Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
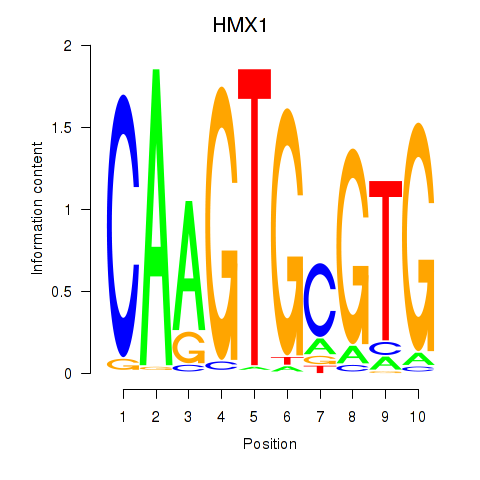
Results for HMX1
Z-value: 0.88
Transcription factors associated with HMX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HMX1
|
ENSG00000215612.5 | H6 family homeobox 1 |
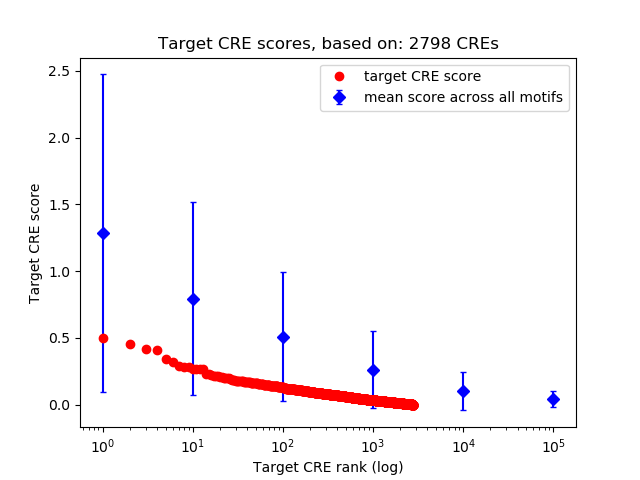
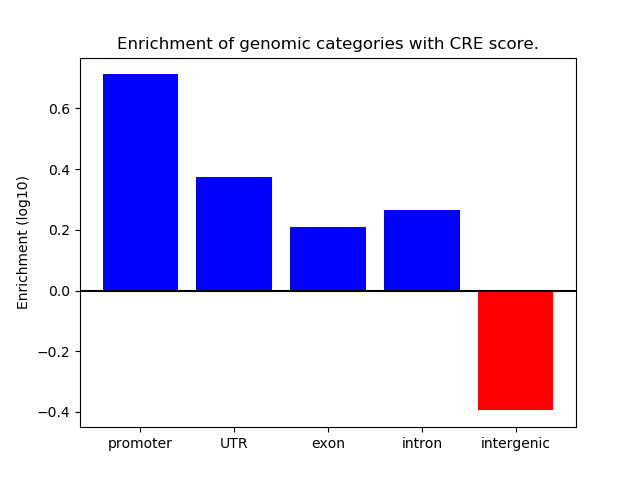
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr4_8873847_8873998 | HMX1 | 379 | 0.884669 | -0.45 | 2.2e-01 | Click! |
| chr4_8873186_8873337 | HMX1 | 87 | 0.976219 | -0.33 | 3.9e-01 | Click! |
| chr4_8874389_8874658 | HMX1 | 980 | 0.604345 | -0.32 | 4.0e-01 | Click! |
| chr4_8920155_8920306 | HMX1 | 46687 | 0.141165 | -0.31 | 4.2e-01 | Click! |
| chr4_8874176_8874327 | HMX1 | 708 | 0.724009 | -0.28 | 4.6e-01 | Click! |
Activity of the HMX1 motif across conditions
Conditions sorted by the z-value of the HMX1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
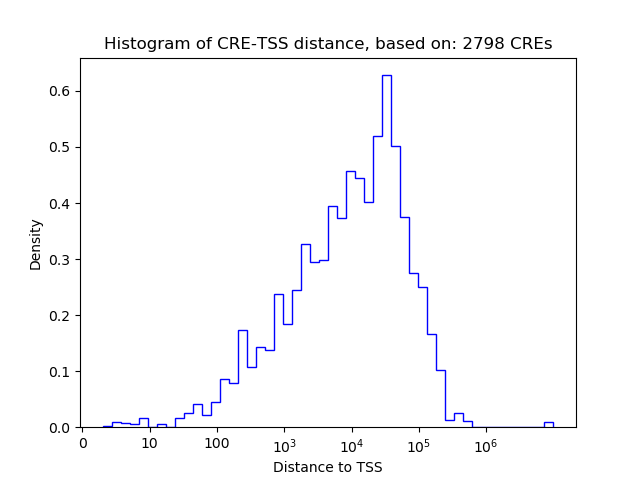
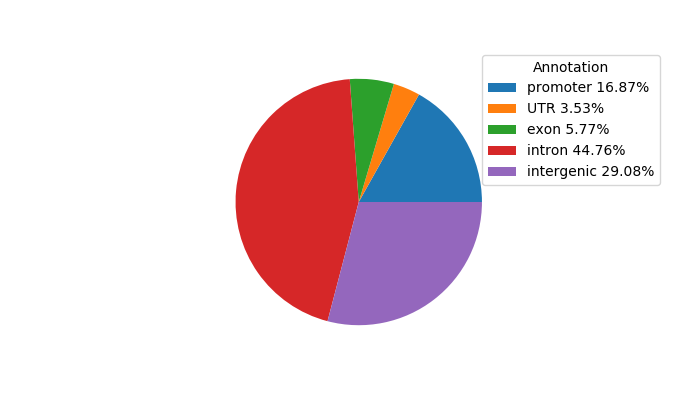
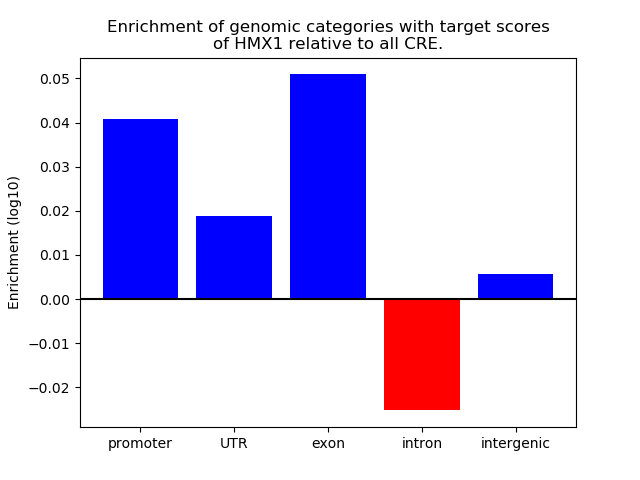
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr16_75284868_75285315 | 0.50 |
BCAR1 |
breast cancer anti-estrogen resistance 1 |
246 |
0.89 |
| chr8_49493164_49493315 | 0.45 |
RP11-770E5.1 |
|
29112 |
0.25 |
| chr22_43658738_43659116 | 0.42 |
SCUBE1 |
signal peptide, CUB domain, EGF-like 1 |
122 |
0.96 |
| chr1_15307701_15307852 | 0.41 |
KAZN |
kazrin, periplakin interacting protein |
35361 |
0.22 |
| chr2_241937276_241937504 | 0.34 |
SNED1 |
sushi, nidogen and EGF-like domains 1 |
865 |
0.49 |
| chr13_114883288_114883480 | 0.32 |
RASA3-IT1 |
RASA3 intronic transcript 1 (non-protein coding) |
9089 |
0.22 |
| chr3_194930783_194931056 | 0.29 |
ENSG00000206600 |
. |
4597 |
0.2 |
| chr12_46797636_46797787 | 0.28 |
SLC38A2 |
solute carrier family 38, member 2 |
31061 |
0.2 |
| chr14_93473458_93473609 | 0.28 |
ITPK1 |
inositol-tetrakisphosphate 1-kinase |
59170 |
0.11 |
| chr19_18770507_18770658 | 0.27 |
KLHL26 |
kelch-like family member 26 |
22715 |
0.1 |
| chr10_44790945_44791096 | 0.27 |
RP11-20J15.3 |
|
1863 |
0.46 |
| chr7_137643245_137643895 | 0.27 |
AC022173.2 |
|
5476 |
0.23 |
| chr7_130603215_130603366 | 0.27 |
ENSG00000226380 |
. |
40992 |
0.17 |
| chr19_55953317_55953576 | 0.23 |
SHISA7 |
shisa family member 7 |
258 |
0.81 |
| chr22_19651364_19651581 | 0.23 |
SEPT5 |
septin 5 |
50515 |
0.11 |
| chr17_55361115_55361288 | 0.22 |
MSI2 |
musashi RNA-binding protein 2 |
2004 |
0.44 |
| chr4_8262898_8263049 | 0.22 |
HTRA3 |
HtrA serine peptidase 3 |
8519 |
0.22 |
| chr11_65307867_65308057 | 0.21 |
LTBP3 |
latent transforming growth factor beta binding protein 3 |
39 |
0.95 |
| chr10_124220042_124220595 | 0.21 |
HTRA1 |
HtrA serine peptidase 1 |
723 |
0.66 |
| chr1_156357812_156357984 | 0.21 |
RHBG |
Rh family, B glycoprotein (gene/pseudogene) |
18895 |
0.1 |
| chr20_48920639_48920885 | 0.20 |
CEBPB |
CCAAT/enhancer binding protein (C/EBP), beta |
113386 |
0.05 |
| chr15_91234173_91234396 | 0.20 |
RP11-387D10.2 |
|
25996 |
0.12 |
| chr4_26461958_26462109 | 0.20 |
CCKAR |
cholecystokinin A receptor |
30051 |
0.2 |
| chr15_68911722_68911873 | 0.20 |
CORO2B |
coronin, actin binding protein, 2B |
2918 |
0.38 |
| chr5_100760236_100760486 | 0.20 |
ENSG00000264318 |
. |
157173 |
0.04 |
| chr4_75174057_75174309 | 0.19 |
EPGN |
epithelial mitogen |
7 |
0.98 |
| chr10_31908093_31908244 | 0.19 |
ENSG00000222412 |
. |
136896 |
0.05 |
| chr10_105397530_105397729 | 0.18 |
SH3PXD2A |
SH3 and PX domains 2A |
23833 |
0.14 |
| chr2_102638617_102638768 | 0.18 |
IL1R2 |
interleukin 1 receptor, type II |
13715 |
0.23 |
| chr6_25296780_25296931 | 0.18 |
ENSG00000207286 |
. |
9094 |
0.17 |
| chr12_88827809_88828036 | 0.18 |
ENSG00000199245 |
. |
3703 |
0.36 |
| chr17_79534326_79534493 | 0.18 |
NPLOC4 |
nuclear protein localization 4 homolog (S. cerevisiae) |
748 |
0.47 |
| chr22_50356250_50357150 | 0.18 |
PIM3 |
pim-3 oncogene |
2539 |
0.27 |
| chr17_31197649_31197800 | 0.18 |
AC084809.2 |
|
6053 |
0.19 |
| chr2_75147104_75147491 | 0.18 |
POLE4 |
polymerase (DNA-directed), epsilon 4, accessory subunit |
38322 |
0.18 |
| chr15_81557784_81557935 | 0.18 |
IL16 |
interleukin 16 |
31395 |
0.18 |
| chr11_68272006_68272464 | 0.17 |
ENSG00000222339 |
. |
884 |
0.48 |
| chr4_143289857_143290479 | 0.17 |
INPP4B |
inositol polyphosphate-4-phosphatase, type II, 105kDa |
62244 |
0.16 |
| chr1_17305422_17305972 | 0.17 |
RP1-37C10.3 |
|
276 |
0.81 |
| chr1_68056614_68056765 | 0.17 |
ENSG00000207504 |
. |
49879 |
0.14 |
| chr20_5519861_5520131 | 0.17 |
RP5-1022P6.5 |
|
5722 |
0.27 |
| chr5_111268378_111268671 | 0.17 |
NREP-AS1 |
NREP antisense RNA 1 |
7226 |
0.27 |
| chr8_108452636_108452787 | 0.17 |
ANGPT1 |
angiopoietin 1 |
54512 |
0.18 |
| chr4_154178117_154178352 | 0.17 |
TRIM2 |
tripartite motif containing 2 |
328 |
0.88 |
| chr1_25098309_25098460 | 0.17 |
ENSG00000238482 |
. |
6086 |
0.23 |
| chr6_25230795_25230946 | 0.16 |
ENSG00000264238 |
. |
27389 |
0.13 |
| chr1_66727440_66727741 | 0.16 |
PDE4B |
phosphodiesterase 4B, cAMP-specific |
4260 |
0.35 |
| chr15_81295813_81296187 | 0.16 |
MESDC1 |
mesoderm development candidate 1 |
2705 |
0.2 |
| chr1_19752185_19752560 | 0.16 |
ENSG00000240490 |
. |
1494 |
0.36 |
| chr6_90842347_90842775 | 0.16 |
ENSG00000222078 |
. |
131336 |
0.05 |
| chr14_91000726_91000954 | 0.16 |
ENSG00000252748 |
. |
30645 |
0.18 |
| chr12_2357520_2357671 | 0.16 |
CACNA1C-IT3 |
CACNA1C intronic transcript 3 (non-protein coding) |
21347 |
0.21 |
| chr9_130559179_130559456 | 0.16 |
FPGS |
folylpolyglutamate synthase |
5837 |
0.09 |
| chr21_44142671_44142822 | 0.16 |
AP001627.1 |
|
19122 |
0.2 |
| chr12_28122414_28123153 | 0.16 |
PTHLH |
parathyroid hormone-like hormone |
197 |
0.95 |
| chr11_76413011_76413162 | 0.16 |
GUCY2EP |
guanylate cyclase 2E, pseudogene |
5599 |
0.18 |
| chr18_12652918_12653086 | 0.16 |
SPIRE1 |
spire-type actin nucleation factor 1 |
82 |
0.96 |
| chr13_21289490_21289641 | 0.16 |
ENSG00000265710 |
. |
11723 |
0.19 |
| chr15_78285915_78286311 | 0.15 |
RP11-114H24.6 |
|
366 |
0.81 |
| chr10_54515051_54515202 | 0.15 |
RP11-556E13.1 |
|
43 |
0.98 |
| chr5_95177892_95178147 | 0.15 |
C5orf27 |
chromosome 5 open reading frame 27 |
9917 |
0.14 |
| chr14_25333651_25333802 | 0.15 |
STXBP6 |
syntaxin binding protein 6 (amisyn) |
45043 |
0.18 |
| chr4_146808228_146808388 | 0.15 |
ZNF827 |
zinc finger protein 827 |
51315 |
0.13 |
| chr2_159942894_159943170 | 0.15 |
ENSG00000202029 |
. |
59378 |
0.14 |
| chr7_71335903_71336054 | 0.15 |
ENSG00000201014 |
. |
42863 |
0.21 |
| chr2_74909742_74909893 | 0.15 |
SEMA4F |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4F |
28379 |
0.14 |
| chr18_43169913_43170064 | 0.15 |
SLC14A2 |
solute carrier family 14 (urea transporter), member 2 |
24778 |
0.19 |
| chr17_79486044_79486605 | 0.15 |
RP13-766D20.2 |
|
258 |
0.83 |
| chr16_70736623_70736774 | 0.15 |
VAC14 |
Vac14 homolog (S. cerevisiae) |
2550 |
0.22 |
| chr12_124743142_124743293 | 0.15 |
FAM101A |
family with sequence similarity 101, member A |
30493 |
0.21 |
| chr1_44770285_44770436 | 0.14 |
ERI3 |
ERI1 exoribonuclease family member 3 |
15062 |
0.17 |
| chr6_74388914_74389065 | 0.14 |
CD109 |
CD109 molecule |
16519 |
0.17 |
| chr6_2901195_2901808 | 0.14 |
SERPINB9 |
serpin peptidase inhibitor, clade B (ovalbumin), member 9 |
2013 |
0.31 |
| chr13_32740029_32740180 | 0.14 |
FRY |
furry homolog (Drosophila) |
98701 |
0.07 |
| chr7_50207733_50207942 | 0.14 |
AC020743.2 |
|
25418 |
0.21 |
| chr17_81146997_81147387 | 0.14 |
METRNL |
meteorin, glial cell differentiation regulator-like |
95198 |
0.08 |
| chr21_43448021_43448283 | 0.14 |
ZBTB21 |
zinc finger and BTB domain containing 21 |
17656 |
0.18 |
| chr1_39716504_39716655 | 0.14 |
RP11-420K8.1 |
|
6742 |
0.2 |
| chr6_10426428_10427175 | 0.14 |
TFAP2A |
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha) |
6930 |
0.18 |
| chr8_38710177_38710328 | 0.14 |
RP11-723D22.3 |
|
6295 |
0.19 |
| chr1_200194675_200194826 | 0.14 |
ENSG00000221403 |
. |
80788 |
0.1 |
| chr2_42983831_42983982 | 0.14 |
OXER1 |
oxoeicosanoid (OXE) receptor 1 |
7495 |
0.23 |
| chr19_47237211_47237388 | 0.14 |
ENSG00000222614 |
. |
2270 |
0.15 |
| chr3_190299152_190299367 | 0.14 |
IL1RAP |
interleukin 1 receptor accessory protein |
18024 |
0.25 |
| chr1_235097341_235097798 | 0.14 |
ENSG00000239690 |
. |
57636 |
0.14 |
| chr1_59900021_59900172 | 0.14 |
FGGY |
FGGY carbohydrate kinase domain containing |
119494 |
0.07 |
| chr17_38248024_38248175 | 0.14 |
NR1D1 |
nuclear receptor subfamily 1, group D, member 1 |
8879 |
0.12 |
| chr19_3670862_3671279 | 0.14 |
AC004637.1 |
|
1510 |
0.26 |
| chr5_158282825_158282976 | 0.14 |
CTD-2363C16.1 |
|
127114 |
0.06 |
| chr15_29262245_29262396 | 0.14 |
RP13-126C7.1 |
|
7347 |
0.23 |
| chr6_16685139_16685441 | 0.13 |
RP1-151F17.1 |
|
76079 |
0.11 |
| chr10_44311022_44311286 | 0.13 |
ZNF32 |
zinc finger protein 32 |
166850 |
0.03 |
| chr21_46708568_46708801 | 0.13 |
ENSG00000215447 |
. |
717 |
0.48 |
| chr11_2402016_2402820 | 0.13 |
CD81 |
CD81 molecule |
1272 |
0.29 |
| chr2_85033994_85034221 | 0.13 |
DUXAP1 |
double homeobox A pseudogene 1 |
56214 |
0.12 |
| chr9_80912339_80912560 | 0.13 |
PSAT1 |
phosphoserine aminotransferase 1 |
390 |
0.91 |
| chr1_22238850_22239001 | 0.13 |
HSPG2 |
heparan sulfate proteoglycan 2 |
16122 |
0.15 |
| chr6_150217225_150217449 | 0.13 |
RAET1E |
retinoic acid early transcript 1E |
142 |
0.93 |
| chr16_48998835_48999027 | 0.13 |
ENSG00000222170 |
. |
233219 |
0.02 |
| chr13_51917193_51917492 | 0.13 |
SERPINE3 |
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 3 |
2174 |
0.24 |
| chr3_23817746_23817897 | 0.13 |
ENSG00000238672 |
. |
8719 |
0.2 |
| chr16_55518647_55518798 | 0.13 |
MMP2 |
matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase) |
3253 |
0.29 |
| chr7_40583846_40584108 | 0.13 |
AC004988.1 |
|
2550 |
0.44 |
| chr10_134678084_134678235 | 0.13 |
TTC40 |
tetratricopeptide repeat domain 40 |
6923 |
0.27 |
| chr16_1351228_1351529 | 0.12 |
UBE2I |
ubiquitin-conjugating enzyme E2I |
7503 |
0.09 |
| chr2_241927031_241927182 | 0.12 |
AC104809.2 |
|
1617 |
0.27 |
| chr10_63517329_63517611 | 0.12 |
RP11-63A2.2 |
|
72324 |
0.11 |
| chr7_66342878_66343029 | 0.12 |
ENSG00000212568 |
. |
13222 |
0.18 |
| chr5_72968550_72968701 | 0.12 |
ARHGEF28 |
Rho guanine nucleotide exchange factor (GEF) 28 |
12032 |
0.23 |
| chr2_227835908_227836059 | 0.12 |
ENSG00000212391 |
. |
2145 |
0.4 |
| chr1_223149184_223149335 | 0.12 |
DISP1 |
dispatched homolog 1 (Drosophila) |
47493 |
0.16 |
| chr9_94439990_94440141 | 0.12 |
ENSG00000266855 |
. |
41532 |
0.21 |
| chr1_213957450_213957633 | 0.12 |
PROX1-AS1 |
PROX1 antisense RNA 1 |
56597 |
0.17 |
| chr4_58013971_58014122 | 0.12 |
IGFBP7-AS1 |
IGFBP7 antisense RNA 1 |
31265 |
0.16 |
| chr5_158235274_158235531 | 0.12 |
CTD-2363C16.1 |
|
174612 |
0.03 |
| chr3_185536101_185536453 | 0.12 |
IGF2BP2 |
insulin-like growth factor 2 mRNA binding protein 2 |
2572 |
0.33 |
| chr20_31332968_31333119 | 0.12 |
COMMD7 |
COMM domain containing 7 |
1240 |
0.47 |
| chr3_129274795_129275075 | 0.12 |
PLXND1 |
plexin D1 |
1131 |
0.43 |
| chr12_63155759_63156101 | 0.12 |
ENSG00000200296 |
. |
88751 |
0.08 |
| chr22_20776716_20777046 | 0.12 |
ENSG00000207343 |
. |
5692 |
0.12 |
| chr2_109790847_109791252 | 0.12 |
ENSG00000264934 |
. |
33005 |
0.2 |
| chr11_123365057_123365208 | 0.12 |
GRAMD1B |
GRAM domain containing 1B |
31212 |
0.19 |
| chr5_39399112_39399365 | 0.12 |
DAB2 |
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
4736 |
0.29 |
| chr22_17436557_17436992 | 0.12 |
IGKV1OR22-1 |
immunoglobulin kappa variable 1/OR22-1 (pseudogene) |
21881 |
0.12 |
| chr12_64430033_64430184 | 0.12 |
SRGAP1 |
SLIT-ROBO Rho GTPase activating protein 1 |
53803 |
0.12 |
| chr15_74693347_74693692 | 0.12 |
SEMA7A |
semaphorin 7A, GPI membrane anchor (John Milton Hagen blood group) |
32482 |
0.11 |
| chr5_177664514_177664665 | 0.12 |
PHYKPL |
5-phosphohydroxy-L-lysine phospho-lyase |
4803 |
0.23 |
| chr11_72567214_72567365 | 0.12 |
FCHSD2 |
FCH and double SH3 domains 2 |
12731 |
0.14 |
| chr4_84482718_84482982 | 0.12 |
AGPAT9 |
1-acylglycerol-3-phosphate O-acyltransferase 9 |
25286 |
0.2 |
| chr12_57541215_57541414 | 0.12 |
RP11-545N8.3 |
|
2 |
0.96 |
| chr19_15363610_15363765 | 0.12 |
EPHX3 |
epoxide hydrolase 3 |
19441 |
0.15 |
| chr9_37998944_37999499 | 0.12 |
ENSG00000251745 |
. |
62456 |
0.11 |
| chr8_103741270_103741446 | 0.11 |
ENSG00000266799 |
. |
4589 |
0.27 |
| chr6_157292558_157292709 | 0.11 |
ARID1B |
AT rich interactive domain 1B (SWI1-like) |
70126 |
0.13 |
| chrY_280293_280444 | 0.11 |
NA |
NA |
> 106 |
NA |
| chr21_40378291_40378490 | 0.11 |
ENSG00000272015 |
. |
111681 |
0.06 |
| chr19_13510955_13511106 | 0.11 |
ENSG00000252177 |
. |
24203 |
0.21 |
| chr15_33396303_33396531 | 0.11 |
FMN1 |
formin 1 |
36054 |
0.21 |
| chr10_31074135_31074422 | 0.11 |
RP11-330O11.3 |
|
48038 |
0.15 |
| chr18_501099_501250 | 0.11 |
COLEC12 |
collectin sub-family member 12 |
452 |
0.84 |
| chr3_13919923_13920246 | 0.11 |
WNT7A |
wingless-type MMTV integration site family, member 7A |
1534 |
0.46 |
| chr17_59385442_59385593 | 0.11 |
RP11-332H18.3 |
|
74629 |
0.08 |
| chr2_219153794_219153945 | 0.11 |
TMBIM1 |
transmembrane BAX inhibitor motif containing 1 |
848 |
0.43 |
| chr6_2738396_2738547 | 0.11 |
MYLK4 |
myosin light chain kinase family, member 4 |
12683 |
0.21 |
| chrX_330295_330446 | 0.11 |
PPP2R3B |
protein phosphatase 2, regulatory subunit B'', beta |
4409 |
0.28 |
| chr3_120280599_120280750 | 0.11 |
NDUFB4 |
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 4, 15kDa |
34482 |
0.19 |
| chr5_150860683_150860842 | 0.11 |
ENSG00000200227 |
. |
3624 |
0.2 |
| chr13_52212499_52212650 | 0.11 |
ENSG00000242893 |
. |
44223 |
0.11 |
| chr3_99378896_99379047 | 0.11 |
COL8A1 |
collagen, type VIII, alpha 1 |
21517 |
0.22 |
| chr15_26056887_26057057 | 0.11 |
ENSG00000199214 |
. |
6158 |
0.21 |
| chr17_65371836_65372120 | 0.11 |
PITPNC1 |
phosphatidylinositol transfer protein, cytoplasmic 1 |
1597 |
0.31 |
| chr1_151519925_151520338 | 0.11 |
ENSG00000207606 |
. |
1859 |
0.2 |
| chr12_5670265_5670416 | 0.11 |
NTF3 |
neurotrophin 3 |
67042 |
0.14 |
| chr19_1253556_1254162 | 0.11 |
MIDN |
midnolin |
2238 |
0.14 |
| chr11_130576966_130577117 | 0.11 |
C11orf44 |
chromosome 11 open reading frame 44 |
34190 |
0.23 |
| chr14_93735624_93735803 | 0.11 |
BTBD7 |
BTB (POZ) domain containing 7 |
24782 |
0.14 |
| chr1_92048607_92048758 | 0.11 |
CDC7 |
cell division cycle 7 |
81987 |
0.11 |
| chr14_35836199_35836350 | 0.11 |
NFKBIA |
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha |
37072 |
0.17 |
| chr12_65671554_65671852 | 0.11 |
MSRB3 |
methionine sulfoxide reductase B3 |
720 |
0.71 |
| chr11_19223388_19223615 | 0.11 |
CSRP3 |
cysteine and glycine-rich protein 3 (cardiac LIM protein) |
88 |
0.97 |
| chr12_26166979_26167130 | 0.11 |
RASSF8 |
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8 |
2385 |
0.36 |
| chr11_13963298_13963495 | 0.11 |
ENSG00000201856 |
. |
34259 |
0.23 |
| chr4_126235290_126235471 | 0.11 |
FAT4 |
FAT atypical cadherin 4 |
2174 |
0.43 |
| chr12_53891051_53891202 | 0.11 |
MAP3K12 |
mitogen-activated protein kinase kinase kinase 12 |
2114 |
0.17 |
| chr1_32043471_32043622 | 0.11 |
TINAGL1 |
tubulointerstitial nephritis antigen-like 1 |
1407 |
0.32 |
| chr2_177866064_177866215 | 0.11 |
ENSG00000206866 |
. |
71321 |
0.11 |
| chr5_112039665_112039816 | 0.11 |
APC |
adenomatous polyposis coli |
3455 |
0.32 |
| chr2_235869285_235869436 | 0.11 |
SH3BP4 |
SH3-domain binding protein 4 |
8640 |
0.33 |
| chr19_6023241_6023392 | 0.11 |
RFX2 |
regulatory factor X, 2 (influences HLA class II expression) |
15079 |
0.13 |
| chr16_2820651_2820802 | 0.11 |
SRRM2 |
serine/arginine repetitive matrix 2 |
186 |
0.81 |
| chr2_192710795_192710946 | 0.10 |
AC098617.1 |
|
395 |
0.78 |
| chr15_76069617_76069825 | 0.10 |
RP11-24M17.5 |
|
1786 |
0.22 |
| chr6_161607912_161608063 | 0.10 |
AGPAT4 |
1-acylglycerol-3-phosphate O-acyltransferase 4 |
37666 |
0.22 |
| chr1_110346543_110346835 | 0.10 |
EPS8L3 |
EPS8-like 3 |
40040 |
0.11 |
| chr9_129295292_129295840 | 0.10 |
ENSG00000221768 |
. |
2296 |
0.32 |
| chr3_129734295_129734446 | 0.10 |
ENSG00000263767 |
. |
18018 |
0.16 |
| chr4_109095665_109095816 | 0.10 |
ENSG00000232021 |
. |
1165 |
0.55 |
| chr17_70571869_70572020 | 0.10 |
ENSG00000200783 |
. |
88347 |
0.1 |
| chr22_43432132_43432283 | 0.10 |
AL022476.2 |
|
2384 |
0.28 |
| chr2_13061451_13061602 | 0.10 |
ENSG00000264370 |
. |
184033 |
0.03 |
| chr10_95898235_95898386 | 0.10 |
ENSG00000206631 |
. |
17192 |
0.16 |
| chr5_134716177_134716328 | 0.10 |
H2AFY |
H2A histone family, member Y |
18649 |
0.15 |
| chr3_46531027_46531567 | 0.10 |
LTF |
lactotransferrin |
4573 |
0.17 |
| chr10_60517984_60518135 | 0.10 |
BICC1 |
bicaudal C homolog 1 (Drosophila) |
35236 |
0.23 |
| chr11_46302438_46302589 | 0.10 |
CREB3L1 |
cAMP responsive element binding protein 3-like 1 |
3285 |
0.22 |
| chr1_64504016_64504167 | 0.10 |
ENSG00000207190 |
. |
9419 |
0.22 |
| chr2_217497778_217498102 | 0.10 |
IGFBP2 |
insulin-like growth factor binding protein 2, 36kDa |
178 |
0.95 |
| chr8_42398189_42398666 | 0.10 |
SLC20A2 |
solute carrier family 20 (phosphate transporter), member 2 |
1358 |
0.33 |
| chr3_128267003_128267154 | 0.10 |
C3orf27 |
chromosome 3 open reading frame 27 |
27851 |
0.16 |
| chr13_30497904_30498125 | 0.10 |
LINC00572 |
long intergenic non-protein coding RNA 572 |
2774 |
0.39 |
| chr2_46189186_46189337 | 0.10 |
PRKCE |
protein kinase C, epsilon |
38780 |
0.21 |
| chr21_32546644_32546955 | 0.10 |
TIAM1 |
T-cell lymphoma invasion and metastasis 1 |
44260 |
0.19 |
| chr16_87840292_87840843 | 0.10 |
RP4-536B24.2 |
|
29571 |
0.14 |
| chr16_90086077_90086472 | 0.10 |
GAS8 |
growth arrest-specific 8 |
135 |
0.66 |
| chr10_80206914_80207065 | 0.10 |
ENSG00000201393 |
. |
79725 |
0.12 |
| chr6_140981400_140981551 | 0.10 |
ENSG00000221336 |
. |
375 |
0.91 |
| chr1_20091412_20091563 | 0.10 |
TMCO4 |
transmembrane and coiled-coil domains 4 |
34350 |
0.14 |
| chr20_40032823_40033217 | 0.10 |
EMILIN3 |
elastin microfibril interfacer 3 |
37553 |
0.16 |
| chr3_16472704_16472855 | 0.10 |
RFTN1 |
raftlin, lipid raft linker 1 |
51593 |
0.14 |
| chr17_65833889_65834040 | 0.10 |
ENSG00000265086 |
. |
6097 |
0.18 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0042819 | pyridoxine metabolic process(GO:0008614) pyridoxine biosynthetic process(GO:0008615) vitamin B6 metabolic process(GO:0042816) vitamin B6 biosynthetic process(GO:0042819) |
| 0.0 | 0.1 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.0 | 0.4 | GO:0007512 | adult heart development(GO:0007512) |
| 0.0 | 0.1 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) |
| 0.0 | 0.1 | GO:0014834 | skeletal muscle satellite cell maintenance involved in skeletal muscle regeneration(GO:0014834) |
| 0.0 | 0.1 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.1 | GO:0039656 | modulation by virus of host gene expression(GO:0039656) |
| 0.0 | 0.5 | GO:0048008 | platelet-derived growth factor receptor signaling pathway(GO:0048008) |
| 0.0 | 0.1 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.1 | GO:0090400 | stress-induced premature senescence(GO:0090400) |
| 0.0 | 0.1 | GO:0070977 | organ maturation(GO:0048799) bone maturation(GO:0070977) |
| 0.0 | 0.2 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 | 0.1 | GO:0009698 | phenylpropanoid metabolic process(GO:0009698) coumarin metabolic process(GO:0009804) |
| 0.0 | 0.0 | GO:0003171 | atrioventricular valve development(GO:0003171) atrioventricular valve morphogenesis(GO:0003181) |
| 0.0 | 0.1 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.0 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 | 0.0 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.0 | GO:0002023 | reduction of food intake in response to dietary excess(GO:0002023) |
| 0.0 | 0.1 | GO:0070670 | response to interleukin-4(GO:0070670) |
| 0.0 | 0.0 | GO:0070254 | mucus secretion(GO:0070254) |
| 0.0 | 0.1 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.0 | 0.0 | GO:0010803 | regulation of tumor necrosis factor-mediated signaling pathway(GO:0010803) |
| 0.0 | 0.0 | GO:0046813 | receptor-mediated virion attachment to host cell(GO:0046813) |
| 0.0 | 0.0 | GO:0050703 | interleukin-1 alpha secretion(GO:0050703) |
| 0.0 | 0.1 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 0.0 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.0 | GO:0022605 | oogenesis stage(GO:0022605) |
| 0.0 | 0.0 | GO:0043578 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.0 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.0 | 0.1 | GO:0021548 | pons development(GO:0021548) |
| 0.0 | 0.0 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 | 0.0 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.0 | GO:0071364 | cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.0 | 0.1 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.0 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0052813 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.1 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.1 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.1 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.0 | 0.1 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.1 | GO:0000987 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.0 | 0.1 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.1 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.1 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.1 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.0 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.0 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.0 | 0.0 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.0 | 0.0 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 0.0 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.0 | 0.2 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.1 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.0 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.0 | 0.1 | GO:0003785 | actin monomer binding(GO:0003785) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.0 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.0 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |