Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
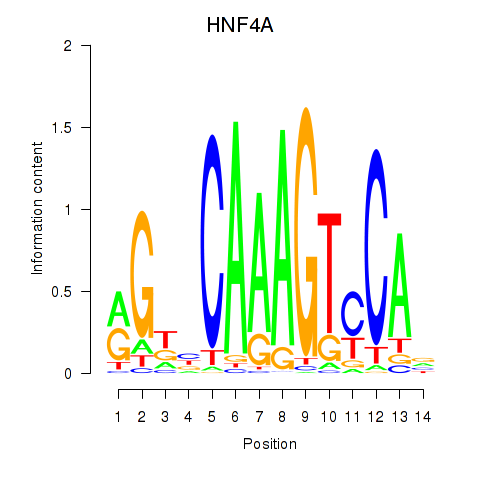
Results for HNF4A
Z-value: 0.44
Transcription factors associated with HNF4A
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HNF4A
|
ENSG00000101076.12 | hepatocyte nuclear factor 4 alpha |
Activity of the HNF4A motif across conditions
Conditions sorted by the z-value of the HNF4A motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
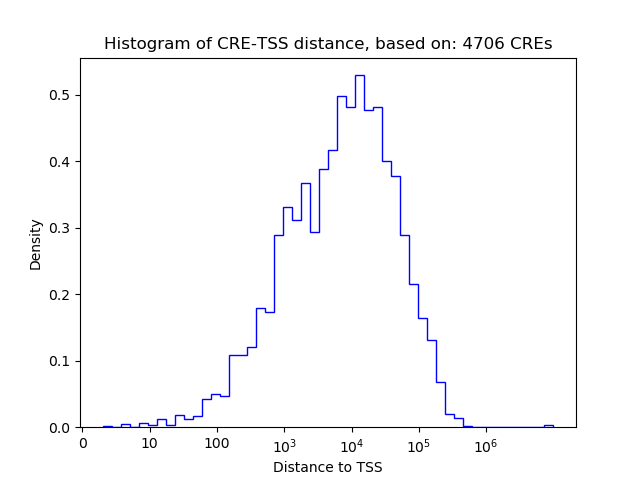
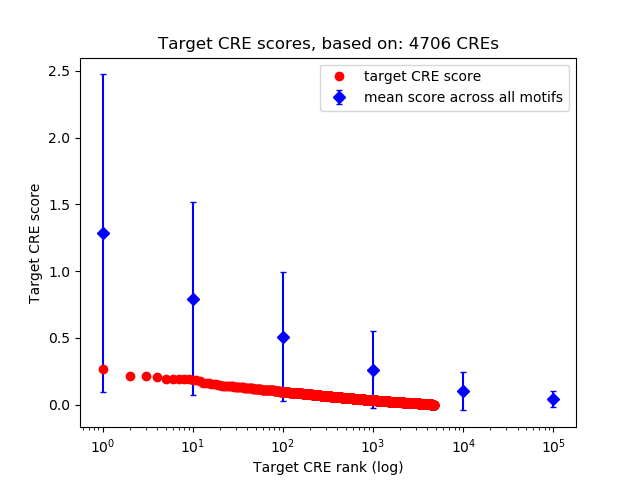
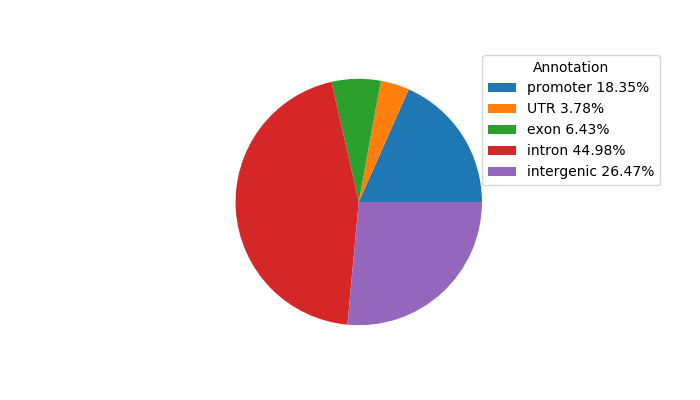
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr9_134530412_134530563 | 0.27 |
RAPGEF1 |
Rap guanine nucleotide exchange factor (GEF) 1 |
29152 |
0.19 |
| chr16_4714562_4714838 | 0.22 |
MGRN1 |
mahogunin ring finger 1, E3 ubiquitin protein ligase |
14299 |
0.11 |
| chr20_55975594_55975873 | 0.22 |
RP4-800J21.3 |
|
7615 |
0.16 |
| chr17_4617422_4617848 | 0.21 |
ARRB2 |
arrestin, beta 2 |
1249 |
0.24 |
| chr3_46134626_46134827 | 0.20 |
XCR1 |
chemokine (C motif) receptor 1 |
65492 |
0.1 |
| chr11_48055584_48055738 | 0.19 |
AC103828.1 |
|
18254 |
0.2 |
| chr7_37431183_37431614 | 0.19 |
ENSG00000200113 |
. |
27833 |
0.16 |
| chr17_4615435_4615799 | 0.19 |
ARRB2 |
arrestin, beta 2 |
1623 |
0.18 |
| chr1_153915425_153915874 | 0.19 |
DENND4B |
DENN/MADD domain containing 4B |
917 |
0.37 |
| chr5_140949949_140950149 | 0.18 |
CTD-2024I7.13 |
|
12171 |
0.11 |
| chr10_129679513_129679717 | 0.18 |
CLRN3 |
clarin 3 |
11596 |
0.22 |
| chr15_40744962_40745272 | 0.18 |
RP11-64K12.9 |
|
1721 |
0.2 |
| chr3_45735106_45735260 | 0.17 |
SACM1L |
SAC1 suppressor of actin mutations 1-like (yeast) |
4232 |
0.2 |
| chr21_46337383_46337695 | 0.16 |
ITGB2 |
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
3231 |
0.13 |
| chr19_54900734_54900885 | 0.16 |
LAIR1 |
leukocyte-associated immunoglobulin-like receptor 1 |
18644 |
0.09 |
| chr19_9968157_9968686 | 0.16 |
OLFM2 |
olfactomedin 2 |
423 |
0.71 |
| chr9_36503743_36503988 | 0.16 |
MELK |
maternal embryonic leucine zipper kinase |
68994 |
0.12 |
| chr14_24777170_24777321 | 0.15 |
CIDEB |
cell death-inducing DFFA-like effector b |
224 |
0.75 |
| chr7_106509329_106509549 | 0.15 |
PIK3CG |
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit gamma |
3515 |
0.33 |
| chr2_162852228_162852540 | 0.15 |
ENSG00000253046 |
. |
77092 |
0.09 |
| chr1_202126750_202127132 | 0.14 |
PTPN7 |
protein tyrosine phosphatase, non-receptor type 7 |
2178 |
0.23 |
| chr19_41260629_41260873 | 0.14 |
SNRPA |
small nuclear ribonucleoprotein polypeptide A |
3637 |
0.13 |
| chr3_18766132_18766453 | 0.14 |
ENSG00000228956 |
. |
20944 |
0.29 |
| chr5_126095615_126095836 | 0.14 |
ENSG00000252185 |
. |
4616 |
0.26 |
| chr6_137674077_137674372 | 0.14 |
IFNGR1 |
interferon gamma receptor 1 |
133638 |
0.05 |
| chr2_8617155_8617315 | 0.14 |
AC011747.7 |
|
198661 |
0.03 |
| chr2_113932343_113932666 | 0.14 |
AC016683.5 |
|
410 |
0.68 |
| chr1_92056374_92056622 | 0.14 |
CDC7 |
cell division cycle 7 |
89803 |
0.09 |
| chr9_116348054_116348205 | 0.14 |
RGS3 |
regulator of G-protein signaling 3 |
3777 |
0.25 |
| chr17_35292636_35292860 | 0.14 |
RP11-445F12.1 |
|
1173 |
0.31 |
| chr3_184033730_184033944 | 0.14 |
EIF4G1 |
eukaryotic translation initiation factor 4 gamma, 1 |
143 |
0.91 |
| chr16_81485634_81485842 | 0.13 |
CMIP |
c-Maf inducing protein |
6963 |
0.27 |
| chr14_72020524_72020893 | 0.13 |
SIPA1L1 |
signal-induced proliferation-associated 1 like 1 |
24666 |
0.25 |
| chr5_75745118_75745695 | 0.13 |
IQGAP2 |
IQ motif containing GTPase activating protein 2 |
45157 |
0.18 |
| chr1_224546493_224546706 | 0.13 |
CNIH4 |
cornichon family AMPA receptor auxiliary protein 4 |
2004 |
0.26 |
| chr6_24941337_24941588 | 0.13 |
FAM65B |
family with sequence similarity 65, member B |
5274 |
0.26 |
| chr9_95792448_95792761 | 0.13 |
FGD3 |
FYVE, RhoGEF and PH domain containing 3 |
15262 |
0.18 |
| chr9_95794742_95794893 | 0.13 |
FGD3 |
FYVE, RhoGEF and PH domain containing 3 |
17475 |
0.17 |
| chr15_60855427_60855584 | 0.13 |
RORA |
RAR-related orphan receptor A |
29235 |
0.16 |
| chr5_64776318_64776785 | 0.13 |
ADAMTS6 |
ADAM metallopeptidase with thrombospondin type 1 motif, 6 |
1153 |
0.57 |
| chr12_6572982_6573330 | 0.12 |
VAMP1 |
vesicle-associated membrane protein 1 (synaptobrevin 1) |
6655 |
0.09 |
| chr12_6888873_6889024 | 0.12 |
ENSG00000244532 |
. |
4379 |
0.08 |
| chr2_74729541_74729763 | 0.12 |
LBX2-AS1 |
LBX2 antisense RNA 1 |
70 |
0.79 |
| chr19_1073176_1073340 | 0.12 |
HMHA1 |
histocompatibility (minor) HA-1 |
1746 |
0.19 |
| chr12_105044364_105044605 | 0.12 |
ENSG00000264295 |
. |
59073 |
0.13 |
| chr17_7454664_7454994 | 0.12 |
TNFSF12 |
tumor necrosis factor (ligand) superfamily, member 12 |
2358 |
0.08 |
| chr21_36258213_36258674 | 0.12 |
RUNX1 |
runt-related transcription factor 1 |
1037 |
0.69 |
| chr8_145547493_145547689 | 0.12 |
DGAT1 |
diacylglycerol O-acyltransferase 1 |
2746 |
0.11 |
| chr3_13052264_13052559 | 0.12 |
IQSEC1 |
IQ motif and Sec7 domain 1 |
23875 |
0.24 |
| chr5_175122013_175122238 | 0.12 |
ENSG00000200648 |
. |
8776 |
0.21 |
| chr19_30171757_30172032 | 0.12 |
PLEKHF1 |
pleckstrin homology domain containing, family F (with FYVE domain) member 1 |
14113 |
0.21 |
| chr1_212428395_212428714 | 0.12 |
PPP2R5A |
protein phosphatase 2, regulatory subunit B', alpha |
30325 |
0.16 |
| chrX_65235385_65235647 | 0.12 |
ENSG00000207939 |
. |
3196 |
0.26 |
| chr2_143940127_143940582 | 0.12 |
RP11-190J23.1 |
|
10613 |
0.27 |
| chr7_7856072_7856264 | 0.11 |
RPA3-AS1 |
RPA3 antisense RNA 1 |
44176 |
0.16 |
| chr8_8937758_8938041 | 0.11 |
ENSG00000239078 |
. |
7873 |
0.16 |
| chr10_11258698_11258849 | 0.11 |
RP3-323N1.2 |
|
45434 |
0.15 |
| chr1_160599867_160600089 | 0.11 |
SLAMF1 |
signaling lymphocytic activation molecule family member 1 |
16833 |
0.15 |
| chr7_77110919_77111130 | 0.11 |
PTPN12 |
protein tyrosine phosphatase, non-receptor type 12 |
55568 |
0.13 |
| chr10_11252064_11252436 | 0.11 |
RP3-323N1.2 |
|
38911 |
0.17 |
| chrX_9542077_9542228 | 0.11 |
TBL1X |
transducin (beta)-like 1X-linked |
39169 |
0.22 |
| chr5_93057250_93057545 | 0.11 |
POU5F2 |
POU domain class 5, transcription factor 2 |
19946 |
0.2 |
| chr12_9142243_9142548 | 0.11 |
KLRG1 |
killer cell lectin-like receptor subfamily G, member 1 |
178 |
0.94 |
| chr2_99272389_99272594 | 0.11 |
MGAT4A |
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A |
7445 |
0.22 |
| chr17_46533902_46534274 | 0.11 |
ENSG00000206805 |
. |
22371 |
0.11 |
| chr3_160474831_160475164 | 0.11 |
PPM1L |
protein phosphatase, Mg2+/Mn2+ dependent, 1L |
1001 |
0.51 |
| chr1_162532046_162532364 | 0.11 |
UAP1 |
UDP-N-acteylglucosamine pyrophosphorylase 1 |
882 |
0.43 |
| chr1_226912901_226913052 | 0.11 |
ITPKB |
inositol-trisphosphate 3-kinase B |
12183 |
0.22 |
| chr6_53658174_53658326 | 0.11 |
RP13-476E20.1 |
|
822 |
0.56 |
| chr12_15127651_15127896 | 0.11 |
PDE6H |
phosphodiesterase 6H, cGMP-specific, cone, gamma |
1817 |
0.32 |
| chr14_106322040_106322191 | 0.11 |
ENSG00000265612 |
. |
1626 |
0.06 |
| chr11_117821389_117821783 | 0.11 |
TMPRSS13 |
transmembrane protease, serine 13 |
21412 |
0.15 |
| chr1_155605830_155606100 | 0.11 |
MSTO1 |
misato 1, mitochondrial distribution and morphology regulator |
22953 |
0.12 |
| chr1_32718399_32718550 | 0.11 |
LCK |
lymphocyte-specific protein tyrosine kinase |
1599 |
0.2 |
| chr10_94832980_94833144 | 0.11 |
CYP26A1 |
cytochrome P450, family 26, subfamily A, polypeptide 1 |
170 |
0.95 |
| chr3_51707561_51707749 | 0.11 |
TEX264 |
testis expressed 264 |
1744 |
0.3 |
| chr2_202060286_202060438 | 0.11 |
CASP10 |
caspase 10, apoptosis-related cysteine peptidase |
12444 |
0.15 |
| chr1_25321785_25321969 | 0.11 |
ENSG00000264371 |
. |
28117 |
0.16 |
| chr2_172883080_172883231 | 0.11 |
METAP1D |
methionyl aminopeptidase type 1D (mitochondrial) |
18665 |
0.21 |
| chr1_40402911_40403113 | 0.11 |
ENSG00000207356 |
. |
7658 |
0.14 |
| chr8_96220596_96220747 | 0.11 |
C8orf37 |
chromosome 8 open reading frame 37 |
60758 |
0.12 |
| chr14_22564266_22564545 | 0.11 |
ENSG00000238634 |
. |
46482 |
0.18 |
| chr8_27189433_27189735 | 0.11 |
PTK2B |
protein tyrosine kinase 2 beta |
5244 |
0.23 |
| chr9_128024635_128025037 | 0.10 |
GAPVD1 |
GTPase activating protein and VPS9 domains 1 |
646 |
0.67 |
| chr1_31255677_31255986 | 0.10 |
LAPTM5 |
lysosomal protein transmembrane 5 |
25164 |
0.14 |
| chr22_24555074_24555225 | 0.10 |
CABIN1 |
calcineurin binding protein 1 |
3343 |
0.22 |
| chr15_70601259_70601472 | 0.10 |
ENSG00000200216 |
. |
115790 |
0.07 |
| chr11_118084739_118084975 | 0.10 |
AMICA1 |
adhesion molecule, interacts with CXADR antigen 1 |
749 |
0.59 |
| chr13_72338862_72339013 | 0.10 |
DACH1 |
dachshund homolog 1 (Drosophila) |
101970 |
0.09 |
| chr2_65281994_65282176 | 0.10 |
CEP68 |
centrosomal protein 68kDa |
1421 |
0.39 |
| chr1_211526495_211526797 | 0.10 |
TRAF5 |
TNF receptor-associated factor 5 |
6940 |
0.27 |
| chr18_5238615_5238823 | 0.10 |
C18orf42 |
chromosome 18 open reading frame 42 |
41217 |
0.18 |
| chr2_87300266_87300417 | 0.10 |
PLGLB1 |
plasminogen-like B1 |
51366 |
0.16 |
| chr1_208017036_208017239 | 0.10 |
ENSG00000203709 |
. |
41269 |
0.16 |
| chr3_52231039_52231190 | 0.10 |
ALAS1 |
aminolevulinate, delta-, synthase 1 |
988 |
0.41 |
| chr5_169698686_169698837 | 0.10 |
LCP2 |
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa) |
4430 |
0.26 |
| chr1_207999994_208000291 | 0.10 |
ENSG00000203709 |
. |
24274 |
0.21 |
| chr12_11697430_11697755 | 0.10 |
ENSG00000251747 |
. |
1821 |
0.33 |
| chr8_79716918_79717078 | 0.10 |
IL7 |
interleukin 7 |
165 |
0.97 |
| chr3_169741071_169741222 | 0.10 |
GPR160 |
G protein-coupled receptor 160 |
14571 |
0.14 |
| chr17_4933015_4933230 | 0.10 |
SLC52A1 |
solute carrier family 52 (riboflavin transporter), member 1 |
5599 |
0.09 |
| chr14_66403868_66404061 | 0.10 |
CTD-2014B16.3 |
Uncharacterized protein |
67277 |
0.13 |
| chr1_167435126_167435452 | 0.10 |
RP11-104L21.2 |
|
7391 |
0.22 |
| chr10_104417543_104417777 | 0.10 |
TRIM8 |
tripartite motif containing 8 |
13016 |
0.16 |
| chr19_39823552_39823765 | 0.10 |
GMFG |
glia maturation factor, gamma |
2987 |
0.12 |
| chrX_48661069_48661290 | 0.09 |
HDAC6 |
histone deacetylase 6 |
222 |
0.87 |
| chr3_52236468_52236805 | 0.09 |
ALAS1 |
aminolevulinate, delta-, synthase 1 |
4012 |
0.14 |
| chr9_36805830_36805981 | 0.09 |
ENSG00000266255 |
. |
17691 |
0.2 |
| chr11_71815272_71815474 | 0.09 |
ENSG00000238768 |
. |
253 |
0.76 |
| chr22_33888140_33888291 | 0.09 |
ENSG00000266012 |
. |
55560 |
0.15 |
| chr9_137274667_137274850 | 0.09 |
ENSG00000263897 |
. |
3501 |
0.29 |
| chr1_101536071_101536299 | 0.09 |
RP11-421L21.3 |
|
2031 |
0.33 |
| chr16_617336_617619 | 0.09 |
PIGQ |
phosphatidylinositol glycan anchor biosynthesis, class Q |
468 |
0.38 |
| chr2_48463667_48463988 | 0.09 |
ENSG00000201010 |
. |
19113 |
0.24 |
| chr1_206292279_206292571 | 0.09 |
C1orf186 |
chromosome 1 open reading frame 186 |
3778 |
0.21 |
| chr19_47856648_47856799 | 0.09 |
DHX34 |
DEAH (Asp-Glu-Ala-His) box polypeptide 34 |
4185 |
0.17 |
| chr19_49839783_49840418 | 0.09 |
CD37 |
CD37 molecule |
372 |
0.72 |
| chr13_107145729_107145927 | 0.09 |
EFNB2 |
ephrin-B2 |
41634 |
0.2 |
| chr15_70878037_70878229 | 0.09 |
UACA |
uveal autoantigen with coiled-coil domains and ankyrin repeats |
116487 |
0.06 |
| chr8_142277219_142277381 | 0.09 |
RP11-10J21.3 |
Uncharacterized protein |
12636 |
0.14 |
| chr17_73073275_73073426 | 0.09 |
SLC16A5 |
solute carrier family 16 (monocarboxylate transporter), member 5 |
10472 |
0.08 |
| chr1_26861688_26862188 | 0.09 |
RPS6KA1 |
ribosomal protein S6 kinase, 90kDa, polypeptide 1 |
4119 |
0.17 |
| chr2_68979555_68979777 | 0.09 |
ARHGAP25 |
Rho GTPase activating protein 25 |
17652 |
0.23 |
| chr3_44038499_44038650 | 0.09 |
ENSG00000252980 |
. |
74005 |
0.11 |
| chr21_39756313_39756535 | 0.09 |
KCNJ15 |
potassium inwardly-rectifying channel, subfamily J, member 15 |
87576 |
0.09 |
| chr6_111444189_111444398 | 0.09 |
ENSG00000200926 |
. |
31758 |
0.15 |
| chr15_41184746_41184897 | 0.09 |
VPS18 |
vacuolar protein sorting 18 homolog (S. cerevisiae) |
1807 |
0.22 |
| chr3_67733685_67733836 | 0.09 |
SUCLG2 |
succinate-CoA ligase, GDP-forming, beta subunit |
28722 |
0.26 |
| chr15_58817759_58817910 | 0.09 |
RP11-50C13.2 |
|
4324 |
0.21 |
| chr4_7055273_7055505 | 0.09 |
TADA2B |
transcriptional adaptor 2B |
309 |
0.83 |
| chrX_129226501_129226837 | 0.09 |
ELF4 |
E74-like factor 4 (ets domain transcription factor) |
17667 |
0.2 |
| chr20_57728060_57728211 | 0.09 |
ZNF831 |
zinc finger protein 831 |
37940 |
0.17 |
| chr7_128789871_128790074 | 0.09 |
TSPAN33 |
tetraspanin 33 |
5260 |
0.16 |
| chr22_37696437_37696674 | 0.09 |
CYTH4 |
cytohesin 4 |
9454 |
0.17 |
| chr19_3604778_3604929 | 0.09 |
TBXA2R |
thromboxane A2 receptor |
1805 |
0.18 |
| chr5_10448843_10449058 | 0.09 |
ROPN1L |
rhophilin associated tail protein 1-like |
6962 |
0.15 |
| chr7_38385634_38385842 | 0.09 |
AMPH |
amphiphysin |
116975 |
0.06 |
| chr1_31220827_31220980 | 0.09 |
ENSG00000264773 |
. |
8824 |
0.16 |
| chr7_21892794_21892945 | 0.09 |
DNAH11 |
dynein, axonemal, heavy chain 11 |
2089 |
0.43 |
| chr16_15733947_15734127 | 0.09 |
KIAA0430 |
KIAA0430 |
2308 |
0.2 |
| chr19_3338824_3339112 | 0.09 |
NFIC |
nuclear factor I/C (CCAAT-binding transcription factor) |
20593 |
0.16 |
| chr14_65173299_65173599 | 0.09 |
PLEKHG3 |
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
2134 |
0.36 |
| chr1_25944948_25945154 | 0.09 |
MAN1C1 |
mannosidase, alpha, class 1C, member 1 |
710 |
0.7 |
| chr6_90868295_90868522 | 0.09 |
BACH2 |
BTB and CNC homology 1, basic leucine zipper transcription factor 2 |
138053 |
0.04 |
| chrX_23970141_23970292 | 0.09 |
CXorf58 |
chromosome X open reading frame 58 |
41716 |
0.14 |
| chr1_27847356_27847507 | 0.09 |
RP1-159A19.4 |
|
4885 |
0.2 |
| chr17_38737904_38738640 | 0.09 |
CCR7 |
chemokine (C-C motif) receptor 7 |
16548 |
0.15 |
| chr16_30198438_30198649 | 0.09 |
RP11-455F5.5 |
|
2265 |
0.12 |
| chr9_36842411_36842562 | 0.09 |
RP11-344B23.2 |
|
14066 |
0.17 |
| chr19_35959595_35959758 | 0.09 |
FFAR2 |
free fatty acid receptor 2 |
19059 |
0.1 |
| chr7_7855098_7855249 | 0.09 |
RPA3-AS1 |
RPA3 antisense RNA 1 |
43181 |
0.16 |
| chr19_39810438_39810644 | 0.09 |
CTC-246B18.8 |
|
957 |
0.35 |
| chr13_109148510_109148798 | 0.09 |
MYO16 |
myosin XVI |
99846 |
0.08 |
| chr4_103434283_103434434 | 0.09 |
NFKB1 |
nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 |
9826 |
0.21 |
| chr10_115012524_115012675 | 0.09 |
ENSG00000238380 |
. |
100585 |
0.08 |
| chr1_157124834_157125040 | 0.09 |
ETV3 |
ets variant 3 |
16671 |
0.22 |
| chr22_31674547_31674798 | 0.09 |
PIK3IP1 |
phosphoinositide-3-kinase interacting protein 1 |
13709 |
0.1 |
| chr16_85935649_85935800 | 0.08 |
IRF8 |
interferon regulatory factor 8 |
571 |
0.8 |
| chr9_136241871_136242236 | 0.08 |
SURF4 |
surfeit 4 |
861 |
0.28 |
| chr16_57176887_57177289 | 0.08 |
CPNE2 |
copine II |
23977 |
0.12 |
| chr4_146100468_146100690 | 0.08 |
OTUD4 |
OTU domain containing 4 |
184 |
0.96 |
| chr11_65585057_65585265 | 0.08 |
SNX32 |
sorting nexin 32 |
15951 |
0.08 |
| chr11_64163174_64163364 | 0.08 |
ENSG00000221273 |
. |
27195 |
0.09 |
| chr10_73511536_73512081 | 0.08 |
C10orf54 |
chromosome 10 open reading frame 54 |
5579 |
0.21 |
| chr2_178103295_178103446 | 0.08 |
NFE2L2 |
nuclear factor, erythroid 2-like 2 |
6155 |
0.12 |
| chr21_36412195_36412690 | 0.08 |
RUNX1 |
runt-related transcription factor 1 |
9020 |
0.32 |
| chr17_4846040_4846191 | 0.08 |
RNF167 |
ring finger protein 167 |
31 |
0.92 |
| chr15_63448303_63448779 | 0.08 |
RPS27L |
ribosomal protein S27-like |
432 |
0.83 |
| chr8_61708524_61708722 | 0.08 |
RP11-33I11.2 |
|
13542 |
0.27 |
| chr17_64444201_64444352 | 0.08 |
RP11-4F22.2 |
|
31304 |
0.2 |
| chr11_104748532_104748683 | 0.08 |
CASP12 |
caspase 12 (gene/pseudogene) |
20534 |
0.2 |
| chr7_30463723_30463874 | 0.08 |
GS1-114I9.1 |
|
490 |
0.84 |
| chr10_14609574_14609811 | 0.08 |
FAM107B |
family with sequence similarity 107, member B |
4337 |
0.29 |
| chr2_43354688_43354839 | 0.08 |
ENSG00000207087 |
. |
36131 |
0.2 |
| chr9_134460225_134460628 | 0.08 |
RAPGEF1 |
Rap guanine nucleotide exchange factor (GEF) 1 |
36799 |
0.13 |
| chr22_28037281_28037432 | 0.08 |
RP11-375H17.1 |
|
75112 |
0.12 |
| chr11_67042731_67042882 | 0.08 |
ADRBK1 |
adrenergic, beta, receptor kinase 1 |
8854 |
0.11 |
| chr2_219867277_219867428 | 0.08 |
AC097468.4 |
|
415 |
0.61 |
| chr3_111263954_111264147 | 0.08 |
CD96 |
CD96 molecule |
3053 |
0.33 |
| chr6_15421901_15422177 | 0.08 |
JARID2 |
jumonji, AT rich interactive domain 2 |
20950 |
0.24 |
| chr11_45000351_45000502 | 0.08 |
TP53I11 |
tumor protein p53 inducible protein 11 |
27818 |
0.22 |
| chr2_26634964_26635159 | 0.08 |
DRC1 |
dynein regulatory complex subunit 1 homolog (Chlamydomonas) |
10277 |
0.21 |
| chr1_46008468_46008619 | 0.08 |
AKR1A1 |
aldo-keto reductase family 1, member A1 (aldehyde reductase) |
7672 |
0.14 |
| chr14_98635190_98635420 | 0.08 |
ENSG00000222066 |
. |
162782 |
0.04 |
| chr14_39571430_39571948 | 0.08 |
SEC23A |
Sec23 homolog A (S. cerevisiae) |
590 |
0.75 |
| chr2_106494435_106494674 | 0.08 |
AC009505.2 |
|
20921 |
0.22 |
| chr15_55549033_55549184 | 0.08 |
RAB27A |
RAB27A, member RAS oncogene family |
7875 |
0.22 |
| chr8_66858855_66859137 | 0.08 |
DNAJC5B |
DnaJ (Hsp40) homolog, subfamily C, member 5 beta |
74799 |
0.11 |
| chr6_99938862_99939169 | 0.08 |
USP45 |
ubiquitin specific peptidase 45 |
8274 |
0.18 |
| chr1_178996379_178996671 | 0.08 |
FAM20B |
family with sequence similarity 20, member B |
1494 |
0.48 |
| chr10_11193217_11193852 | 0.08 |
CELF2 |
CUGBP, Elav-like family member 2 |
13459 |
0.2 |
| chr12_121422324_121422475 | 0.08 |
HNF1A-AS1 |
HNF1A antisense RNA 1 |
3631 |
0.15 |
| chr14_74035706_74035986 | 0.08 |
ACOT2 |
acyl-CoA thioesterase 2 |
83 |
0.95 |
| chr3_185542303_185542454 | 0.08 |
IGF2BP2 |
insulin-like growth factor 2 mRNA binding protein 2 |
422 |
0.87 |
| chr19_13212328_13212569 | 0.08 |
LYL1 |
lymphoblastic leukemia derived sequence 1 |
1233 |
0.3 |
| chrX_39901530_39901681 | 0.08 |
BCOR |
BCL6 corepressor |
20585 |
0.28 |
| chr2_102978258_102978409 | 0.08 |
IL18R1 |
interleukin 18 receptor 1 |
764 |
0.64 |
| chr2_64440957_64441229 | 0.08 |
AC074289.1 |
|
6984 |
0.28 |
| chr22_20225336_20225601 | 0.08 |
RTN4R |
reticulon 4 receptor |
5739 |
0.14 |
| chr7_8111935_8112159 | 0.08 |
AC006042.6 |
|
41608 |
0.16 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.1 | GO:0016115 | diterpenoid catabolic process(GO:0016103) terpenoid catabolic process(GO:0016115) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.1 | GO:0071218 | cellular response to misfolded protein(GO:0071218) |
| 0.0 | 0.1 | GO:0045341 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) |
| 0.0 | 0.1 | GO:0001911 | negative regulation of leukocyte mediated cytotoxicity(GO:0001911) |
| 0.0 | 0.1 | GO:0010957 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.0 | 0.1 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.0 | 0.1 | GO:0002360 | T cell lineage commitment(GO:0002360) |
| 0.0 | 0.1 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.1 | GO:0050685 | positive regulation of mRNA 3'-end processing(GO:0031442) positive regulation of mRNA processing(GO:0050685) positive regulation of mRNA metabolic process(GO:1903313) |
| 0.0 | 0.1 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.0 | 0.0 | GO:0043441 | acetoacetic acid biosynthetic process(GO:0043441) |
| 0.0 | 0.1 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.1 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) regulation of store-operated calcium channel activity(GO:1901339) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.1 | GO:0031061 | negative regulation of histone methylation(GO:0031061) |
| 0.0 | 0.0 | GO:0061030 | epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.0 | 0.0 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.1 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.0 | GO:0060242 | contact inhibition(GO:0060242) |
| 0.0 | 0.1 | GO:0045409 | negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 0.0 | 0.1 | GO:0034122 | negative regulation of toll-like receptor signaling pathway(GO:0034122) |
| 0.0 | 0.0 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.1 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.1 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.0 | GO:0043218 | compact myelin(GO:0043218) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.1 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.1 | GO:0019958 | C-X-C chemokine binding(GO:0019958) interleukin-8 binding(GO:0019959) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.1 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.1 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.0 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.1 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.0 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.0 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.0 | 0.1 | GO:0031701 | angiotensin receptor binding(GO:0031701) |
| 0.0 | 0.0 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.1 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.1 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.1 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) inositol trisphosphate kinase activity(GO:0051766) |
| 0.0 | 0.0 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.4 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.1 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |