Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
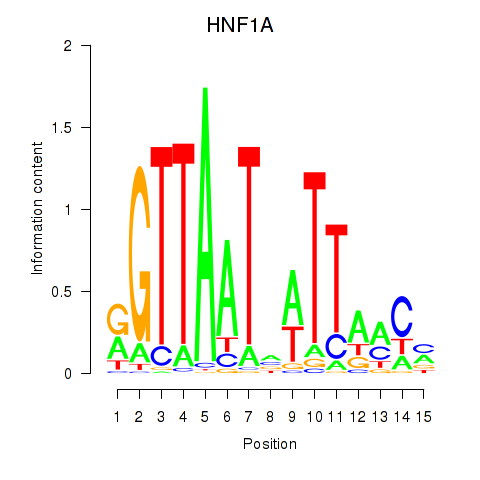
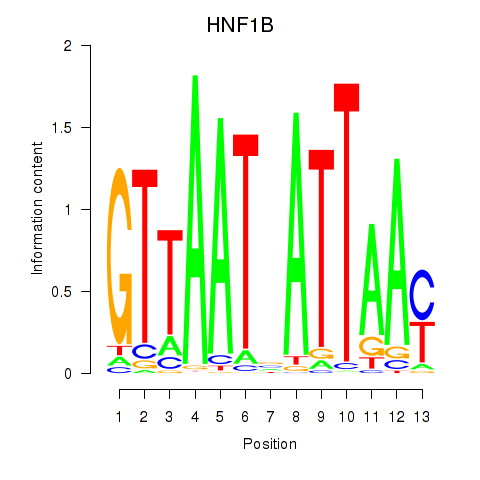
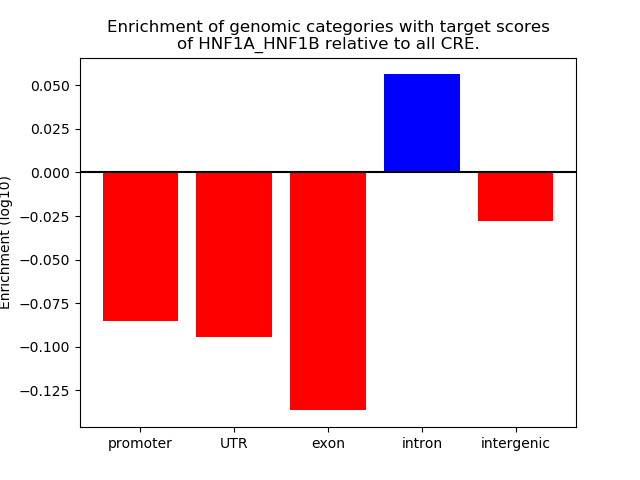
Results for HNF1A_HNF1B
Z-value: 0.80
Transcription factors associated with HNF1A_HNF1B
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HNF1A
|
ENSG00000135100.13 | HNF1 homeobox A |
|
HNF1B
|
ENSG00000108753.8 | HNF1 homeobox B |
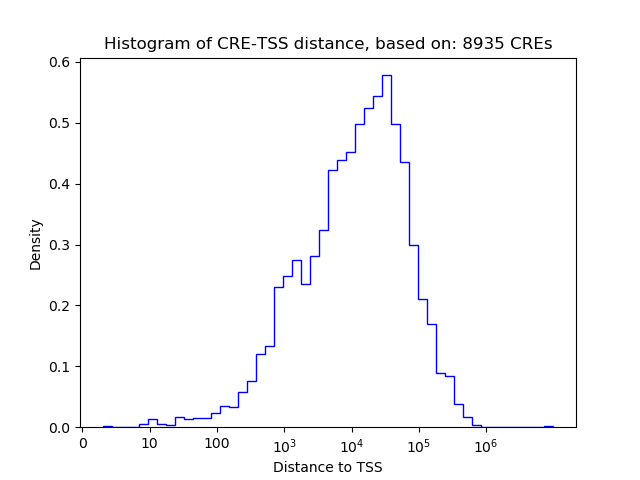
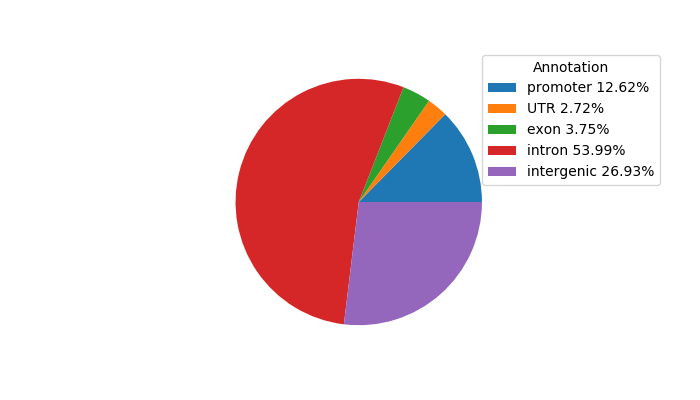
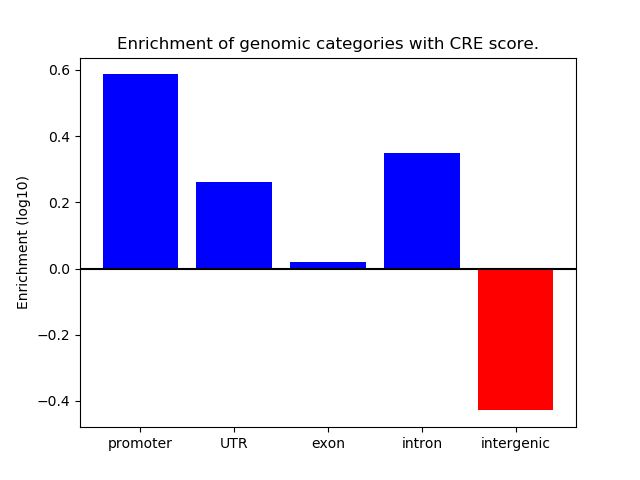
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr17_36135419_36135570 | HNF1B | 30257 | 0.150237 | -0.89 | 1.1e-03 | Click! |
| chr17_36112352_36112607 | HNF1B | 7242 | 0.192190 | 0.80 | 1.0e-02 | Click! |
| chr17_36122844_36122995 | HNF1B | 17682 | 0.173448 | -0.78 | 1.4e-02 | Click! |
| chr17_36121536_36121745 | HNF1B | 16403 | 0.175492 | -0.70 | 3.5e-02 | Click! |
| chr17_36124947_36125098 | HNF1B | 19785 | 0.169880 | -0.63 | 7.1e-02 | Click! |
Activity of the HNF1A_HNF1B motif across conditions
Conditions sorted by the z-value of the HNF1A_HNF1B motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
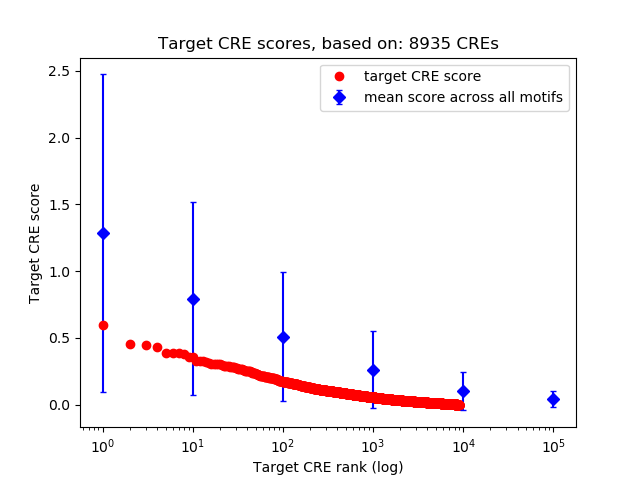
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr14_69405435_69405631 | 0.59 |
ACTN1 |
actinin, alpha 1 |
8732 |
0.24 |
| chr1_68214159_68214518 | 0.46 |
ENSG00000238778 |
. |
23998 |
0.19 |
| chr10_97849004_97849155 | 0.45 |
ENTPD1-AS1 |
ENTPD1 antisense RNA 1 |
833 |
0.59 |
| chr9_20320271_20320422 | 0.43 |
ENSG00000221744 |
. |
25344 |
0.21 |
| chr1_181121299_181121450 | 0.39 |
IER5 |
immediate early response 5 |
63736 |
0.13 |
| chr7_37071372_37071523 | 0.39 |
ELMO1-AS1 |
ELMO1 antisense RNA 1 |
34046 |
0.17 |
| chr10_29924859_29925052 | 0.39 |
SVIL |
supervillin |
1054 |
0.57 |
| chr18_68086526_68086677 | 0.38 |
ENSG00000264059 |
. |
57142 |
0.13 |
| chr14_50691962_50692201 | 0.36 |
SOS2 |
son of sevenless homolog 2 (Drosophila) |
5946 |
0.22 |
| chr3_42066075_42066314 | 0.36 |
ULK4 |
unc-51 like kinase 4 |
62272 |
0.12 |
| chr11_16945886_16946397 | 0.33 |
PLEKHA7 |
pleckstrin homology domain containing, family A member 7 |
43236 |
0.13 |
| chr4_114394825_114395109 | 0.33 |
CAMK2D |
calcium/calmodulin-dependent protein kinase II delta |
40031 |
0.16 |
| chr6_40925892_40926043 | 0.32 |
UNC5CL |
unc-5 homolog C (C. elegans)-like |
76907 |
0.08 |
| chr3_58057429_58057580 | 0.32 |
FLNB |
filamin B, beta |
6552 |
0.27 |
| chr2_24269080_24269320 | 0.31 |
ENSG00000222940 |
. |
495 |
0.58 |
| chr3_183016185_183016744 | 0.31 |
B3GNT5 |
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5 |
33332 |
0.16 |
| chr7_38254764_38254915 | 0.31 |
STARD3NL |
STARD3 N-terminal like |
36842 |
0.23 |
| chr4_124571341_124571610 | 0.31 |
SPRY1 |
sprouty homolog 1, antagonist of FGF signaling (Drosophila) |
250352 |
0.02 |
| chr18_805710_806203 | 0.30 |
YES1 |
v-yes-1 Yamaguchi sarcoma viral oncogene homolog 1 |
6283 |
0.16 |
| chr8_70745558_70745875 | 0.30 |
SLCO5A1 |
solute carrier organic anion transporter family, member 5A1 |
91 |
0.91 |
| chr3_188599582_188599733 | 0.29 |
TPRG1 |
tumor protein p63 regulated 1 |
65346 |
0.14 |
| chr5_121498494_121498645 | 0.29 |
CTC-441N14.1 |
|
6859 |
0.2 |
| chr11_117695326_117695570 | 0.29 |
FXYD2 |
FXYD domain containing ion transport regulator 2 |
11 |
0.97 |
| chr17_47440332_47440604 | 0.29 |
ZNF652 |
zinc finger protein 652 |
633 |
0.61 |
| chr12_27487334_27487485 | 0.29 |
ARNTL2 |
aryl hydrocarbon receptor nuclear translocator-like 2 |
1421 |
0.49 |
| chr18_10169648_10169799 | 0.29 |
ENSG00000263630 |
. |
164529 |
0.03 |
| chr3_81777225_81777376 | 0.29 |
GBE1 |
glucan (1,4-alpha-), branching enzyme 1 |
15480 |
0.31 |
| chr4_24606533_24606684 | 0.28 |
DHX15 |
DEAH (Asp-Glu-Ala-His) box helicase 15 |
20435 |
0.22 |
| chrX_119689036_119689385 | 0.28 |
CUL4B |
cullin 4B |
4588 |
0.21 |
| chr8_49322257_49322678 | 0.28 |
ENSG00000252710 |
. |
101877 |
0.08 |
| chr6_74512837_74512988 | 0.28 |
RP11-553A21.3 |
|
107058 |
0.06 |
| chr4_169408783_169408934 | 0.27 |
DDX60L |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 60-like |
7193 |
0.2 |
| chr11_124595039_124595293 | 0.27 |
NRGN |
neurogranin (protein kinase C substrate, RC3) |
14576 |
0.1 |
| chr17_67498171_67498584 | 0.27 |
MAP2K6 |
mitogen-activated protein kinase kinase 6 |
30 |
0.99 |
| chr7_45927984_45928135 | 0.26 |
IGFBP1 |
insulin-like growth factor binding protein 1 |
37 |
0.98 |
| chr4_38387684_38387835 | 0.26 |
ENSG00000221495 |
. |
147525 |
0.04 |
| chr15_73927173_73927324 | 0.26 |
NPTN |
neuroplastin |
773 |
0.72 |
| chr15_64636500_64636651 | 0.26 |
ENSG00000252774 |
. |
1837 |
0.25 |
| chr15_64927439_64927681 | 0.26 |
ENSG00000207223 |
. |
17527 |
0.14 |
| chr6_154640196_154640597 | 0.25 |
IPCEF1 |
interaction protein for cytohesin exchange factors 1 |
10819 |
0.3 |
| chr3_192534775_192534926 | 0.25 |
FGF12 |
fibroblast growth factor 12 |
49297 |
0.16 |
| chr16_12707264_12707435 | 0.25 |
CTD-3037G24.3 |
|
2448 |
0.33 |
| chr2_208337340_208337637 | 0.25 |
CREB1 |
cAMP responsive element binding protein 1 |
56973 |
0.13 |
| chr3_183563309_183563581 | 0.25 |
PARL |
presenilin associated, rhomboid-like |
1383 |
0.34 |
| chr10_98623724_98623875 | 0.24 |
LCOR |
ligand dependent nuclear receptor corepressor |
31062 |
0.15 |
| chr3_172312196_172312406 | 0.24 |
AC007919.2 |
HCG1787166; PRO1163; Uncharacterized protein |
49182 |
0.13 |
| chr20_1921396_1921547 | 0.24 |
RP4-684O24.5 |
|
6421 |
0.23 |
| chr4_79489691_79489842 | 0.24 |
ANXA3 |
annexin A3 |
14692 |
0.23 |
| chr2_230793147_230793445 | 0.23 |
TRIP12 |
thyroid hormone receptor interactor 12 |
5341 |
0.14 |
| chr15_67419351_67419566 | 0.23 |
SMAD3 |
SMAD family member 3 |
983 |
0.62 |
| chrX_17581294_17581554 | 0.23 |
NHS-AS1 |
NHS antisense RNA 1 |
5916 |
0.25 |
| chr10_63718660_63718935 | 0.23 |
ENSG00000221272 |
. |
32020 |
0.21 |
| chr7_32538317_32538480 | 0.23 |
AVL9 |
AVL9 homolog (S. cerevisiase) |
3076 |
0.29 |
| chr18_13491175_13491326 | 0.22 |
RP11-53B2.2 |
|
4789 |
0.11 |
| chr10_111816096_111816353 | 0.22 |
ADD3 |
adducin 3 (gamma) |
48502 |
0.14 |
| chr2_44518783_44519008 | 0.22 |
SLC3A1 |
solute carrier family 3 (amino acid transporter heavy chain), member 1 |
6261 |
0.21 |
| chr5_142554814_142554965 | 0.22 |
ARHGAP26-IT1 |
ARHGAP26 intronic transcript 1 (non-protein coding) |
17176 |
0.24 |
| chr13_114881324_114881475 | 0.22 |
RASA3-IT1 |
RASA3 intronic transcript 1 (non-protein coding) |
7104 |
0.23 |
| chr15_85937202_85937353 | 0.22 |
ENSG00000251891 |
. |
4162 |
0.24 |
| chr6_108971832_108971983 | 0.22 |
FOXO3 |
forkhead box O3 |
5642 |
0.32 |
| chr1_120174414_120175022 | 0.21 |
ZNF697 |
zinc finger protein 697 |
15678 |
0.18 |
| chr5_64500241_64500433 | 0.21 |
ENSG00000207439 |
. |
81141 |
0.11 |
| chr5_133387222_133387373 | 0.21 |
VDAC1 |
voltage-dependent anion channel 1 |
46473 |
0.14 |
| chr16_81030908_81031149 | 0.21 |
CMC2 |
C-x(9)-C motif containing 2 |
1236 |
0.36 |
| chr2_43986548_43986990 | 0.21 |
ENSG00000252490 |
. |
12720 |
0.19 |
| chr12_25648509_25648660 | 0.21 |
IFLTD1 |
intermediate filament tail domain containing 1 |
24311 |
0.23 |
| chr12_46867328_46867682 | 0.21 |
SLC38A2 |
solute carrier family 38, member 2 |
100855 |
0.08 |
| chr7_22627428_22627634 | 0.21 |
AC002480.4 |
|
1793 |
0.36 |
| chr9_14251173_14251324 | 0.21 |
NFIB |
nuclear factor I/B |
56764 |
0.15 |
| chrX_129243328_129243479 | 0.21 |
ELF4 |
E74-like factor 4 (ets domain transcription factor) |
933 |
0.6 |
| chr6_132654884_132655035 | 0.20 |
MOXD1 |
monooxygenase, DBH-like 1 |
41245 |
0.19 |
| chr18_66372478_66372744 | 0.20 |
TMX3 |
thioredoxin-related transmembrane protein 3 |
9683 |
0.22 |
| chr17_71040137_71040288 | 0.20 |
SLC39A11 |
solute carrier family 39, member 11 |
45691 |
0.17 |
| chr21_34905841_34905992 | 0.20 |
GART |
phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase |
8504 |
0.13 |
| chr1_200865216_200865405 | 0.20 |
C1orf106 |
chromosome 1 open reading frame 106 |
1361 |
0.42 |
| chr1_68643879_68644030 | 0.20 |
ENSG00000221203 |
. |
5339 |
0.24 |
| chr9_74377924_74378411 | 0.20 |
TMEM2 |
transmembrane protein 2 |
5135 |
0.32 |
| chr11_86431921_86432072 | 0.20 |
ME3 |
malic enzyme 3, NADP(+)-dependent, mitochondrial |
48318 |
0.17 |
| chr2_169387098_169387498 | 0.20 |
ENSG00000265694 |
. |
52155 |
0.13 |
| chr9_3359879_3360030 | 0.19 |
RFX3 |
regulatory factor X, 3 (influences HLA class II expression) |
13190 |
0.3 |
| chr6_117818099_117818250 | 0.19 |
DCBLD1 |
discoidin, CUB and LCCL domain containing 1 |
14349 |
0.17 |
| chr4_76584048_76584199 | 0.19 |
ENSG00000201644 |
. |
3274 |
0.22 |
| chr15_69850121_69850272 | 0.19 |
ENSG00000207119 |
. |
99708 |
0.06 |
| chr3_18177094_18177245 | 0.19 |
RP11-158G18.1 |
|
273958 |
0.02 |
| chr9_22104098_22104249 | 0.19 |
CDKN2B-AS1 |
CDKN2B antisense RNA 1 |
9504 |
0.26 |
| chr7_47613250_47613541 | 0.19 |
TNS3 |
tensin 3 |
7834 |
0.3 |
| chr2_109203925_109204163 | 0.19 |
LIMS1 |
LIM and senescent cell antigen-like domains 1 |
723 |
0.74 |
| chr2_30336938_30337089 | 0.19 |
AC016907.3 |
|
22186 |
0.18 |
| chr10_89142685_89142836 | 0.18 |
NUTM2D |
NUT family member 2D |
18014 |
0.21 |
| chr15_38549505_38549656 | 0.18 |
SPRED1 |
sprouty-related, EVH1 domain containing 1 |
4198 |
0.36 |
| chr2_202270772_202271289 | 0.18 |
ENSG00000202008 |
. |
17206 |
0.15 |
| chr6_16367315_16367466 | 0.18 |
ENSG00000265642 |
. |
61364 |
0.14 |
| chr10_30388154_30388305 | 0.18 |
KIAA1462 |
KIAA1462 |
39776 |
0.21 |
| chr1_183812842_183812993 | 0.18 |
RGL1 |
ral guanine nucleotide dissociation stimulator-like 1 |
38627 |
0.18 |
| chr3_183266299_183266524 | 0.18 |
KLHL6-AS1 |
KLHL6 antisense RNA 1 |
112 |
0.96 |
| chr5_130601387_130601538 | 0.18 |
CDC42SE2 |
CDC42 small effector 2 |
1669 |
0.51 |
| chr21_36265275_36265757 | 0.18 |
RUNX1 |
runt-related transcription factor 1 |
3429 |
0.38 |
| chr1_20916896_20917047 | 0.17 |
CDA |
cytidine deaminase |
1530 |
0.37 |
| chr7_135015211_135015362 | 0.17 |
STRA8 |
stimulated by retinoic acid 8 |
98555 |
0.06 |
| chr6_130645263_130645558 | 0.17 |
SAMD3 |
sterile alpha motif domain containing 3 |
41160 |
0.17 |
| chr10_104103101_104103252 | 0.17 |
NFKB2 |
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) |
50691 |
0.08 |
| chr3_71634599_71634767 | 0.17 |
FOXP1 |
forkhead box P1 |
1543 |
0.42 |
| chr8_121080549_121080727 | 0.17 |
COL14A1 |
collagen, type XIV, alpha 1 |
37178 |
0.21 |
| chr7_155601790_155601976 | 0.17 |
SHH |
sonic hedgehog |
117 |
0.97 |
| chr6_2103859_2104525 | 0.17 |
GMDS |
GDP-mannose 4,6-dehydratase |
72033 |
0.13 |
| chr4_100007595_100007746 | 0.17 |
ADH5 |
alcohol dehydrogenase 5 (class III), chi polypeptide |
1303 |
0.38 |
| chr3_156252924_156253075 | 0.17 |
RP11-305K5.1 |
|
10963 |
0.22 |
| chr13_27132620_27132950 | 0.17 |
WASF3 |
WAS protein family, member 3 |
898 |
0.72 |
| chr9_27094212_27094497 | 0.17 |
TEK |
TEK tyrosine kinase, endothelial |
14785 |
0.23 |
| chr9_21543413_21543564 | 0.17 |
MIR31HG |
MIR31 host gene (non-protein coding) |
16180 |
0.15 |
| chr2_233432775_233432926 | 0.17 |
EIF4E2 |
eukaryotic translation initiation factor 4E family member 2 |
5934 |
0.11 |
| chrX_39967563_39967714 | 0.17 |
BCOR |
BCL6 corepressor |
10982 |
0.31 |
| chr6_43730965_43731135 | 0.17 |
VEGFA |
vascular endothelial growth factor A |
6900 |
0.18 |
| chr7_116228829_116229027 | 0.17 |
AC006159.4 |
|
17640 |
0.18 |
| chr12_65700883_65701407 | 0.16 |
MSRB3 |
methionine sulfoxide reductase B3 |
19510 |
0.22 |
| chr9_123960312_123961002 | 0.16 |
RAB14 |
RAB14, member RAS oncogene family |
3490 |
0.19 |
| chr13_20769719_20769870 | 0.16 |
GJB2 |
gap junction protein, beta 2, 26kDa |
2757 |
0.25 |
| chr11_27417260_27417411 | 0.16 |
CCDC34 |
coiled-coil domain containing 34 |
31920 |
0.18 |
| chr1_234953186_234953405 | 0.16 |
ENSG00000201638 |
. |
20425 |
0.25 |
| chr15_71346441_71346592 | 0.16 |
THSD4 |
thrombospondin, type I, domain containing 4 |
42775 |
0.16 |
| chr5_133939031_133939182 | 0.16 |
SAR1B |
SAR1 homolog B (S. cerevisiae) |
9670 |
0.17 |
| chr18_452761_452993 | 0.16 |
RP11-720L2.2 |
|
28461 |
0.17 |
| chr6_12343879_12344030 | 0.16 |
EDN1 |
endothelin 1 |
53358 |
0.16 |
| chr3_183947384_183948074 | 0.16 |
VWA5B2 |
von Willebrand factor A domain containing 5B2 |
488 |
0.65 |
| chr3_152153870_152154021 | 0.16 |
ENSG00000201217 |
. |
13116 |
0.21 |
| chr5_95683555_95683706 | 0.16 |
PCSK1 |
proprotein convertase subtilisin/kexin type 1 |
84234 |
0.09 |
| chr6_25311839_25311990 | 0.16 |
ENSG00000207286 |
. |
24153 |
0.15 |
| chr11_111999683_111999892 | 0.16 |
IL18 |
interleukin 18 (interferon-gamma-inducing factor) |
35010 |
0.08 |
| chr2_29339694_29340047 | 0.16 |
CLIP4 |
CAP-GLY domain containing linker protein family, member 4 |
1191 |
0.44 |
| chr7_42302451_42302602 | 0.16 |
GLI3 |
GLI family zinc finger 3 |
25868 |
0.28 |
| chr14_58748062_58749029 | 0.16 |
RP11-349A22.5 |
|
5196 |
0.12 |
| chr3_185431049_185431200 | 0.16 |
C3orf65 |
chromosome 3 open reading frame 65 |
44 |
0.98 |
| chr20_34380481_34380768 | 0.16 |
PHF20 |
PHD finger protein 20 |
20676 |
0.13 |
| chr1_33414688_33414839 | 0.16 |
RNF19B |
ring finger protein 19B |
15523 |
0.12 |
| chr12_54383579_54383730 | 0.16 |
HOXC6 |
homeobox C6 |
754 |
0.31 |
| chr7_30836266_30836417 | 0.16 |
AC004691.5 |
|
6685 |
0.2 |
| chr7_110872801_110872972 | 0.16 |
ENSG00000238922 |
. |
123473 |
0.06 |
| chr7_139473808_139474066 | 0.16 |
HIPK2 |
homeodomain interacting protein kinase 2 |
3580 |
0.27 |
| chr4_41179011_41179162 | 0.16 |
APBB2 |
amyloid beta (A4) precursor protein-binding, family B, member 2 |
37389 |
0.15 |
| chr8_17220869_17221020 | 0.16 |
MTMR7 |
myotubularin related protein 7 |
49892 |
0.15 |
| chr15_85455088_85455239 | 0.15 |
ENSG00000207037 |
. |
26590 |
0.13 |
| chr8_38029592_38029743 | 0.15 |
BAG4 |
BCL2-associated athanogene 4 |
4439 |
0.11 |
| chr6_136760823_136760974 | 0.15 |
MAP7 |
microtubule-associated protein 7 |
27115 |
0.21 |
| chr3_48733781_48734150 | 0.15 |
IP6K2 |
inositol hexakisphosphate kinase 2 |
950 |
0.42 |
| chr6_46458096_46458346 | 0.15 |
RCAN2 |
regulator of calcineurin 2 |
878 |
0.55 |
| chr4_38084935_38085086 | 0.15 |
TBC1D1 |
TBC1 (tre-2/USP6, BUB2, cdc16) domain family, member 1 |
37518 |
0.21 |
| chr6_169615833_169615984 | 0.15 |
XXyac-YX65C7_A.2 |
|
2559 |
0.38 |
| chr1_8156252_8156403 | 0.15 |
ERRFI1 |
ERBB receptor feedback inhibitor 1 |
69959 |
0.1 |
| chr10_71529428_71529579 | 0.15 |
COL13A1 |
collagen, type XIII, alpha 1 |
32141 |
0.17 |
| chr5_88717098_88717249 | 0.15 |
MEF2C-AS1 |
MEF2C antisense RNA 1 |
2549 |
0.45 |
| chr6_144586213_144586364 | 0.15 |
UTRN |
utrophin |
20549 |
0.24 |
| chr6_52415213_52415364 | 0.14 |
TRAM2 |
translocation associated membrane protein 2 |
26425 |
0.21 |
| chr6_2020825_2021145 | 0.14 |
GMDS |
GDP-mannose 4,6-dehydratase |
155240 |
0.04 |
| chr3_189743016_189743167 | 0.14 |
ENSG00000265045 |
. |
88632 |
0.08 |
| chr8_129031222_129031516 | 0.14 |
ENSG00000222501 |
. |
8512 |
0.18 |
| chr1_183483781_183483932 | 0.14 |
ENSG00000265042 |
. |
3957 |
0.25 |
| chr19_12722676_12723040 | 0.14 |
ZNF791 |
zinc finger protein 791 |
1090 |
0.26 |
| chr10_91092779_91093035 | 0.14 |
IFIT3 |
interferon-induced protein with tetratricopeptide repeats 3 |
666 |
0.62 |
| chr5_61564187_61564338 | 0.14 |
KIF2A |
kinesin heavy chain member 2A |
37727 |
0.2 |
| chr8_125266983_125267210 | 0.14 |
ENSG00000206724 |
. |
5791 |
0.28 |
| chr12_79098466_79098617 | 0.14 |
SYT1 |
synaptotagmin I |
159232 |
0.04 |
| chr12_56609980_56610131 | 0.14 |
RNF41 |
ring finger protein 41 |
5430 |
0.07 |
| chr10_11602345_11602496 | 0.14 |
USP6NL |
USP6 N-terminal like |
13316 |
0.25 |
| chr5_134707593_134707838 | 0.14 |
H2AFY |
H2A histone family, member Y |
27186 |
0.14 |
| chr3_135710315_135710559 | 0.14 |
PPP2R3A |
protein phosphatase 2, regulatory subunit B'', alpha |
25851 |
0.25 |
| chr17_57746657_57747029 | 0.14 |
CLTC |
clathrin, heavy chain (Hc) |
14424 |
0.17 |
| chr17_29889114_29889342 | 0.14 |
ENSG00000221038 |
. |
1685 |
0.21 |
| chr4_169361836_169361987 | 0.14 |
DDX60L |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 60-like |
15299 |
0.2 |
| chr10_33995531_33995682 | 0.14 |
NRP1 |
neuropilin 1 |
370416 |
0.01 |
| chr17_50225184_50225335 | 0.14 |
CA10 |
carbonic anhydrase X |
10164 |
0.3 |
| chr7_17411629_17411780 | 0.14 |
ENSG00000199473 |
. |
1246 |
0.58 |
| chr8_87496135_87496286 | 0.14 |
CPNE3 |
copine III |
849 |
0.58 |
| chrX_149881301_149881600 | 0.14 |
MTMR1 |
myotubularin related protein 1 |
5699 |
0.3 |
| chr12_13054850_13055147 | 0.14 |
GPRC5A |
G protein-coupled receptor, family C, group 5, member A |
7005 |
0.15 |
| chr10_31255665_31255816 | 0.14 |
ZNF438 |
zinc finger protein 438 |
17615 |
0.26 |
| chr7_89974617_89974768 | 0.14 |
GTPBP10 |
GTP-binding protein 10 (putative) |
1287 |
0.45 |
| chr11_14375636_14375787 | 0.14 |
RRAS2 |
related RAS viral (r-ras) oncogene homolog 2 |
335 |
0.93 |
| chr4_123795832_123795983 | 0.14 |
RP11-170N16.3 |
|
9960 |
0.18 |
| chr6_143193207_143193806 | 0.14 |
HIVEP2 |
human immunodeficiency virus type I enhancer binding protein 2 |
35322 |
0.21 |
| chr11_113186702_113186853 | 0.14 |
TTC12 |
tetratricopeptide repeat domain 12 |
186 |
0.92 |
| chr1_27039284_27039619 | 0.14 |
ARID1A |
AT rich interactive domain 1A (SWI-like) |
14748 |
0.14 |
| chr20_19795494_19795742 | 0.13 |
AL121761.2 |
Uncharacterized protein |
56939 |
0.12 |
| chr15_71006405_71006665 | 0.13 |
UACA |
uveal autoantigen with coiled-coil domains and ankyrin repeats |
10921 |
0.26 |
| chr3_100112860_100113011 | 0.13 |
LNP1 |
leukemia NUP98 fusion partner 1 |
7102 |
0.2 |
| chr3_150936070_150936221 | 0.13 |
GPR171 |
G protein-coupled receptor 171 |
15157 |
0.14 |
| chr13_98908338_98908489 | 0.13 |
RP11-573N10.1 |
|
22509 |
0.17 |
| chr18_42313110_42313658 | 0.13 |
SETBP1 |
SET binding protein 1 |
36255 |
0.24 |
| chr10_11060742_11061158 | 0.13 |
CELF2 |
CUGBP, Elav-like family member 2 |
844 |
0.61 |
| chr9_127428096_127428247 | 0.13 |
MIR181A2HG |
MIR181A2 host gene (non-protein coding) |
7425 |
0.17 |
| chr4_185709233_185709384 | 0.13 |
ACSL1 |
acyl-CoA synthetase long-chain family member 1 |
14281 |
0.18 |
| chr12_889095_889427 | 0.13 |
ENSG00000221439 |
. |
1038 |
0.58 |
| chr2_173952072_173952223 | 0.13 |
MLTK |
Mitogen-activated protein kinase kinase kinase MLT |
3200 |
0.29 |
| chr18_55480395_55480546 | 0.13 |
ATP8B1 |
ATPase, aminophospholipid transporter, class I, type 8B, member 1 |
10143 |
0.24 |
| chr15_52627311_52627462 | 0.13 |
MYO5A |
myosin VA (heavy chain 12, myoxin) |
16193 |
0.2 |
| chr3_161027813_161027964 | 0.13 |
SPTSSB |
serine palmitoyltransferase, small subunit B |
61401 |
0.14 |
| chr14_89019667_89019818 | 0.13 |
PTPN21 |
protein tyrosine phosphatase, non-receptor type 21 |
1335 |
0.33 |
| chr6_144171782_144171933 | 0.13 |
LTV1 |
LTV1 homolog (S. cerevisiae) |
7376 |
0.24 |
| chr2_98566489_98566757 | 0.13 |
ENSG00000238719 |
. |
37043 |
0.17 |
| chr10_3848729_3848956 | 0.13 |
KLF6 |
Kruppel-like factor 6 |
21369 |
0.22 |
| chr10_131555762_131556161 | 0.13 |
RP11-109A6.2 |
|
56140 |
0.14 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.1 | 0.3 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 | 0.1 | GO:0044413 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.0 | 0.2 | GO:0060291 | long-term synaptic potentiation(GO:0060291) |
| 0.0 | 0.1 | GO:0060594 | mammary gland specification(GO:0060594) |
| 0.0 | 0.2 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.2 | GO:0009649 | entrainment of circadian clock(GO:0009649) |
| 0.0 | 0.2 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 | 0.1 | GO:0001866 | NK T cell proliferation(GO:0001866) |
| 0.0 | 0.1 | GO:0060426 | lung vasculature development(GO:0060426) |
| 0.0 | 0.1 | GO:0046185 | aldehyde catabolic process(GO:0046185) |
| 0.0 | 0.1 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.1 | GO:0022030 | cerebral cortex radial glia guided migration(GO:0021801) telencephalon glial cell migration(GO:0022030) |
| 0.0 | 0.1 | GO:0001547 | antral ovarian follicle growth(GO:0001547) |
| 0.0 | 0.1 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 0.1 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.1 | GO:0071692 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.1 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.0 | 0.1 | GO:0006655 | phosphatidylglycerol biosynthetic process(GO:0006655) cardiolipin metabolic process(GO:0032048) cardiolipin biosynthetic process(GO:0032049) phosphatidylglycerol metabolic process(GO:0046471) |
| 0.0 | 0.1 | GO:0006065 | UDP-glucuronate biosynthetic process(GO:0006065) |
| 0.0 | 0.0 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.0 | 0.1 | GO:0033603 | positive regulation of dopamine secretion(GO:0033603) |
| 0.0 | 0.1 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.0 | 0.0 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.0 | 0.1 | GO:0006216 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.0 | 0.0 | GO:0007132 | meiotic metaphase I(GO:0007132) |
| 0.0 | 0.1 | GO:0010988 | regulation of low-density lipoprotein particle clearance(GO:0010988) |
| 0.0 | 0.0 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.0 | 0.1 | GO:0009128 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.0 | 0.0 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.1 | GO:0006188 | IMP biosynthetic process(GO:0006188) IMP metabolic process(GO:0046040) |
| 0.0 | 0.1 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.0 | 0.1 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.0 | 0.0 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.0 | 0.0 | GO:0022038 | corpus callosum development(GO:0022038) |
| 0.0 | 0.0 | GO:0048859 | formation of anatomical boundary(GO:0048859) |
| 0.0 | 0.0 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.0 | GO:0071803 | regulation of podosome assembly(GO:0071801) positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.1 | GO:0046149 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.0 | 0.0 | GO:0072583 | synaptic vesicle recycling(GO:0036465) synaptic vesicle endocytosis(GO:0048488) clathrin-mediated endocytosis(GO:0072583) |
| 0.0 | 0.0 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) |
| 0.0 | 0.0 | GO:0090273 | regulation of somatostatin secretion(GO:0090273) |
| 0.0 | 0.1 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.0 | 0.1 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.0 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.0 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 0.0 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.0 | 0.0 | GO:0045627 | positive regulation of T-helper 1 cell differentiation(GO:0045627) |
| 0.0 | 0.0 | GO:0000101 | sulfur amino acid transport(GO:0000101) L-cystine transport(GO:0015811) |
| 0.0 | 0.0 | GO:0072112 | renal filtration cell differentiation(GO:0061318) glomerular epithelium development(GO:0072010) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.0 | 0.1 | GO:0008347 | glial cell migration(GO:0008347) |
| 0.0 | 0.0 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.0 | GO:0001302 | replicative cell aging(GO:0001302) |
| 0.0 | 0.1 | GO:0016264 | gap junction assembly(GO:0016264) |
| 0.0 | 0.0 | GO:0055064 | cellular chloride ion homeostasis(GO:0030644) chloride ion homeostasis(GO:0055064) |
| 0.0 | 0.0 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.0 | 0.0 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.0 | GO:0060457 | negative regulation of digestive system process(GO:0060457) |
| 0.0 | 0.0 | GO:0007386 | compartment pattern specification(GO:0007386) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.1 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.2 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.2 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.1 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.1 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.0 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.0 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.1 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 0.0 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.0 | GO:0005954 | calcium- and calmodulin-dependent protein kinase complex(GO:0005954) |
| 0.0 | 0.1 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 0.1 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.1 | 0.2 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.1 | GO:0001159 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.0 | 0.1 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.1 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.2 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.1 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.1 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.1 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.0 | 0.2 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.1 | GO:0015925 | galactosidase activity(GO:0015925) |
| 0.0 | 0.2 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.1 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.1 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.0 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.1 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.1 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.0 | 0.3 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.1 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.1 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.1 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.2 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.1 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.1 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.0 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.0 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.0 | 0.0 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.1 | GO:0000832 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) |
| 0.0 | 0.0 | GO:0004470 | malic enzyme activity(GO:0004470) |
| 0.0 | 0.0 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.0 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.0 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.0 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.0 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.1 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.0 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.0 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.3 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.2 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.1 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.1 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |