Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
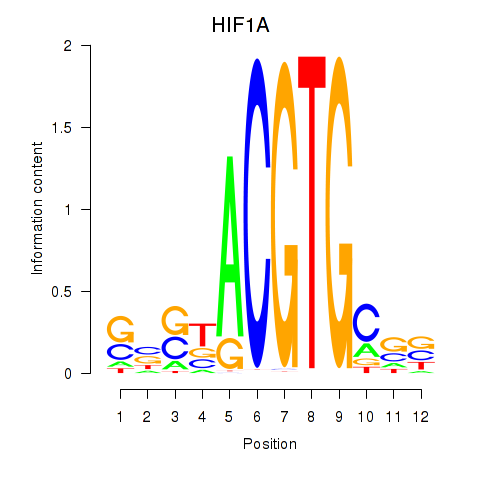
Results for HIF1A
Z-value: 0.76
Transcription factors associated with HIF1A
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HIF1A
|
ENSG00000100644.12 | hypoxia inducible factor 1 subunit alpha |
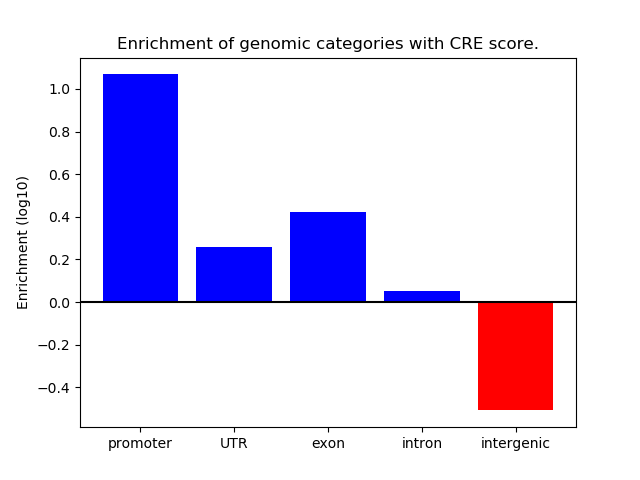
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr14_62183724_62184123 | HIF1A | 19583 | 0.216505 | -0.86 | 2.6e-03 | Click! |
| chr14_62176844_62177091 | HIF1A | 12627 | 0.234024 | -0.85 | 3.3e-03 | Click! |
| chr14_62169999_62170295 | HIF1A | 5807 | 0.260531 | -0.82 | 7.1e-03 | Click! |
| chr14_62182699_62182850 | HIF1A | 18434 | 0.219529 | -0.80 | 1.0e-02 | Click! |
| chr14_62173062_62173362 | HIF1A | 8872 | 0.244524 | -0.76 | 1.7e-02 | Click! |
Activity of the HIF1A motif across conditions
Conditions sorted by the z-value of the HIF1A motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
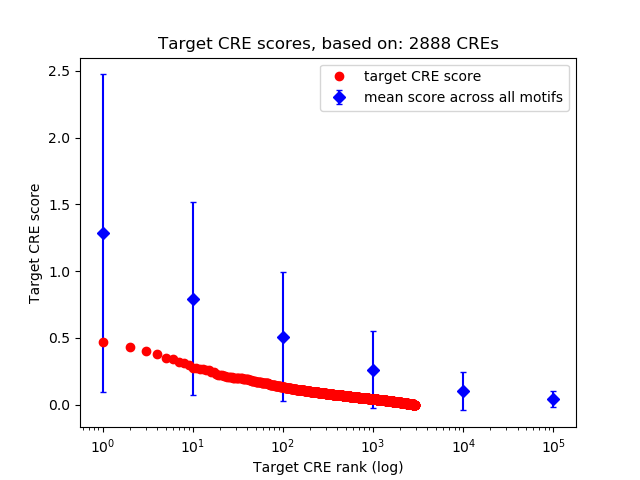
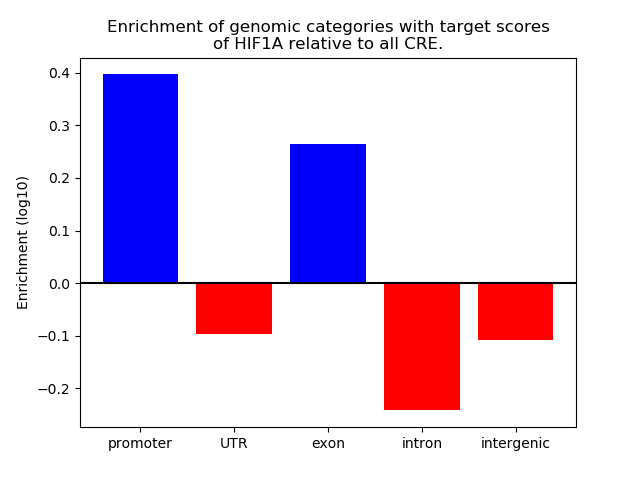
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr18_10726488_10726639 | 0.47 |
PIEZO2 |
piezo-type mechanosensitive ion channel component 2 |
21935 |
0.2 |
| chr2_74648839_74648990 | 0.43 |
WDR54 |
WD repeat domain 54 |
57 |
0.67 |
| chr8_61973816_61973967 | 0.40 |
CLVS1 |
clavesin 1 |
4174 |
0.36 |
| chr4_82136461_82136814 | 0.38 |
PRKG2 |
protein kinase, cGMP-dependent, type II |
419 |
0.89 |
| chr9_93956113_93956298 | 0.35 |
AUH |
AU RNA binding protein/enoyl-CoA hydratase |
167972 |
0.04 |
| chr9_138999867_139000258 | 0.34 |
C9orf69 |
chromosome 9 open reading frame 69 |
10058 |
0.19 |
| chr11_133920937_133921088 | 0.32 |
JAM3 |
junctional adhesion molecule 3 |
17808 |
0.22 |
| chr19_42391298_42391529 | 0.31 |
ARHGEF1 |
Rho guanine nucleotide exchange factor (GEF) 1 |
2898 |
0.16 |
| chr18_43913575_43913778 | 0.30 |
RNF165 |
ring finger protein 165 |
255 |
0.95 |
| chr5_11384831_11385016 | 0.28 |
CTNND2 |
catenin (cadherin-associated protein), delta 2 |
204000 |
0.03 |
| chr1_65533556_65533707 | 0.27 |
ENSG00000199135 |
. |
9440 |
0.19 |
| chrX_77359735_77359886 | 0.27 |
PGK1 |
phosphoglycerate kinase 1 |
58 |
0.98 |
| chr16_68022948_68023099 | 0.27 |
DUS2 |
dihydrouridine synthase 2 |
1348 |
0.22 |
| chr10_81002057_81002208 | 0.26 |
ZMIZ1 |
zinc finger, MIZ-type containing 1 |
63843 |
0.13 |
| chr15_90606260_90606411 | 0.26 |
ZNF710 |
zinc finger protein 710 |
4911 |
0.16 |
| chr18_51795667_51795818 | 0.24 |
POLI |
polymerase (DNA directed) iota |
32 |
0.98 |
| chr7_44303433_44303584 | 0.24 |
CAMK2B |
calcium/calmodulin-dependent protein kinase II beta |
21603 |
0.17 |
| chr19_41328238_41328415 | 0.23 |
EGLN2 |
egl-9 family hypoxia-inducible factor 2 |
15178 |
0.12 |
| chr8_72755880_72756041 | 0.23 |
RP11-383H13.1 |
Protein LOC100132891; cDNA FLJ53548 |
164 |
0.88 |
| chr5_43397167_43397386 | 0.22 |
CCL28 |
chemokine (C-C motif) ligand 28 |
90 |
0.97 |
| chr4_187112479_187112630 | 0.22 |
AC110771.1 |
Uncharacterized protein |
72 |
0.56 |
| chr3_100240423_100240574 | 0.22 |
TMEM45A |
transmembrane protein 45A |
28994 |
0.2 |
| chr3_133576348_133576499 | 0.21 |
RAB6B |
RAB6B, member RAS oncogene family |
37140 |
0.15 |
| chr13_27936429_27936610 | 0.21 |
ENSG00000252247 |
. |
18636 |
0.15 |
| chr9_140051094_140051245 | 0.21 |
ENSG00000263697 |
. |
12553 |
0.06 |
| chr16_85590036_85590187 | 0.20 |
GSE1 |
Gse1 coiled-coil protein |
54904 |
0.13 |
| chr21_39850220_39850518 | 0.20 |
ERG |
v-ets avian erythroblastosis virus E26 oncogene homolog |
19976 |
0.28 |
| chr16_3766966_3767153 | 0.20 |
TRAP1 |
TNF receptor-associated protein 1 |
455 |
0.82 |
| chr7_95100446_95100597 | 0.20 |
ASB4 |
ankyrin repeat and SOCS box containing 4 |
14692 |
0.17 |
| chr2_232505608_232505920 | 0.20 |
ENSG00000239202 |
. |
5220 |
0.2 |
| chr6_139695018_139695224 | 0.20 |
CITED2 |
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 |
229 |
0.96 |
| chr8_126614315_126614538 | 0.20 |
ENSG00000266452 |
. |
157619 |
0.04 |
| chr14_34407515_34407805 | 0.20 |
EGLN3 |
egl-9 family hypoxia-inducible factor 3 |
12372 |
0.25 |
| chr19_2288228_2288485 | 0.20 |
LINGO3 |
leucine rich repeat and Ig domain containing 3 |
3667 |
0.1 |
| chr17_75317312_75317463 | 0.20 |
SEPT9 |
septin 9 |
992 |
0.6 |
| chr5_10746814_10746997 | 0.20 |
CTD-2154B17.4 |
|
14272 |
0.22 |
| chr1_36040729_36040916 | 0.20 |
RP4-728D4.2 |
|
1474 |
0.29 |
| chr17_38511467_38511733 | 0.20 |
CTD-2267D19.3 |
Uncharacterized protein |
5307 |
0.11 |
| chr8_130722541_130722692 | 0.19 |
GSDMC |
gasdermin C |
76518 |
0.1 |
| chr19_55670752_55670903 | 0.19 |
CTD-2587H24.5 |
|
520 |
0.5 |
| chr14_64804805_64805119 | 0.19 |
ESR2 |
estrogen receptor 2 (ER beta) |
132 |
0.76 |
| chr9_134901891_134902042 | 0.18 |
MED27 |
mediator complex subunit 27 |
53287 |
0.16 |
| chr4_2044269_2044420 | 0.18 |
C4orf48 |
chromosome 4 open reading frame 48 |
567 |
0.49 |
| chr8_145698238_145698394 | 0.18 |
FOXH1 |
forkhead box H1 |
3402 |
0.07 |
| chr20_25058615_25058863 | 0.18 |
VSX1 |
visual system homeobox 1 |
4028 |
0.24 |
| chr20_30659418_30659569 | 0.18 |
HCK |
hemopoietic cell kinase |
19429 |
0.11 |
| chr17_18893756_18893907 | 0.18 |
FAM83G |
family with sequence similarity 83, member G |
12630 |
0.12 |
| chr16_30472246_30472632 | 0.18 |
ENSG00000202476 |
. |
7021 |
0.06 |
| chr3_127308866_127309017 | 0.18 |
TPRA1 |
transmembrane protein, adipocyte asscociated 1 |
601 |
0.74 |
| chr6_160238565_160238716 | 0.17 |
PNLDC1 |
poly(A)-specific ribonuclease (PARN)-like domain containing 1 |
17334 |
0.12 |
| chr15_50474903_50475054 | 0.17 |
ATP8B4 |
ATPase, class I, type 8B, member 4 |
36 |
0.91 |
| chr17_8090528_8090815 | 0.17 |
ENSG00000266638 |
. |
178 |
0.86 |
| chr14_74079365_74079641 | 0.17 |
ACOT6 |
acyl-CoA thioesterase 6 |
1854 |
0.22 |
| chr20_61050006_61050157 | 0.17 |
GATA5 |
GATA binding protein 5 |
945 |
0.53 |
| chr18_78007225_78007376 | 0.17 |
PARD6G |
par-6 family cell polarity regulator gamma |
1871 |
0.44 |
| chr17_7592540_7592816 | 0.17 |
WRAP53 |
WD repeat containing, antisense to TP53 |
883 |
0.32 |
| chr1_236064109_236064608 | 0.17 |
ENSG00000206803 |
. |
14357 |
0.16 |
| chr1_28052713_28052864 | 0.17 |
FAM76A |
family with sequence similarity 76, member A |
220 |
0.9 |
| chr2_28767791_28768074 | 0.17 |
PLB1 |
phospholipase B1 |
3435 |
0.31 |
| chr17_79067593_79067744 | 0.17 |
BAIAP2 |
BAI1-associated protein 2 |
3711 |
0.14 |
| chr9_136023976_136024127 | 0.17 |
RALGDS |
ral guanine nucleotide dissociation stimulator |
670 |
0.63 |
| chr22_50721428_50721579 | 0.16 |
PLXNB2 |
plexin B2 |
194 |
0.87 |
| chr6_603340_603491 | 0.16 |
HUS1B |
HUS1 checkpoint homolog b (S. pombe) |
53548 |
0.15 |
| chr7_1513917_1514083 | 0.16 |
INTS1 |
integrator complex subunit 1 |
1214 |
0.4 |
| chr3_182101523_182101674 | 0.16 |
RP11-225N10.1 |
|
409584 |
0.01 |
| chr5_169172919_169173070 | 0.16 |
DOCK2 |
dedicator of cytokinesis 2 |
13727 |
0.24 |
| chr6_26087576_26087727 | 0.16 |
HFE |
hemochromatosis |
16 |
0.94 |
| chr2_240168955_240169228 | 0.16 |
HDAC4 |
histone deacetylase 4 |
56339 |
0.1 |
| chr2_99954133_99954284 | 0.15 |
EIF5B |
eukaryotic translation initiation factor 5B |
392 |
0.77 |
| chr15_63664697_63665270 | 0.15 |
CA12 |
carbonic anhydrase XII |
9051 |
0.26 |
| chr8_122558149_122558407 | 0.15 |
ENSG00000263525 |
. |
3043 |
0.37 |
| chr3_10057184_10057732 | 0.15 |
EMC3 |
ER membrane protein complex subunit 3 |
4658 |
0.11 |
| chr8_131011679_131011830 | 0.15 |
ENSG00000253043 |
. |
5093 |
0.19 |
| chr10_134402672_134402855 | 0.15 |
INPP5A |
inositol polyphosphate-5-phosphatase, 40kDa |
18667 |
0.23 |
| chr16_2391058_2391263 | 0.15 |
ABCA3 |
ATP-binding cassette, sub-family A (ABC1), member 3 |
413 |
0.73 |
| chr19_19539011_19539215 | 0.15 |
GATAD2A |
GATA zinc finger domain containing 2A |
777 |
0.55 |
| chr16_1584078_1584229 | 0.15 |
TMEM204 |
transmembrane protein 204 |
579 |
0.41 |
| chr10_88975874_88976033 | 0.15 |
NUTM2A |
NUT family member 2A |
9252 |
0.21 |
| chr3_30398539_30398690 | 0.15 |
ENSG00000199927 |
. |
52587 |
0.18 |
| chr7_2979039_2979190 | 0.15 |
AC004906.3 |
|
4555 |
0.23 |
| chr22_47170259_47170421 | 0.14 |
TBC1D22A |
TBC1 domain family, member 22A |
516 |
0.79 |
| chr2_208675165_208675316 | 0.14 |
FZD5 |
frizzled family receptor 5 |
40953 |
0.12 |
| chr19_47951061_47951212 | 0.14 |
SLC8A2 |
solute carrier family 8 (sodium/calcium exchanger), member 2 |
23984 |
0.12 |
| chr1_3514916_3515113 | 0.14 |
MEGF6 |
multiple EGF-like-domains 6 |
13045 |
0.13 |
| chr11_2223807_2223958 | 0.14 |
ENSG00000265258 |
. |
29589 |
0.1 |
| chr1_247336437_247336716 | 0.14 |
ZNF124 |
zinc finger protein 124 |
1258 |
0.39 |
| chr16_67424084_67424235 | 0.14 |
ENSG00000239194 |
. |
594 |
0.54 |
| chr9_132250210_132250368 | 0.14 |
ENSG00000264298 |
. |
9454 |
0.22 |
| chr1_234614717_234614988 | 0.14 |
TARBP1 |
TAR (HIV-1) RNA binding protein 1 |
3 |
0.98 |
| chr6_85483668_85483897 | 0.14 |
TBX18 |
T-box 18 |
9545 |
0.3 |
| chr17_39656063_39656224 | 0.14 |
AC019349.5 |
|
1092 |
0.28 |
| chr5_178016736_178016887 | 0.14 |
COL23A1 |
collagen, type XXIII, alpha 1 |
745 |
0.68 |
| chr18_22930035_22930188 | 0.14 |
ZNF521 |
zinc finger protein 521 |
1046 |
0.66 |
| chr17_26878964_26879115 | 0.14 |
UNC119 |
unc-119 homolog (C. elegans) |
219 |
0.84 |
| chr17_80175257_80175581 | 0.14 |
RP13-516M14.2 |
|
3316 |
0.13 |
| chr20_62339731_62339882 | 0.13 |
ZGPAT |
zinc finger, CCCH-type with G patch domain |
364 |
0.48 |
| chr1_155164076_155164322 | 0.13 |
ENSG00000271748 |
. |
769 |
0.29 |
| chr14_62000458_62000638 | 0.13 |
RP11-47I22.4 |
|
4702 |
0.22 |
| chr15_75931759_75932122 | 0.13 |
IMP3 |
IMP3, U3 small nucleolar ribonucleoprotein, homolog (yeast) |
612 |
0.55 |
| chr17_78801730_78801881 | 0.13 |
RP11-28G8.1 |
|
22373 |
0.2 |
| chr11_119455747_119455948 | 0.13 |
RP11-196E1.3 |
|
23288 |
0.19 |
| chr1_41964550_41964701 | 0.13 |
EDN2 |
endothelin 2 |
14283 |
0.2 |
| chr10_47668348_47668587 | 0.13 |
ANTXRL |
anthrax toxin receptor-like |
10218 |
0.2 |
| chr17_39656944_39657095 | 0.13 |
AC019349.5 |
|
216 |
0.84 |
| chr8_49427273_49427424 | 0.13 |
RP11-770E5.1 |
|
36779 |
0.23 |
| chr9_124308087_124308238 | 0.13 |
ENSG00000252940 |
. |
6007 |
0.19 |
| chr10_73731046_73731197 | 0.13 |
CHST3 |
carbohydrate (chondroitin 6) sulfotransferase 3 |
6998 |
0.26 |
| chr20_23064521_23064672 | 0.13 |
CD93 |
CD93 molecule |
2381 |
0.27 |
| chr17_8533868_8534019 | 0.13 |
MYH10 |
myosin, heavy chain 10, non-muscle |
92 |
0.98 |
| chrX_130925857_130926008 | 0.13 |
ENSG00000200587 |
. |
147052 |
0.04 |
| chr2_109745417_109745568 | 0.13 |
SH3RF3 |
SH3 domain containing ring finger 3 |
312 |
0.92 |
| chr3_53184032_53184571 | 0.13 |
PRKCD |
protein kinase C, delta |
5724 |
0.19 |
| chr11_45001370_45001623 | 0.13 |
TP53I11 |
tumor protein p53 inducible protein 11 |
28888 |
0.22 |
| chr12_25055485_25055776 | 0.12 |
BCAT1 |
branched chain amino-acid transaminase 1, cytosolic |
379 |
0.48 |
| chr2_26990361_26990533 | 0.12 |
SLC35F6 |
solute carrier family 35, member F6 |
3290 |
0.23 |
| chr3_57935477_57935765 | 0.12 |
SLMAP |
sarcolemma associated protein |
32929 |
0.19 |
| chr1_6355119_6355270 | 0.12 |
GPR153 |
G protein-coupled receptor 153 |
34159 |
0.1 |
| chr19_8553452_8553653 | 0.12 |
HNRNPM |
heterogeneous nuclear ribonucleoprotein M |
2510 |
0.18 |
| chr11_2187663_2187887 | 0.12 |
TH |
tyrosine hydroxylase |
1561 |
0.23 |
| chr2_60579152_60579304 | 0.12 |
ENSG00000200807 |
. |
32512 |
0.19 |
| chr13_114525150_114525301 | 0.12 |
GAS6-AS1 |
GAS6 antisense RNA 1 |
6622 |
0.22 |
| chr1_150039467_150039618 | 0.12 |
VPS45 |
vacuolar protein sorting 45 homolog (S. cerevisiae) |
103 |
0.96 |
| chr19_41349423_41349599 | 0.12 |
CYP2A6 |
cytochrome P450, family 2, subfamily A, polypeptide 6 |
6841 |
0.14 |
| chr4_103609194_103609711 | 0.12 |
MANBA |
mannosidase, beta A, lysosomal |
72638 |
0.1 |
| chr16_48570088_48570239 | 0.12 |
RP11-44I10.3 |
|
23409 |
0.17 |
| chr8_101322815_101323530 | 0.12 |
RNF19A |
ring finger protein 19A, RBR E3 ubiquitin protein ligase |
617 |
0.78 |
| chr6_157977560_157977711 | 0.12 |
ENSG00000266617 |
. |
27471 |
0.21 |
| chrX_49030494_49030789 | 0.12 |
PLP2 |
proteolipid protein 2 (colonic epithelium-enriched) |
2368 |
0.13 |
| chr8_11565915_11566066 | 0.12 |
GATA4 |
GATA binding protein 4 |
248 |
0.92 |
| chr19_5827352_5827630 | 0.12 |
NRTN |
neurturin |
3678 |
0.1 |
| chr15_83875350_83875501 | 0.12 |
HDGFRP3 |
Hepatoma-derived growth factor-related protein 3 |
1345 |
0.43 |
| chr16_1968651_1968802 | 0.12 |
HS3ST6 |
heparan sulfate (glucosamine) 3-O-sulfotransferase 6 |
285 |
0.74 |
| chr19_56015578_56015806 | 0.12 |
SSC5D |
scavenger receptor cysteine rich domain containing (5 domains) |
10434 |
0.08 |
| chr14_34529991_34530222 | 0.12 |
EGLN3-AS1 |
EGLN3 antisense RNA 1 |
20803 |
0.25 |
| chr3_145879067_145879218 | 0.12 |
PLOD2 |
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2 |
188 |
0.96 |
| chr2_105489675_105489826 | 0.12 |
LINC01159 |
long intergenic non-protein coding RNA 1159 |
697 |
0.61 |
| chr16_57037843_57038089 | 0.12 |
NLRC5 |
NLR family, CARD domain containing 5 |
201 |
0.92 |
| chr8_130709802_130710031 | 0.11 |
GSDMC |
gasdermin C |
89218 |
0.08 |
| chr22_50122646_50122797 | 0.11 |
ENSG00000223142 |
. |
9604 |
0.22 |
| chr1_224032718_224032869 | 0.11 |
TP53BP2 |
tumor protein p53 binding protein, 2 |
839 |
0.72 |
| chrX_107019456_107019640 | 0.11 |
TSC22D3 |
TSC22 domain family, member 3 |
60 |
0.96 |
| chr3_197476743_197476894 | 0.11 |
FYTTD1 |
forty-two-three domain containing 1 |
197 |
0.56 |
| chr3_124880542_124880693 | 0.11 |
ENSG00000264986 |
. |
10221 |
0.19 |
| chr9_135045081_135045232 | 0.11 |
NTNG2 |
netrin G2 |
7822 |
0.25 |
| chr19_10859737_10859950 | 0.11 |
DNM2 |
dynamin 2 |
30606 |
0.09 |
| chr22_26565763_26565931 | 0.11 |
SEZ6L |
seizure related 6 homolog (mouse)-like |
223 |
0.94 |
| chr1_154474981_154475132 | 0.11 |
TDRD10 |
tudor domain containing 10 |
361 |
0.54 |
| chr7_148762266_148762471 | 0.11 |
ZNF786 |
zinc finger protein 786 |
25429 |
0.11 |
| chr2_20647190_20647341 | 0.11 |
RHOB |
ras homolog family member B |
430 |
0.84 |
| chr8_29420044_29420195 | 0.11 |
RP4-676L2.1 |
|
209432 |
0.02 |
| chr17_75369794_75369945 | 0.11 |
SEPT9 |
septin 9 |
450 |
0.8 |
| chr16_2521832_2521983 | 0.11 |
NTN3 |
netrin 3 |
407 |
0.61 |
| chr11_74952604_74952755 | 0.11 |
TPBGL |
trophoblast glycoprotein-like |
729 |
0.6 |
| chr3_143321015_143321295 | 0.11 |
SLC9A9-AS2 |
SLC9A9 antisense RNA 2 |
220975 |
0.02 |
| chr9_20621148_20621336 | 0.11 |
MLLT3 |
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
630 |
0.75 |
| chr20_39765712_39765863 | 0.11 |
PLCG1 |
phospholipase C, gamma 1 |
90 |
0.94 |
| chr3_193689544_193689695 | 0.11 |
RP11-135A1.2 |
|
159371 |
0.04 |
| chr11_33279873_33280030 | 0.11 |
HIPK3 |
homeodomain interacting protein kinase 3 |
74 |
0.98 |
| chr14_75389794_75389999 | 0.11 |
RPS6KL1 |
ribosomal protein S6 kinase-like 1 |
58 |
0.97 |
| chr3_14444833_14444984 | 0.11 |
SLC6A6 |
solute carrier family 6 (neurotransmitter transporter), member 6 |
788 |
0.69 |
| chr17_38187925_38188076 | 0.11 |
ENSG00000238793 |
. |
4102 |
0.13 |
| chr3_169041201_169041352 | 0.11 |
MECOM |
MDS1 and EVI1 complex locus |
54250 |
0.16 |
| chr7_2019784_2019935 | 0.11 |
MAD1L1 |
MAD1 mitotic arrest deficient-like 1 (yeast) |
567 |
0.84 |
| chr17_76137750_76137971 | 0.11 |
C17orf99 |
chromosome 17 open reading frame 99 |
4574 |
0.12 |
| chr17_17631785_17632060 | 0.11 |
RAI1-AS1 |
RAI1 antisense RNA 1 |
42213 |
0.1 |
| chr1_9942861_9943012 | 0.11 |
CTNNBIP1 |
catenin, beta interacting protein 1 |
4702 |
0.17 |
| chr9_137276901_137277082 | 0.11 |
ENSG00000263897 |
. |
5734 |
0.25 |
| chr2_232478276_232478563 | 0.11 |
C2orf57 |
chromosome 2 open reading frame 57 |
20844 |
0.15 |
| chr16_79046532_79046733 | 0.11 |
RP11-319G9.5 |
|
18390 |
0.19 |
| chr7_105710732_105710883 | 0.11 |
SYPL1 |
synaptophysin-like 1 |
27501 |
0.22 |
| chr12_124985454_124985605 | 0.11 |
NCOR2 |
nuclear receptor corepressor 2 |
5731 |
0.33 |
| chr6_1703338_1703561 | 0.11 |
FOXC1 |
forkhead box C1 |
92768 |
0.09 |
| chr7_28076620_28076771 | 0.11 |
JAZF1 |
JAZF zinc finger 1 |
16277 |
0.26 |
| chr9_135116163_135116314 | 0.11 |
ENSG00000221105 |
. |
25976 |
0.2 |
| chr10_80831884_80832062 | 0.11 |
ZMIZ1 |
zinc finger, MIZ-type containing 1 |
3181 |
0.3 |
| chr6_158401711_158402013 | 0.11 |
SYNJ2 |
synaptojanin 2 |
1026 |
0.54 |
| chr11_1851555_1851968 | 0.11 |
SYT8 |
synaptotagmin VIII |
922 |
0.39 |
| chr15_99211968_99212377 | 0.11 |
IGF1R |
insulin-like growth factor 1 receptor |
19899 |
0.21 |
| chr2_202098532_202098973 | 0.11 |
CASP8 |
caspase 8, apoptosis-related cysteine peptidase |
523 |
0.78 |
| chr13_28540704_28540855 | 0.10 |
CDX2 |
caudal type homeobox 2 |
4497 |
0.16 |
| chr16_25704360_25704511 | 0.10 |
HS3ST4 |
heparan sulfate (glucosamine) 3-O-sulfotransferase 4 |
1088 |
0.69 |
| chr16_11391591_11391742 | 0.10 |
ENSG00000238409 |
. |
4599 |
0.09 |
| chr16_81542300_81542584 | 0.10 |
CMIP |
c-Maf inducing protein |
13488 |
0.27 |
| chr8_142216214_142216365 | 0.10 |
DENND3 |
DENN/MADD domain containing 3 |
13859 |
0.16 |
| chr11_76381975_76382126 | 0.10 |
LRRC32 |
leucine rich repeat containing 32 |
259 |
0.91 |
| chr21_38593352_38593525 | 0.10 |
ENSG00000263969 |
. |
5532 |
0.17 |
| chr14_77416963_77417114 | 0.10 |
ENSG00000266553 |
. |
17070 |
0.2 |
| chr15_50549728_50549936 | 0.10 |
HDC |
histidine decarboxylase |
5794 |
0.16 |
| chr16_770731_770926 | 0.10 |
FAM173A |
family with sequence similarity 173, member A |
147 |
0.85 |
| chr7_83690336_83690549 | 0.10 |
SEMA3A |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
133775 |
0.05 |
| chr16_66519004_66519282 | 0.10 |
AC132186.1 |
CDNA FLJ27243 fis, clone SYN08134; Uncharacterized protein |
604 |
0.62 |
| chr6_158993431_158993582 | 0.10 |
TMEM181 |
transmembrane protein 181 |
36038 |
0.15 |
| chr18_77267409_77267811 | 0.10 |
AC018445.1 |
Uncharacterized protein |
8447 |
0.28 |
| chr21_40386580_40386797 | 0.10 |
ENSG00000272015 |
. |
119979 |
0.05 |
| chr22_37414309_37414730 | 0.10 |
TST |
thiosulfate sulfurtransferase (rhodanese) |
989 |
0.3 |
| chr2_218354093_218354244 | 0.10 |
ENSG00000251982 |
. |
237889 |
0.02 |
| chr14_93526704_93527068 | 0.10 |
ITPK1 |
inositol-tetrakisphosphate 1-kinase |
5817 |
0.21 |
| chr1_44680378_44680549 | 0.10 |
DMAP1 |
DNA methyltransferase 1 associated protein 1 |
950 |
0.55 |
| chr9_100396157_100396308 | 0.10 |
TSTD2 |
thiosulfate sulfurtransferase (rhodanese)-like domain containing 2 |
270 |
0.53 |
| chr4_965118_965269 | 0.10 |
DGKQ |
diacylglycerol kinase, theta 110kDa |
1917 |
0.24 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0060586 | multicellular organismal iron ion homeostasis(GO:0060586) |
| 0.0 | 0.2 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.0 | 0.1 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.1 | GO:0045343 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) |
| 0.0 | 0.1 | GO:0015691 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.0 | 0.1 | GO:0046628 | positive regulation of insulin receptor signaling pathway(GO:0046628) positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.0 | 0.1 | GO:0071364 | cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.0 | 0.1 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.1 | GO:0036336 | dendritic cell chemotaxis(GO:0002407) dendritic cell migration(GO:0036336) |
| 0.0 | 0.1 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.1 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.1 | GO:0006927 | obsolete transformed cell apoptotic process(GO:0006927) |
| 0.0 | 0.1 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.1 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.0 | 0.1 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.1 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.0 | 0.1 | GO:0042416 | dopamine biosynthetic process(GO:0042416) |
| 0.0 | 0.0 | GO:0002538 | arachidonic acid metabolite production involved in inflammatory response(GO:0002538) leukotriene production involved in inflammatory response(GO:0002540) |
| 0.0 | 0.0 | GO:0003078 | obsolete regulation of natriuresis(GO:0003078) |
| 0.0 | 0.1 | GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0043619) |
| 0.0 | 0.1 | GO:0072201 | negative regulation of mesenchymal cell proliferation(GO:0072201) |
| 0.0 | 0.2 | GO:0034405 | response to fluid shear stress(GO:0034405) |
| 0.0 | 0.1 | GO:0007028 | cytoplasm organization(GO:0007028) |
| 0.0 | 0.1 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.0 | 0.1 | GO:0061029 | eyelid development in camera-type eye(GO:0061029) |
| 0.0 | 0.0 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.0 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.0 | 0.0 | GO:0045905 | translational frameshifting(GO:0006452) positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.1 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.0 | GO:0072338 | allantoin metabolic process(GO:0000255) creatinine metabolic process(GO:0046449) cellular lactam metabolic process(GO:0072338) |
| 0.0 | 0.1 | GO:0090400 | stress-induced premature senescence(GO:0090400) |
| 0.0 | 0.2 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.0 | 0.1 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.1 | GO:0009649 | entrainment of circadian clock(GO:0009649) |
| 0.0 | 0.0 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.0 | GO:0032352 | positive regulation of hormone metabolic process(GO:0032352) positive regulation of hormone biosynthetic process(GO:0046886) |
| 0.0 | 0.0 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.0 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.0 | GO:0032203 | telomere formation via telomerase(GO:0032203) |
| 0.0 | 0.1 | GO:0014721 | voluntary skeletal muscle contraction(GO:0003010) twitch skeletal muscle contraction(GO:0014721) |
| 0.0 | 0.0 | GO:0032823 | regulation of natural killer cell differentiation(GO:0032823) positive regulation of natural killer cell differentiation(GO:0032825) |
| 0.0 | 0.1 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 0.0 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.1 | GO:0032354 | response to follicle-stimulating hormone(GO:0032354) |
| 0.0 | 0.1 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.0 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) semicircular canal development(GO:0060872) |
| 0.0 | 0.0 | GO:0021798 | forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.0 | 0.0 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.0 | GO:0001766 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 0.0 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 0.0 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.0 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 | 0.0 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 0.1 | GO:0070265 | necrotic cell death(GO:0070265) |
| 0.0 | 0.0 | GO:0030011 | maintenance of cell polarity(GO:0030011) |
| 0.0 | 0.0 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.0 | 0.0 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.1 | GO:0010452 | histone H3-K36 methylation(GO:0010452) |
| 0.0 | 0.0 | GO:0051594 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.0 | 0.1 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.1 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.0 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.0 | 0.0 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.0 | 0.0 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.1 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0031264 | death-inducing signaling complex(GO:0031264) |
| 0.0 | 0.1 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 0.1 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.1 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.1 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.4 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.1 | GO:0031312 | extrinsic component of organelle membrane(GO:0031312) |
| 0.0 | 0.0 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.0 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.0 | 0.0 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.1 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.1 | 0.2 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.1 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.0 | 0.3 | GO:0004690 | cyclic nucleotide-dependent protein kinase activity(GO:0004690) |
| 0.0 | 0.2 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.1 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.1 | GO:0001846 | opsonin binding(GO:0001846) |
| 0.0 | 0.1 | GO:0050698 | proteoglycan sulfotransferase activity(GO:0050698) |
| 0.0 | 0.1 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 0.1 | GO:0042808 | obsolete neuronal Cdc2-like kinase binding(GO:0042808) |
| 0.0 | 0.1 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.1 | GO:0001012 | RNA polymerase II regulatory region sequence-specific DNA binding(GO:0000977) RNA polymerase II regulatory region DNA binding(GO:0001012) |
| 0.0 | 0.1 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.0 | 0.1 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.1 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.0 | 0.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.0 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.0 | 0.0 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.0 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.0 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.0 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.0 | GO:0031708 | bombesin receptor binding(GO:0031705) endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.1 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.1 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.0 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.1 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.0 | 0.0 | GO:0050656 | 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.0 | 0.1 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.0 | 0.0 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.2 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.1 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.0 | GO:0034187 | obsolete apolipoprotein E binding(GO:0034187) |
| 0.0 | 0.1 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.0 | GO:0045118 | azole transporter activity(GO:0045118) azole transmembrane transporter activity(GO:1901474) |
| 0.0 | 0.0 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.1 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.1 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 0.0 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.0 | GO:0034617 | tetrahydrobiopterin binding(GO:0034617) |
| 0.0 | 0.1 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.0 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.0 | GO:0030151 | molybdenum ion binding(GO:0030151) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.2 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME SIGNALING BY ERBB2 | Genes involved in Signaling by ERBB2 |
| 0.0 | 0.3 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.2 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 0.2 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.1 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.1 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |