GO:0018027) |
| 0.0 | 0.0 | GO:0003078 | obsolete regulation of natriuresis(GO:0003078) |
| 0.0 | 0.1 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.0 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.0 | GO:0005954 | calcium- and calmodulin-dependent protein kinase complex(GO:0005954) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.1 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.1 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.1 | GO:0043142 | single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 0.0 | 0.1 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.0 | 0.0 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |