Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
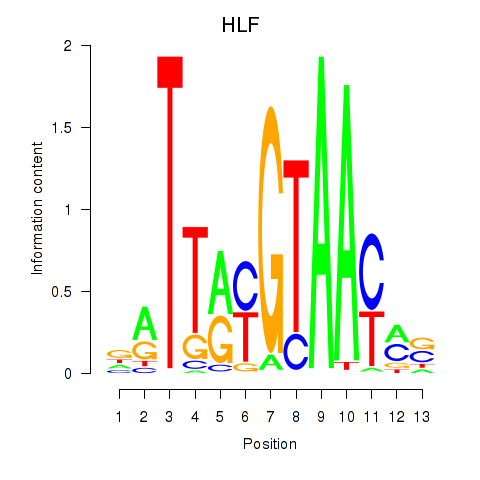
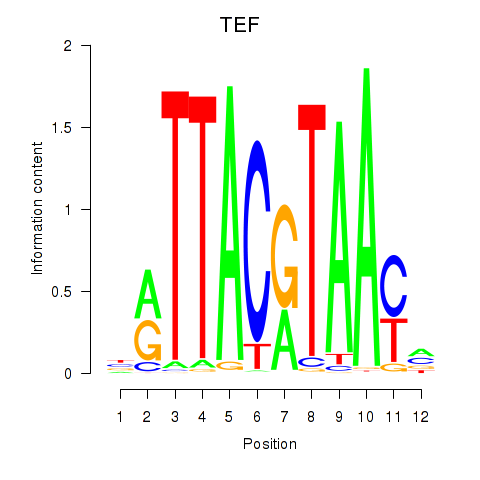
Results for HLF_TEF
Z-value: 1.57
Transcription factors associated with HLF_TEF
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HLF
|
ENSG00000108924.9 | HLF transcription factor, PAR bZIP family member |
|
TEF
|
ENSG00000167074.10 | TEF transcription factor, PAR bZIP family member |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr17_53340851_53341002 | HLF | 1447 | 0.489344 | -0.66 | 5.1e-02 | Click! |
| chr17_53346103_53346254 | HLF | 1001 | 0.610099 | 0.65 | 5.9e-02 | Click! |
| chr17_53341084_53341235 | HLF | 1214 | 0.550873 | -0.64 | 6.2e-02 | Click! |
| chr17_53245033_53245184 | HLF | 97265 | 0.078470 | 0.63 | 6.9e-02 | Click! |
| chr17_53315545_53315826 | HLF | 26688 | 0.212619 | 0.49 | 1.8e-01 | Click! |
| chr22_41763864_41764015 | TEF | 602 | 0.672897 | 0.67 | 5.1e-02 | Click! |
| chr22_41763473_41763758 | TEF | 278 | 0.881960 | 0.61 | 8.0e-02 | Click! |
| chr22_41776460_41776636 | TEF | 1385 | 0.339077 | 0.55 | 1.2e-01 | Click! |
| chr22_41762881_41763188 | TEF | 303 | 0.867458 | 0.54 | 1.4e-01 | Click! |
| chr22_41809018_41809169 | TEF | 30154 | 0.108285 | 0.53 | 1.4e-01 | Click! |
Activity of the HLF_TEF motif across conditions
Conditions sorted by the z-value of the HLF_TEF motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
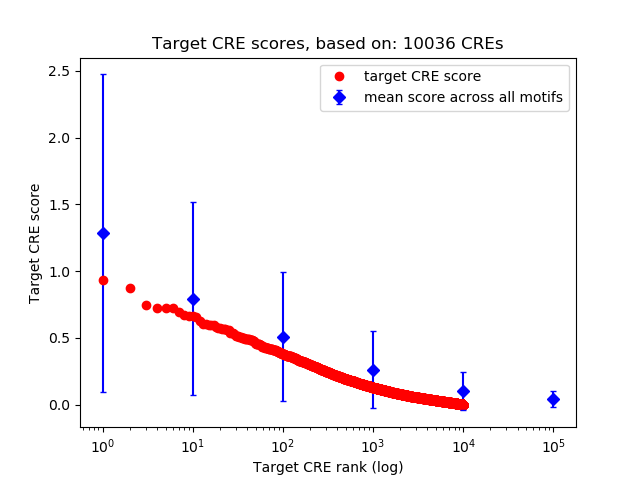
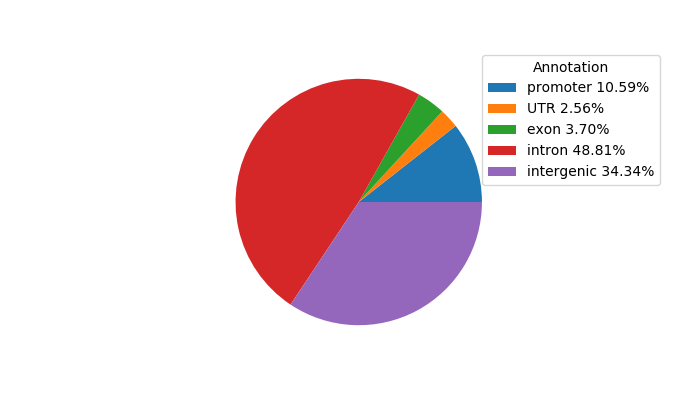
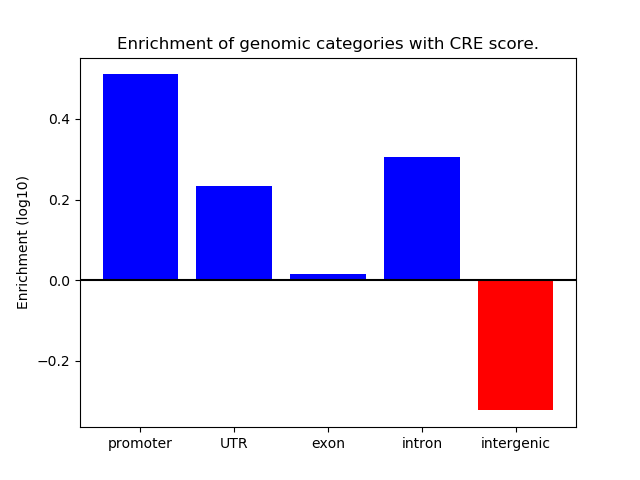
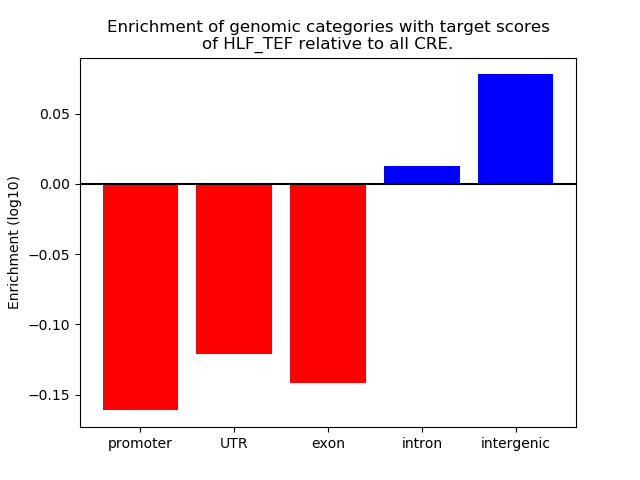
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr15_71006405_71006665 | 0.94 |
UACA |
uveal autoantigen with coiled-coil domains and ankyrin repeats |
10921 |
0.26 |
| chr3_65930585_65930845 | 0.88 |
ENSG00000264716 |
. |
3793 |
0.23 |
| chr12_48163883_48164256 | 0.75 |
RAPGEF3 |
Rap guanine nucleotide exchange factor (GEF) 3 |
748 |
0.49 |
| chr11_112027840_112028079 | 0.73 |
IL18 |
interleukin 18 (interferon-gamma-inducing factor) |
6838 |
0.11 |
| chr21_40386888_40387293 | 0.73 |
ENSG00000272015 |
. |
120381 |
0.05 |
| chr8_61896258_61896409 | 0.72 |
CLVS1 |
clavesin 1 |
73384 |
0.12 |
| chr6_132513964_132514185 | 0.70 |
ENSG00000265669 |
. |
77671 |
0.11 |
| chr10_22723256_22723780 | 0.67 |
RP11-301N24.3 |
|
73417 |
0.1 |
| chr6_5206187_5206422 | 0.67 |
ENSG00000264541 |
. |
18726 |
0.19 |
| chr10_64380892_64381169 | 0.66 |
ZNF365 |
zinc finger protein 365 |
22503 |
0.22 |
| chr20_20840342_20840493 | 0.65 |
ENSG00000264361 |
. |
121043 |
0.06 |
| chr4_38822000_38822151 | 0.63 |
TLR6 |
toll-like receptor 6 |
9085 |
0.16 |
| chr4_48226743_48226894 | 0.60 |
TEC |
tec protein tyrosine kinase |
45063 |
0.14 |
| chr10_22284671_22284822 | 0.60 |
DNAJC1 |
DnaJ (Hsp40) homolog, subfamily C, member 1 |
7582 |
0.28 |
| chr12_48340702_48340853 | 0.60 |
VDR |
vitamin D (1,25- dihydroxyvitamin D3) receptor |
3946 |
0.19 |
| chr4_77303492_77303643 | 0.60 |
ENSG00000199857 |
. |
19662 |
0.14 |
| chr5_145375816_145375967 | 0.60 |
SH3RF2 |
SH3 domain containing ring finger 2 |
58451 |
0.13 |
| chr6_14766197_14766507 | 0.58 |
ENSG00000206960 |
. |
119586 |
0.07 |
| chr13_40025518_40025722 | 0.58 |
ENSG00000238408 |
. |
4267 |
0.34 |
| chr3_64034181_64034364 | 0.58 |
RP11-129B22.1 |
|
19368 |
0.1 |
| chr12_8043408_8043946 | 0.57 |
SLC2A14 |
solute carrier family 2 (facilitated glucose transporter), member 14 |
67 |
0.95 |
| chr8_741942_742093 | 0.57 |
ERICH1-AS1 |
ERICH1 antisense RNA 1 |
54275 |
0.14 |
| chr4_140967735_140967886 | 0.57 |
RP11-392B6.1 |
|
81359 |
0.1 |
| chr12_65599568_65599732 | 0.56 |
LEMD3 |
LEM domain containing 3 |
36299 |
0.17 |
| chr21_43352747_43353275 | 0.56 |
C2CD2 |
C2 calcium-dependent domain containing 2 |
6212 |
0.19 |
| chr6_18427728_18428016 | 0.54 |
ENSG00000238458 |
. |
25589 |
0.2 |
| chr12_102250942_102251233 | 0.53 |
ENSG00000252863 |
. |
14492 |
0.12 |
| chr7_133224861_133225123 | 0.53 |
EXOC4 |
exocyst complex component 4 |
36224 |
0.24 |
| chr1_214598693_214598940 | 0.53 |
PTPN14 |
protein tyrosine phosphatase, non-receptor type 14 |
39330 |
0.19 |
| chr2_219020666_219020817 | 0.52 |
CXCR1 |
chemokine (C-X-C motif) receptor 1 |
10977 |
0.14 |
| chr6_41680623_41680774 | 0.52 |
TFEB |
transcription factor EB |
7020 |
0.13 |
| chr3_124500151_124500305 | 0.51 |
ITGB5-AS1 |
ITGB5 antisense RNA 1 |
226 |
0.93 |
| chr3_185466059_185466210 | 0.51 |
ENSG00000265470 |
. |
19558 |
0.2 |
| chr6_11536168_11536406 | 0.51 |
TMEM170B |
transmembrane protein 170B |
1651 |
0.46 |
| chr13_51939096_51939247 | 0.50 |
RP11-24B19.3 |
|
1592 |
0.28 |
| chr5_148365723_148365874 | 0.50 |
RP11-44B19.1 |
|
15895 |
0.18 |
| chr10_101288694_101289161 | 0.50 |
RP11-129J12.2 |
|
1727 |
0.34 |
| chr14_97292729_97292880 | 0.49 |
VRK1 |
vaccinia related kinase 1 |
26421 |
0.26 |
| chr6_135061273_135061513 | 0.49 |
ALDH8A1 |
aldehyde dehydrogenase 8 family, member A1 |
188906 |
0.03 |
| chr19_6103075_6103226 | 0.49 |
CTB-66B24.1 |
|
6183 |
0.15 |
| chr8_104293348_104293656 | 0.49 |
RP11-318M2.2 |
|
4628 |
0.2 |
| chr1_246271383_246271534 | 0.49 |
RP11-36N20.1 |
|
470 |
0.86 |
| chr9_70615947_70616098 | 0.49 |
AL591479.1 |
Uncharacterized protein |
30567 |
0.22 |
| chr9_42221074_42221225 | 0.48 |
RP11-216M21.7 |
|
145943 |
0.04 |
| chr4_15081864_15082159 | 0.48 |
ENSG00000199420 |
. |
13707 |
0.24 |
| chr5_14544889_14545095 | 0.48 |
FAM105A |
family with sequence similarity 105, member A |
36892 |
0.19 |
| chr20_10286775_10287859 | 0.48 |
ENSG00000211588 |
. |
55561 |
0.13 |
| chr6_34255201_34255352 | 0.48 |
C6orf1 |
chromosome 6 open reading frame 1 |
38029 |
0.15 |
| chr7_92456741_92457310 | 0.46 |
CDK6 |
cyclin-dependent kinase 6 |
6206 |
0.23 |
| chr21_45005885_45006040 | 0.46 |
HSF2BP |
heat shock transcription factor 2 binding protein |
72063 |
0.1 |
| chr18_24326449_24326698 | 0.45 |
ENSG00000265369 |
. |
57080 |
0.13 |
| chr2_216838775_216839059 | 0.45 |
MREG |
melanoregulin |
39429 |
0.18 |
| chr12_89700664_89700866 | 0.45 |
ENSG00000238302 |
. |
24703 |
0.22 |
| chr16_69775420_69775717 | 0.45 |
NOB1 |
NIN1/RPN12 binding protein 1 homolog (S. cerevisiae) |
13275 |
0.1 |
| chr2_26622798_26622949 | 0.45 |
DRC1 |
dynein regulatory complex subunit 1 homolog (Chlamydomonas) |
1911 |
0.36 |
| chr9_45472222_45472373 | 0.45 |
AL354718.1 |
Uncharacterized protein |
30755 |
0.25 |
| chr16_81642292_81642617 | 0.45 |
CMIP |
c-Maf inducing protein |
36509 |
0.21 |
| chr2_169542981_169543171 | 0.44 |
ENSG00000199348 |
. |
80070 |
0.09 |
| chr19_42947007_42947215 | 0.43 |
CXCL17 |
chemokine (C-X-C motif) ligand 17 |
89 |
0.95 |
| chr18_54524017_54524317 | 0.43 |
RP11-383D22.1 |
|
17034 |
0.22 |
| chr9_80359497_80359648 | 0.43 |
GNAQ |
guanine nucleotide binding protein (G protein), q polypeptide |
78343 |
0.1 |
| chr3_106466004_106466155 | 0.43 |
ENSG00000200361 |
. |
58554 |
0.17 |
| chr12_121158667_121159015 | 0.43 |
ENSG00000265455 |
. |
2155 |
0.16 |
| chr2_145130083_145130281 | 0.43 |
GTDC1 |
glycosyltransferase-like domain containing 1 |
40099 |
0.19 |
| chr11_110349991_110350329 | 0.43 |
FDX1 |
ferredoxin 1 |
49553 |
0.18 |
| chr14_75122953_75123104 | 0.43 |
AREL1 |
apoptosis resistant E3 ubiquitin protein ligase 1 |
19890 |
0.15 |
| chr1_61719110_61719389 | 0.42 |
NFIA |
nuclear factor I/A |
98873 |
0.08 |
| chr2_210337378_210337560 | 0.42 |
MAP2 |
microtubule-associated protein 2 |
48687 |
0.19 |
| chr7_41516088_41516239 | 0.42 |
INHBA-AS1 |
INHBA antisense RNA 1 |
217351 |
0.02 |
| chr12_116436351_116436502 | 0.42 |
MED13L |
mediator complex subunit 13-like |
15186 |
0.22 |
| chr14_91163076_91163339 | 0.42 |
RP11-661G16.2 |
|
470 |
0.81 |
| chr9_3299512_3299760 | 0.42 |
RFX3 |
regulatory factor X, 3 (influences HLA class II expression) |
28898 |
0.26 |
| chr12_71842305_71842456 | 0.42 |
TSPAN8 |
tetraspanin 8 |
6702 |
0.24 |
| chr5_88120771_88120961 | 0.42 |
MEF2C |
myocyte enhancer factor 2C |
709 |
0.77 |
| chr7_149450583_149450734 | 0.41 |
ZNF467 |
zinc finger protein 467 |
19650 |
0.19 |
| chr6_116690381_116690532 | 0.41 |
DSE |
dermatan sulfate epimerase |
554 |
0.78 |
| chr7_92437724_92438202 | 0.41 |
CDK6 |
cyclin-dependent kinase 6 |
25268 |
0.19 |
| chr20_49026246_49026397 | 0.41 |
ENSG00000244376 |
. |
19699 |
0.22 |
| chr11_12088585_12089097 | 0.41 |
MICAL2 |
microtubule associated monooxygenase, calponin and LIM domain containing 2 |
26702 |
0.22 |
| chr6_37008004_37008155 | 0.41 |
COX6A1P2 |
cytochrome c oxidase subunit VIa polypeptide 1 pseudogene 2 |
4528 |
0.22 |
| chr2_172380569_172381265 | 0.41 |
CYBRD1 |
cytochrome b reductase 1 |
1328 |
0.53 |
| chr2_178283039_178283431 | 0.41 |
AGPS |
alkylglycerone phosphate synthase |
25739 |
0.14 |
| chr3_141248480_141249204 | 0.41 |
RASA2-IT1 |
RASA2 intronic transcript 1 (non-protein coding) |
4867 |
0.27 |
| chr16_71760597_71760829 | 0.40 |
PHLPP2 |
PH domain and leucine rich repeat protein phosphatase 2 |
2109 |
0.19 |
| chr4_118955423_118955622 | 0.40 |
NDST3 |
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 3 |
22 |
0.99 |
| chr17_19172351_19172531 | 0.40 |
EPN2 |
epsin 2 |
11208 |
0.12 |
| chr10_21311864_21312015 | 0.40 |
NEBL |
nebulette |
47007 |
0.18 |
| chr6_15463967_15464118 | 0.40 |
JARID2 |
jumonji, AT rich interactive domain 2 |
62953 |
0.13 |
| chr15_67094989_67095410 | 0.39 |
SMAD6 |
SMAD family member 6 |
91164 |
0.08 |
| chr1_241705552_241705751 | 0.39 |
KMO |
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
9946 |
0.22 |
| chr5_148181532_148181683 | 0.39 |
ADRB2 |
adrenoceptor beta 2, surface |
24549 |
0.23 |
| chr3_87849755_87849906 | 0.39 |
ENSG00000200703 |
. |
58031 |
0.15 |
| chr1_207203760_207203911 | 0.39 |
C1orf116 |
chromosome 1 open reading frame 116 |
2257 |
0.21 |
| chr1_85154249_85154907 | 0.39 |
SSX2IP |
synovial sarcoma, X breakpoint 2 interacting protein |
1394 |
0.5 |
| chr5_90677472_90677623 | 0.39 |
ARRDC3 |
arrestin domain containing 3 |
1629 |
0.47 |
| chr17_47393434_47393962 | 0.38 |
ENSG00000251964 |
. |
9565 |
0.15 |
| chr5_134702384_134702535 | 0.38 |
C5orf66 |
chromosome 5 open reading frame 66 |
28830 |
0.14 |
| chr7_131156247_131156398 | 0.38 |
RP11-180C16.1 |
|
26131 |
0.24 |
| chr19_32837185_32838232 | 0.38 |
ZNF507 |
zinc finger protein 507 |
1083 |
0.51 |
| chr14_97886866_97887017 | 0.38 |
ENSG00000240730 |
. |
109569 |
0.08 |
| chr12_94997883_94998034 | 0.38 |
TMCC3 |
transmembrane and coiled-coil domain family 3 |
11879 |
0.23 |
| chr3_128964455_128964687 | 0.38 |
COPG1 |
coatomer protein complex, subunit gamma 1 |
3878 |
0.19 |
| chrX_92616111_92616262 | 0.38 |
NAP1L3 |
nucleosome assembly protein 1-like 3 |
312381 |
0.01 |
| chrX_15874840_15875069 | 0.38 |
AP1S2 |
adaptor-related protein complex 1, sigma 2 subunit |
1900 |
0.39 |
| chr12_106673629_106673780 | 0.37 |
ENSG00000238609 |
. |
3063 |
0.23 |
| chr12_70079938_70080089 | 0.37 |
BEST3 |
bestrophin 3 |
3043 |
0.26 |
| chr12_66289441_66289878 | 0.37 |
RP11-366L20.2 |
Uncharacterized protein |
13712 |
0.17 |
| chr7_141611302_141611453 | 0.37 |
OR9N1P |
olfactory receptor, family 9, subfamily N, member 1 pseudogene |
114 |
0.94 |
| chr4_46007135_46007286 | 0.37 |
ENSG00000222257 |
. |
10074 |
0.31 |
| chr8_107978697_107978848 | 0.37 |
ABRA |
actin-binding Rho activating protein |
196299 |
0.03 |
| chr19_46586098_46586249 | 0.37 |
IGFL4 |
IGF-like family member 4 |
5821 |
0.17 |
| chr14_73643539_73643710 | 0.37 |
PSEN1 |
presenilin 1 |
9087 |
0.19 |
| chr2_122405151_122406002 | 0.37 |
CLASP1 |
cytoplasmic linker associated protein 1 |
1476 |
0.32 |
| chr9_127420744_127420996 | 0.36 |
MIR181A2HG |
MIR181A2 host gene (non-protein coding) |
124 |
0.96 |
| chr8_128003052_128003209 | 0.36 |
ENSG00000212451 |
. |
319363 |
0.01 |
| chr3_153850737_153851019 | 0.36 |
ARHGEF26 |
Rho guanine nucleotide exchange factor (GEF) 26 |
11649 |
0.22 |
| chr11_95786586_95786916 | 0.36 |
MTMR2 |
myotubularin related protein 2 |
129292 |
0.05 |
| chr7_38969302_38969595 | 0.36 |
VPS41 |
vacuolar protein sorting 41 homolog (S. cerevisiae) |
288 |
0.94 |
| chr11_92931221_92931372 | 0.36 |
SLC36A4 |
solute carrier family 36 (proton/amino acid symporter), member 4 |
167 |
0.97 |
| chr8_130594633_130594805 | 0.36 |
ENSG00000266387 |
. |
84806 |
0.07 |
| chr14_75323168_75323351 | 0.36 |
PROX2 |
prospero homeobox 2 |
7278 |
0.14 |
| chr4_91278223_91278374 | 0.36 |
ENSG00000252087 |
. |
12976 |
0.31 |
| chr11_75367473_75367624 | 0.36 |
MAP6 |
microtubule-associated protein 6 |
11931 |
0.14 |
| chr13_88727217_88727368 | 0.36 |
ENSG00000223059 |
. |
112883 |
0.08 |
| chr1_21277303_21277509 | 0.36 |
EIF4G3 |
eukaryotic translation initiation factor 4 gamma, 3 |
9295 |
0.23 |
| chr8_67042092_67042243 | 0.36 |
TRIM55 |
tripartite motif containing 55 |
2889 |
0.34 |
| chr3_37860112_37860344 | 0.36 |
ENSG00000235257 |
. |
2673 |
0.3 |
| chr13_72142845_72142996 | 0.36 |
DACH1 |
dachshund homolog 1 (Drosophila) |
297987 |
0.01 |
| chr2_160374836_160375140 | 0.36 |
ENSG00000207117 |
. |
17869 |
0.24 |
| chr4_68312692_68312843 | 0.35 |
ENSG00000206629 |
. |
17295 |
0.26 |
| chr15_52436562_52436713 | 0.35 |
CTD-2184D3.7 |
|
6012 |
0.16 |
| chr6_27570448_27570599 | 0.35 |
ENSG00000206671 |
. |
6332 |
0.22 |
| chr12_1427689_1428011 | 0.35 |
RP5-951N9.2 |
|
67149 |
0.11 |
| chr2_232106617_232106773 | 0.35 |
ARMC9 |
armadillo repeat containing 9 |
6630 |
0.24 |
| chr13_91067405_91067695 | 0.35 |
ENSG00000207858 |
. |
184114 |
0.03 |
| chr5_175893259_175893617 | 0.35 |
ENSG00000221373 |
. |
14675 |
0.12 |
| chr1_90408609_90408825 | 0.35 |
ENSG00000252797 |
. |
44786 |
0.15 |
| chr17_29553936_29554129 | 0.35 |
NF1 |
neurofibromin 1 |
26478 |
0.15 |
| chr6_149727250_149727401 | 0.35 |
SUMO4 |
small ubiquitin-like modifier 4 |
5830 |
0.2 |
| chr11_114034929_114035103 | 0.35 |
ENSG00000221112 |
. |
77365 |
0.09 |
| chr9_22165561_22165745 | 0.34 |
CDKN2B-AS1 |
CDKN2B antisense RNA 1 |
51976 |
0.16 |
| chr1_27425346_27425645 | 0.34 |
SLC9A1 |
solute carrier family 9, subfamily A (NHE1, cation proton antiporter 1), member 1 |
55478 |
0.1 |
| chr8_99248451_99248632 | 0.34 |
ENSG00000252558 |
. |
44000 |
0.14 |
| chr11_116747490_116747649 | 0.34 |
SIK3-IT1 |
SIK3 intronic transcript 1 (non-protein coding) |
13868 |
0.12 |
| chr11_35954304_35954455 | 0.34 |
LDLRAD3 |
low density lipoprotein receptor class A domain containing 3 |
11152 |
0.18 |
| chr16_48364740_48364935 | 0.34 |
LONP2 |
lon peptidase 2, peroxisomal |
3386 |
0.26 |
| chr5_36948326_36948572 | 0.34 |
NIPBL |
Nipped-B homolog (Drosophila) |
71557 |
0.13 |
| chr5_143241658_143241809 | 0.34 |
HMHB1 |
histocompatibility (minor) HB-1 |
50007 |
0.18 |
| chr9_35526012_35526444 | 0.33 |
RUSC2 |
RUN and SH3 domain containing 2 |
12401 |
0.15 |
| chr17_15869540_15869896 | 0.33 |
ADORA2B |
adenosine A2b receptor |
21487 |
0.15 |
| chr2_175018489_175018640 | 0.33 |
OLA1 |
Obg-like ATPase 1 |
64110 |
0.12 |
| chr11_19265860_19266011 | 0.33 |
E2F8 |
E2F transcription factor 8 |
2768 |
0.28 |
| chr2_174026267_174026418 | 0.33 |
MLK7-AS1 |
MLK7 antisense RNA 1 |
45998 |
0.16 |
| chr5_115374031_115374427 | 0.33 |
ENSG00000220996 |
. |
4729 |
0.15 |
| chr3_51654767_51654946 | 0.33 |
RAD54L2 |
RAD54-like 2 (S. cerevisiae) |
8551 |
0.17 |
| chr5_160121113_160121264 | 0.33 |
CTC-529G1.1 |
|
8830 |
0.21 |
| chr1_183430720_183430871 | 0.33 |
SMG7 |
SMG7 nonsense mediated mRNA decay factor |
10711 |
0.21 |
| chr1_212433899_212434288 | 0.33 |
PPP2R5A |
protein phosphatase 2, regulatory subunit B', alpha |
24786 |
0.16 |
| chr21_23887924_23888075 | 0.33 |
ENSG00000263796 |
. |
164993 |
0.04 |
| chr6_106994075_106994866 | 0.33 |
AIM1 |
absent in melanoma 1 |
5441 |
0.23 |
| chr10_77190817_77191121 | 0.33 |
RP11-399K21.10 |
|
377 |
0.87 |
| chr7_105807957_105808108 | 0.33 |
ENSG00000201796 |
. |
40581 |
0.17 |
| chr11_103935040_103935191 | 0.32 |
PDGFD |
platelet derived growth factor D |
27361 |
0.2 |
| chr2_54191777_54192195 | 0.32 |
ACYP2 |
acylphosphatase 2, muscle type |
5989 |
0.21 |
| chr9_74363230_74363515 | 0.32 |
TMEM2 |
transmembrane protein 2 |
19930 |
0.27 |
| chr8_59084985_59085323 | 0.32 |
FAM110B |
family with sequence similarity 110, member B |
178041 |
0.03 |
| chr2_101728983_101729400 | 0.32 |
ENSG00000265860 |
. |
19160 |
0.19 |
| chr5_139532409_139532571 | 0.32 |
CYSTM1 |
cysteine-rich transmembrane module containing 1 |
21737 |
0.12 |
| chr8_131278471_131278622 | 0.32 |
ASAP1 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1 |
19258 |
0.26 |
| chr1_25458423_25458574 | 0.32 |
RP4-706G24.1 |
|
76132 |
0.08 |
| chr3_18787817_18787968 | 0.32 |
ENSG00000228956 |
. |
656 |
0.84 |
| chr11_78101755_78101906 | 0.32 |
RP11-452H21.2 |
|
2723 |
0.27 |
| chr17_58516951_58517102 | 0.32 |
C17orf64 |
chromosome 17 open reading frame 64 |
17156 |
0.18 |
| chr2_135843202_135843353 | 0.32 |
ENSG00000238337 |
. |
19973 |
0.17 |
| chr5_151063563_151063813 | 0.32 |
SPARC |
secreted protein, acidic, cysteine-rich (osteonectin) |
1103 |
0.43 |
| chr9_31590126_31590277 | 0.32 |
ENSG00000252313 |
. |
703355 |
0.0 |
| chr18_29260479_29260630 | 0.32 |
B4GALT6 |
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 6 |
3913 |
0.17 |
| chr7_102452212_102452699 | 0.31 |
ENSG00000252643 |
. |
22597 |
0.16 |
| chr11_8719978_8720129 | 0.31 |
ST5 |
suppression of tumorigenicity 5 |
3851 |
0.13 |
| chr9_297842_297993 | 0.31 |
DOCK8 |
dedicator of cytokinesis 8 |
24847 |
0.18 |
| chrX_135967912_135968067 | 0.31 |
RBMX |
RNA binding motif protein, X-linked |
5066 |
0.21 |
| chr20_47448722_47448873 | 0.31 |
PREX1 |
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 |
4377 |
0.31 |
| chr1_174758122_174758399 | 0.31 |
RABGAP1L |
RAB GTPase activating protein 1-like |
10810 |
0.28 |
| chr5_148645320_148645471 | 0.31 |
AFAP1L1 |
actin filament associated protein 1-like 1 |
6039 |
0.14 |
| chr1_234639139_234639392 | 0.31 |
TARBP1 |
TAR (HIV-1) RNA binding protein 1 |
24416 |
0.19 |
| chr1_9962207_9962358 | 0.31 |
CTNNBIP1 |
catenin, beta interacting protein 1 |
8039 |
0.14 |
| chr12_89777034_89777222 | 0.31 |
DUSP6 |
dual specificity phosphatase 6 |
30080 |
0.2 |
| chr10_73104848_73104999 | 0.31 |
ENSG00000238918 |
. |
19838 |
0.18 |
| chr8_96875884_96876062 | 0.31 |
ENSG00000223297 |
. |
184830 |
0.03 |
| chr4_38077270_38077549 | 0.31 |
TBC1D1 |
TBC1 (tre-2/USP6, BUB2, cdc16) domain family, member 1 |
29917 |
0.24 |
| chr3_171528829_171529427 | 0.31 |
PLD1 |
phospholipase D1, phosphatidylcholine-specific |
844 |
0.52 |
| chr12_77777777_77777928 | 0.31 |
AC073528.1 |
|
172716 |
0.04 |
| chr1_234755566_234755985 | 0.31 |
IRF2BP2 |
interferon regulatory factor 2 binding protein 2 |
10504 |
0.19 |
| chr20_5837496_5837647 | 0.30 |
ENSG00000202380 |
. |
33130 |
0.16 |
| chr2_208734859_208735279 | 0.30 |
AC083900.1 |
|
430 |
0.86 |
| chr2_160470412_160470682 | 0.30 |
AC009506.1 |
|
1260 |
0.4 |
| chr5_127059262_127059522 | 0.30 |
CTC-548H10.2 |
|
59671 |
0.14 |
| chr12_113251939_113252090 | 0.30 |
RPH3A |
rabphilin 3A homolog (mouse) |
14110 |
0.24 |
| chr2_192241823_192241993 | 0.30 |
ENSG00000252130 |
. |
6244 |
0.26 |
| chr12_6292503_6292996 | 0.30 |
CD9 |
CD9 molecule |
16132 |
0.18 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.1 | 0.3 | GO:0030812 | negative regulation of nucleotide catabolic process(GO:0030812) |
| 0.1 | 0.1 | GO:0072367 | regulation of lipid transport by regulation of transcription from RNA polymerase II promoter(GO:0072367) |
| 0.1 | 0.2 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.1 | 0.4 | GO:0045652 | regulation of megakaryocyte differentiation(GO:0045652) regulation of hematopoietic progenitor cell differentiation(GO:1901532) |
| 0.1 | 0.2 | GO:0001714 | endodermal cell fate specification(GO:0001714) regulation of endodermal cell fate specification(GO:0042663) regulation of endodermal cell differentiation(GO:1903224) |
| 0.1 | 0.2 | GO:0070340 | detection of bacterial lipoprotein(GO:0042494) detection of bacterial lipopeptide(GO:0070340) |
| 0.1 | 0.2 | GO:0060058 | apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 0.1 | 0.2 | GO:0071436 | sodium ion export(GO:0071436) |
| 0.1 | 0.2 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.0 | 0.2 | GO:0002138 | retinoic acid biosynthetic process(GO:0002138) diterpenoid biosynthetic process(GO:0016102) |
| 0.0 | 0.2 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 0.2 | GO:0050651 | dermatan sulfate proteoglycan biosynthetic process(GO:0050651) |
| 0.0 | 0.2 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.0 | 0.1 | GO:0051142 | regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) |
| 0.0 | 0.4 | GO:0042119 | neutrophil activation(GO:0042119) |
| 0.0 | 0.1 | GO:0010891 | negative regulation of sequestering of triglyceride(GO:0010891) |
| 0.0 | 0.0 | GO:0048087 | positive regulation of developmental pigmentation(GO:0048087) |
| 0.0 | 0.1 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.1 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.0 | 0.2 | GO:2000831 | corticosteroid hormone secretion(GO:0035930) regulation of steroid hormone secretion(GO:2000831) regulation of corticosteroid hormone secretion(GO:2000846) |
| 0.0 | 0.2 | GO:0045634 | regulation of melanocyte differentiation(GO:0045634) regulation of pigment cell differentiation(GO:0050932) |
| 0.0 | 0.1 | GO:0032959 | inositol trisphosphate biosynthetic process(GO:0032959) |
| 0.0 | 0.1 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.1 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.1 | GO:0070339 | response to bacterial lipoprotein(GO:0032493) response to bacterial lipopeptide(GO:0070339) cellular response to bacterial lipoprotein(GO:0071220) cellular response to bacterial lipopeptide(GO:0071221) |
| 0.0 | 0.1 | GO:0042640 | anagen(GO:0042640) |
| 0.0 | 0.1 | GO:0045410 | positive regulation of interleukin-6 biosynthetic process(GO:0045410) |
| 0.0 | 0.1 | GO:0010566 | regulation of ketone biosynthetic process(GO:0010566) regulation of aldosterone metabolic process(GO:0032344) regulation of aldosterone biosynthetic process(GO:0032347) |
| 0.0 | 0.1 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.1 | GO:0070672 | response to interleukin-15(GO:0070672) |
| 0.0 | 0.1 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.0 | 0.1 | GO:1900115 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.1 | GO:0051877 | pigment granule aggregation in cell center(GO:0051877) |
| 0.0 | 0.1 | GO:0060585 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.0 | 0.0 | GO:0033591 | response to L-ascorbic acid(GO:0033591) |
| 0.0 | 0.1 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.2 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.1 | GO:0046049 | UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.0 | 0.1 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 | 0.2 | GO:0031274 | pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.1 | GO:0060633 | negative regulation of transcription initiation from RNA polymerase II promoter(GO:0060633) negative regulation of DNA-templated transcription, initiation(GO:2000143) |
| 0.0 | 0.1 | GO:0044557 | relaxation of smooth muscle(GO:0044557) relaxation of vascular smooth muscle(GO:0060087) |
| 0.0 | 0.1 | GO:0033131 | regulation of glucokinase activity(GO:0033131) positive regulation of glucokinase activity(GO:0033133) regulation of hexokinase activity(GO:1903299) positive regulation of hexokinase activity(GO:1903301) |
| 0.0 | 0.0 | GO:0071224 | cellular response to peptidoglycan(GO:0071224) |
| 0.0 | 0.1 | GO:0009415 | response to water deprivation(GO:0009414) response to water(GO:0009415) |
| 0.0 | 0.1 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.0 | 0.1 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.0 | 0.1 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 | 0.1 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 0.0 | 0.2 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.1 | GO:0003323 | type B pancreatic cell development(GO:0003323) |
| 0.0 | 0.1 | GO:0070296 | sarcoplasmic reticulum calcium ion transport(GO:0070296) |
| 0.0 | 0.1 | GO:0010903 | negative regulation of very-low-density lipoprotein particle remodeling(GO:0010903) |
| 0.0 | 0.1 | GO:0060426 | lung vasculature development(GO:0060426) |
| 0.0 | 0.2 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.1 | GO:0034086 | maintenance of sister chromatid cohesion(GO:0034086) maintenance of mitotic sister chromatid cohesion(GO:0034088) |
| 0.0 | 0.1 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.1 | GO:0002606 | positive regulation of dendritic cell antigen processing and presentation(GO:0002606) |
| 0.0 | 0.1 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.1 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.0 | 0.1 | GO:0030219 | megakaryocyte differentiation(GO:0030219) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.0 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.1 | GO:0046835 | carbohydrate phosphorylation(GO:0046835) |
| 0.0 | 0.2 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 | 0.1 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.1 | GO:0032528 | microvillus assembly(GO:0030033) microvillus organization(GO:0032528) |
| 0.0 | 0.1 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) positive regulation of signal transduction by p53 class mediator(GO:1901798) |
| 0.0 | 0.2 | GO:0007567 | parturition(GO:0007567) |
| 0.0 | 0.1 | GO:0072112 | renal filtration cell differentiation(GO:0061318) glomerular epithelium development(GO:0072010) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.0 | 0.1 | GO:0048541 | Peyer's patch development(GO:0048541) |
| 0.0 | 0.4 | GO:0006662 | glycerol ether metabolic process(GO:0006662) ether metabolic process(GO:0018904) |
| 0.0 | 0.1 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.0 | 0.1 | GO:0035461 | vitamin transmembrane transport(GO:0035461) |
| 0.0 | 0.1 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.0 | 0.1 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.0 | 0.1 | GO:0002238 | response to molecule of fungal origin(GO:0002238) |
| 0.0 | 0.1 | GO:0045631 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.0 | 0.1 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.1 | GO:0048853 | forebrain morphogenesis(GO:0048853) |
| 0.0 | 0.1 | GO:1903670 | regulation of cell migration involved in sprouting angiogenesis(GO:0090049) regulation of sprouting angiogenesis(GO:1903670) |
| 0.0 | 0.1 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) negative regulation of neural precursor cell proliferation(GO:2000178) |
| 0.0 | 0.1 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.1 | GO:0008615 | pyridoxine metabolic process(GO:0008614) pyridoxine biosynthetic process(GO:0008615) vitamin B6 metabolic process(GO:0042816) vitamin B6 biosynthetic process(GO:0042819) |
| 0.0 | 0.0 | GO:0072393 | microtubule anchoring at microtubule organizing center(GO:0072393) |
| 0.0 | 0.1 | GO:0009649 | entrainment of circadian clock(GO:0009649) |
| 0.0 | 0.1 | GO:0043461 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.0 | 0.1 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.0 | GO:0060398 | regulation of growth hormone receptor signaling pathway(GO:0060398) |
| 0.0 | 0.1 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.0 | 0.0 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.0 | 0.1 | GO:0018101 | protein citrullination(GO:0018101) |
| 0.0 | 0.0 | GO:0060083 | smooth muscle contraction involved in micturition(GO:0060083) |
| 0.0 | 0.1 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.1 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.1 | GO:0001547 | antral ovarian follicle growth(GO:0001547) |
| 0.0 | 0.1 | GO:0032700 | negative regulation of interleukin-17 production(GO:0032700) |
| 0.0 | 0.0 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.2 | GO:0007128 | meiotic prophase I(GO:0007128) |
| 0.0 | 0.0 | GO:0048245 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.0 | 0.0 | GO:0002227 | innate immune response in mucosa(GO:0002227) mucosal immune response(GO:0002385) |
| 0.0 | 0.2 | GO:0045668 | negative regulation of osteoblast differentiation(GO:0045668) |
| 0.0 | 0.1 | GO:1903054 | regulation of extracellular matrix disassembly(GO:0010715) negative regulation of extracellular matrix disassembly(GO:0010716) regulation of extracellular matrix organization(GO:1903053) negative regulation of extracellular matrix organization(GO:1903054) |
| 0.0 | 0.1 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.1 | GO:0043302 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of leukocyte degranulation(GO:0043302) positive regulation of mast cell degranulation(GO:0043306) |
| 0.0 | 0.1 | GO:0006734 | NADH metabolic process(GO:0006734) |
| 0.0 | 0.0 | GO:0042339 | keratan sulfate metabolic process(GO:0042339) |
| 0.0 | 0.1 | GO:0006930 | substrate-dependent cell migration, cell extension(GO:0006930) |
| 0.0 | 0.1 | GO:0051570 | regulation of histone H3-K9 methylation(GO:0051570) |
| 0.0 | 0.0 | GO:0021827 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) substrate-dependent cerebral cortex tangential migration(GO:0021825) postnatal olfactory bulb interneuron migration(GO:0021827) |
| 0.0 | 0.1 | GO:0097061 | dendritic spine morphogenesis(GO:0060997) regulation of dendritic spine morphogenesis(GO:0061001) dendritic spine organization(GO:0097061) |
| 0.0 | 0.0 | GO:0032898 | neurotrophin production(GO:0032898) |
| 0.0 | 0.0 | GO:0060437 | lung growth(GO:0060437) |
| 0.0 | 0.1 | GO:0045077 | negative regulation of interferon-gamma biosynthetic process(GO:0045077) |
| 0.0 | 0.0 | GO:0002513 | tolerance induction to self antigen(GO:0002513) |
| 0.0 | 0.0 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.0 | 0.2 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 0.1 | GO:0032782 | canalicular bile acid transport(GO:0015722) bile acid secretion(GO:0032782) |
| 0.0 | 0.0 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.0 | 0.0 | GO:0090273 | regulation of somatostatin secretion(GO:0090273) |
| 0.0 | 0.1 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.2 | GO:0035336 | long-chain fatty-acyl-CoA metabolic process(GO:0035336) long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.0 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.0 | 0.1 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.0 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 | 0.0 | GO:0060435 | bronchiole development(GO:0060435) |
| 0.0 | 0.1 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.1 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.0 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 | 0.0 | GO:0001865 | NK T cell differentiation(GO:0001865) regulation of NK T cell differentiation(GO:0051136) positive regulation of NK T cell differentiation(GO:0051138) |
| 0.0 | 0.0 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.0 | 0.1 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.0 | 0.0 | GO:1901970 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.0 | 0.0 | GO:0060693 | regulation of branching involved in salivary gland morphogenesis(GO:0060693) |
| 0.0 | 0.2 | GO:0046464 | triglyceride catabolic process(GO:0019433) neutral lipid catabolic process(GO:0046461) acylglycerol catabolic process(GO:0046464) |
| 0.0 | 0.1 | GO:0007635 | chemosensory behavior(GO:0007635) |
| 0.0 | 0.1 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.0 | 0.0 | GO:0051790 | short-chain fatty acid biosynthetic process(GO:0051790) |
| 0.0 | 0.0 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.0 | 0.1 | GO:0045351 | type I interferon biosynthetic process(GO:0045351) |
| 0.0 | 0.0 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 0.1 | GO:0042095 | interferon-gamma biosynthetic process(GO:0042095) regulation of interferon-gamma biosynthetic process(GO:0045072) positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.0 | 0.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.1 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.0 | 0.0 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.0 | 0.0 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.0 | 0.0 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.0 | 0.0 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) |
| 0.0 | 0.0 | GO:0009822 | alkaloid catabolic process(GO:0009822) |
| 0.0 | 0.0 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.0 | 0.1 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.0 | 0.1 | GO:0047497 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.0 | 0.1 | GO:0097435 | extracellular fibril organization(GO:0043206) fibril organization(GO:0097435) |
| 0.0 | 0.1 | GO:0014049 | positive regulation of glutamate secretion(GO:0014049) |
| 0.0 | 0.0 | GO:0052555 | pathogen-associated molecular pattern dependent induction by symbiont of host innate immune response(GO:0052033) positive regulation by symbiont of host innate immune response(GO:0052166) modulation by symbiont of host innate immune response(GO:0052167) pathogen-associated molecular pattern dependent modulation by symbiont of host innate immune response(GO:0052169) pathogen-associated molecular pattern dependent induction by organism of innate immune response of other organism involved in symbiotic interaction(GO:0052257) positive regulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052305) modulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052306) pathogen-associated molecular pattern dependent modulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052308) positive regulation by organism of immune response of other organism involved in symbiotic interaction(GO:0052555) positive regulation by symbiont of host immune response(GO:0052556) |
| 0.0 | 0.1 | GO:0034405 | response to fluid shear stress(GO:0034405) |
| 0.0 | 0.0 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.0 | 0.0 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.0 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.0 | 0.1 | GO:0018027 | peptidyl-lysine dimethylation(GO:0018027) |
| 0.0 | 0.0 | GO:0021903 | rostrocaudal neural tube patterning(GO:0021903) |
| 0.0 | 0.0 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.0 | GO:0007500 | mesodermal cell fate determination(GO:0007500) |
| 0.0 | 0.0 | GO:0030300 | regulation of intestinal cholesterol absorption(GO:0030300) regulation of intestinal absorption(GO:1904478) regulation of intestinal lipid absorption(GO:1904729) |
| 0.0 | 0.0 | GO:0046618 | drug export(GO:0046618) |
| 0.0 | 0.0 | GO:0048143 | astrocyte activation(GO:0048143) |
| 0.0 | 0.0 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.0 | 0.0 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.0 | GO:0097576 | vacuole fusion(GO:0097576) |
| 0.0 | 0.0 | GO:0016999 | antibiotic metabolic process(GO:0016999) antibiotic biosynthetic process(GO:0017000) |
| 0.0 | 0.0 | GO:0042501 | serine phosphorylation of STAT protein(GO:0042501) |
| 0.0 | 0.0 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.0 | GO:0002903 | negative regulation of B cell apoptotic process(GO:0002903) |
| 0.0 | 0.1 | GO:0044091 | membrane biogenesis(GO:0044091) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.1 | 0.2 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.4 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.2 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.0 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.3 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.3 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.2 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.2 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.3 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 0.1 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 0.1 | GO:0070083 | clathrin-sculpted monoamine transport vesicle(GO:0070081) clathrin-sculpted monoamine transport vesicle membrane(GO:0070083) |
| 0.0 | 0.1 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.1 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.1 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.2 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.1 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.1 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.2 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.1 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.1 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 0.4 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.1 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 0.0 | 0.7 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.1 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0045283 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.1 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.1 | GO:0001740 | Barr body(GO:0001740) |
| 0.0 | 0.1 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.1 | GO:0098642 | network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.0 | 0.1 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.0 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.0 | GO:0000805 | X chromosome(GO:0000805) |
| 0.0 | 0.1 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.0 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.1 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.0 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.2 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.2 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.0 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.0 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.1 | GO:0071778 | obsolete WINAC complex(GO:0071778) |
| 0.0 | 0.0 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 0.0 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.1 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.1 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.1 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.1 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.0 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.2 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.1 | GO:0031082 | BLOC complex(GO:0031082) BLOC-1 complex(GO:0031083) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.1 | 0.3 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.1 | 0.3 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.1 | 0.2 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.1 | 0.2 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.1 | 0.3 | GO:0030955 | potassium ion binding(GO:0030955) |
| 0.1 | 0.3 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.1 | 0.2 | GO:0050542 | icosanoid binding(GO:0050542) fatty acid derivative binding(GO:1901567) |
| 0.1 | 0.2 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.1 | 0.3 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.2 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.1 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.3 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.2 | GO:0004904 | interferon receptor activity(GO:0004904) |
| 0.0 | 0.3 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.1 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.1 | GO:0019958 | C-X-C chemokine binding(GO:0019958) interleukin-8 binding(GO:0019959) |
| 0.0 | 0.1 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.2 | GO:0045569 | TRAIL binding(GO:0045569) |
| 0.0 | 0.2 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
| 0.0 | 0.6 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) |
| 0.0 | 0.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.4 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.0 | 0.1 | GO:0001099 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.0 | 0.2 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.1 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.1 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.1 | GO:0016972 | thiol oxidase activity(GO:0016972) |
| 0.0 | 0.4 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.2 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.1 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.1 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.2 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.1 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.0 | 0.1 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.0 | 0.2 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.1 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.0 | 0.2 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.5 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.1 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.0 | 0.1 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.1 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 0.1 | GO:0045118 | azole transporter activity(GO:0045118) azole transmembrane transporter activity(GO:1901474) |
| 0.0 | 0.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.1 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.0 | 0.1 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.0 | 0.1 | GO:0004515 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.0 | 0.1 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.1 | GO:0031708 | bombesin receptor binding(GO:0031705) endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.1 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.1 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.1 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.0 | GO:0008061 | chitin binding(GO:0008061) |
| 0.0 | 0.1 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.0 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 0.0 | 0.0 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.2 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.2 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.1 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.1 | GO:0008035 | high-density lipoprotein particle binding(GO:0008035) |
| 0.0 | 0.1 | GO:0098518 | polynucleotide phosphatase activity(GO:0098518) |
| 0.0 | 0.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.1 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.0 | 0.1 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.0 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.1 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.0 | 0.1 | GO:0030343 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.0 | 0.1 | GO:0042808 | obsolete neuronal Cdc2-like kinase binding(GO:0042808) |
| 0.0 | 0.1 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.0 | 0.1 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.1 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.1 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.0 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.0 | 0.3 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.1 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.2 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.1 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.0 | 0.1 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) inositol trisphosphate phosphatase activity(GO:0046030) |
| 0.0 | 0.1 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 0.1 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 0.1 | GO:0030250 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.0 | 0.0 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.0 | 0.1 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.0 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.0 | 0.1 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.1 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.0 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 0.1 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.0 | 0.0 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.0 | 0.1 | GO:0015065 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.0 | 0.0 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.0 | GO:0034648 | histone demethylase activity (H3-K4 specific)(GO:0032453) histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.0 | 0.1 | GO:0001595 | angiotensin receptor activity(GO:0001595) angiotensin type II receptor activity(GO:0004945) |
| 0.0 | 0.0 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.1 | GO:0034739 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) histone deacetylase activity (H4-K16 specific)(GO:0034739) NAD-dependent histone deacetylase activity (H4-K16 specific)(GO:0046970) |
| 0.0 | 0.1 | GO:0016653 | oxidoreductase activity, acting on NAD(P)H, heme protein as acceptor(GO:0016653) |
| 0.0 | 0.1 | GO:0044390 | ubiquitin conjugating enzyme binding(GO:0031624) ubiquitin-like protein conjugating enzyme binding(GO:0044390) |
| 0.0 | 0.1 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.0 | 0.1 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.0 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.1 | GO:0008329 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.0 | 0.0 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.0 | 0.1 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.0 | 0.1 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.1 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.1 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.1 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.0 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.1 | GO:0022821 | potassium ion antiporter activity(GO:0022821) |
| 0.0 | 0.0 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.0 | 0.1 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.0 | GO:0004738 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.0 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.0 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.0 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.1 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.0 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.2 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 0.0 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.0 | 0.0 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.0 | 0.3 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.0 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 0.0 | GO:0033765 | steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) |
| 0.0 | 0.0 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.0 | 0.0 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.0 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.1 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
| 0.0 | 0.1 | GO:0005451 | monovalent cation:proton antiporter activity(GO:0005451) |
| 0.0 | 0.1 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.0 | 0.1 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.0 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.1 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.1 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.1 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 0.1 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.0 | 0.0 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.0 | 0.1 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.0 | 0.1 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.2 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 0.5 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.0 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.0 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.8 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.0 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.1 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.0 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.3 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.0 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.2 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.2 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.6 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.1 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.0 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 0.0 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.1 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.0 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.0 | 0.3 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.1 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.2 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 0.1 | PID AP1 PATHWAY | AP-1 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | REACTOME SIGNALING BY FGFR | Genes involved in Signaling by FGFR |
| 0.0 | 0.0 | REACTOME BILE ACID AND BILE SALT METABOLISM | Genes involved in Bile acid and bile salt metabolism |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.3 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.0 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.2 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.5 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.0 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.3 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.0 | 0.0 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.0 | 0.3 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.2 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PC | Genes involved in Acyl chain remodelling of PC |
| 0.0 | 0.2 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.2 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.0 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.3 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.5 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.5 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.2 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.0 | 0.2 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.2 | REACTOME BOTULINUM NEUROTOXICITY | Genes involved in Botulinum neurotoxicity |
| 0.0 | 0.1 | REACTOME SCFSKP2 MEDIATED DEGRADATION OF P27 P21 | Genes involved in SCF(Skp2)-mediated degradation of p27/p21 |
| 0.0 | 0.2 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.1 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.0 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.4 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.1 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.0 | REACTOME SCF BETA TRCP MEDIATED DEGRADATION OF EMI1 | Genes involved in SCF-beta-TrCP mediated degradation of Emi1 |
| 0.0 | 0.2 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |