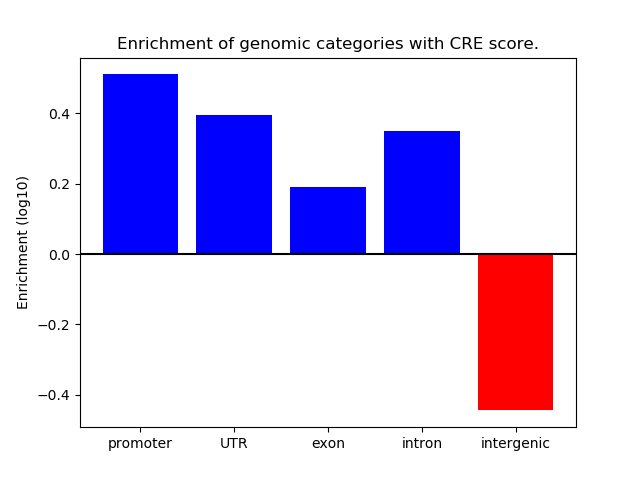
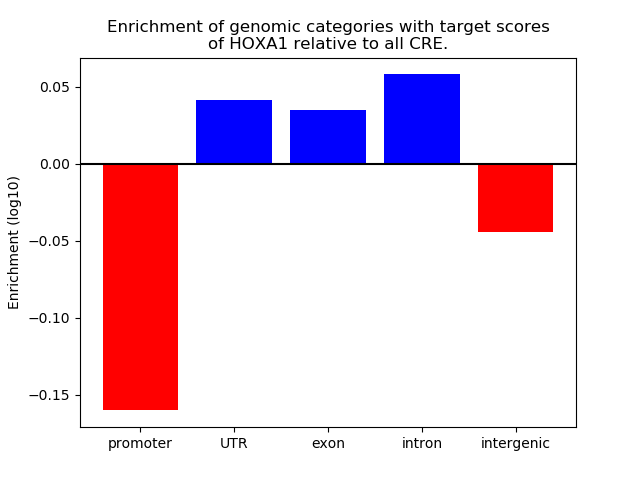
Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
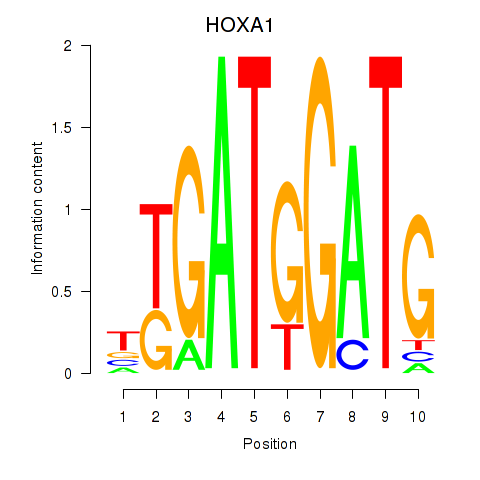
Results for HOXA1
Z-value: 2.43
Transcription factors associated with HOXA1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXA1
|
ENSG00000105991.7 | homeobox A1 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr7_27135370_27135555 | HOXA1 | 131 | 0.571442 | -0.07 | 8.6e-01 | Click! |
Activity of the HOXA1 motif across conditions
Conditions sorted by the z-value of the HOXA1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
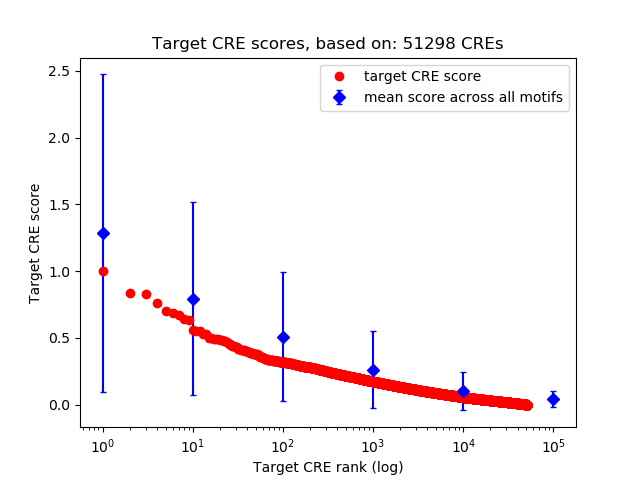
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr3_13065957_13066108 | 1.00 |
IQSEC1 |
IQ motif and Sec7 domain 1 |
37496 |
0.2 |
| chr13_114873900_114874051 | 0.84 |
RASA3-IT1 |
RASA3 intronic transcript 1 (non-protein coding) |
320 |
0.91 |
| chr14_90003505_90003656 | 0.83 |
FOXN3-AS2 |
FOXN3 antisense RNA 2 |
38980 |
0.13 |
| chr7_640540_641424 | 0.76 |
PRKAR1B |
protein kinase, cAMP-dependent, regulatory, type I, beta |
1338 |
0.36 |
| chr22_34263041_34263192 | 0.70 |
LARGE |
like-glycosyltransferase |
5504 |
0.29 |
| chr2_3299611_3299762 | 0.69 |
TSSC1-IT1 |
TSSC1 intronic transcript 1 (non-protein coding) |
5550 |
0.27 |
| chr5_153657458_153657728 | 0.67 |
ENSG00000221070 |
. |
62093 |
0.1 |
| chr1_16492736_16492887 | 0.64 |
RP11-276H7.3 |
|
7050 |
0.12 |
| chr19_381111_381262 | 0.63 |
THEG |
theg spermatid protein |
5173 |
0.15 |
| chr1_11742762_11742913 | 0.56 |
MAD2L2 |
MAD2 mitotic arrest deficient-like 2 (yeast) |
1566 |
0.27 |
| chr6_90778869_90779020 | 0.55 |
ENSG00000222078 |
. |
67719 |
0.11 |
| chr15_52019009_52019407 | 0.55 |
LYSMD2 |
LysM, putative peptidoglycan-binding, domain containing 2 |
11110 |
0.13 |
| chr1_202132763_202133787 | 0.53 |
PTPN7 |
protein tyrosine phosphatase, non-receptor type 7 |
2559 |
0.21 |
| chr2_3299852_3300003 | 0.53 |
TSSC1-IT1 |
TSSC1 intronic transcript 1 (non-protein coding) |
5309 |
0.28 |
| chr10_124180219_124180566 | 0.50 |
ENSG00000265442 |
. |
3911 |
0.21 |
| chr11_95928553_95928846 | 0.50 |
ENSG00000266192 |
. |
145903 |
0.04 |
| chr9_95793129_95793280 | 0.49 |
FGD3 |
FYVE, RhoGEF and PH domain containing 3 |
15862 |
0.17 |
| chr15_101741938_101742937 | 0.49 |
CHSY1 |
chondroitin sulfate synthase 1 |
49700 |
0.12 |
| chr3_108550085_108550421 | 0.49 |
TRAT1 |
T cell receptor associated transmembrane adaptor 1 |
8634 |
0.27 |
| chr4_113437546_113438608 | 0.48 |
NEUROG2 |
neurogenin 2 |
749 |
0.58 |
| chr10_30474293_30474444 | 0.48 |
ENSG00000200887 |
. |
114234 |
0.06 |
| chr11_121336559_121337538 | 0.48 |
RP11-730K11.1 |
|
13326 |
0.24 |
| chr10_14051000_14051172 | 0.47 |
FRMD4A |
FERM domain containing 4A |
554 |
0.8 |
| chr21_32738689_32738840 | 0.47 |
TIAM1 |
T-cell lymphoma invasion and metastasis 1 |
22170 |
0.27 |
| chr17_65830082_65830233 | 0.46 |
BPTF |
bromodomain PHD finger transcription factor |
7899 |
0.17 |
| chr6_42326971_42327197 | 0.45 |
ENSG00000221252 |
. |
53050 |
0.13 |
| chr4_122073193_122073446 | 0.44 |
TNIP3 |
TNFAIP3 interacting protein 3 |
10265 |
0.23 |
| chr2_121372137_121372546 | 0.44 |
ENSG00000201006 |
. |
36508 |
0.21 |
| chr11_102267624_102267962 | 0.44 |
BIRC2 |
baculoviral IAP repeat containing 2 |
49793 |
0.1 |
| chr2_144220343_144220494 | 0.44 |
AC096558.1 |
|
17932 |
0.26 |
| chr11_12832428_12832703 | 0.43 |
RP11-47J17.3 |
|
12649 |
0.21 |
| chr8_67404169_67404359 | 0.42 |
C8orf46 |
chromosome 8 open reading frame 46 |
1179 |
0.48 |
| chr1_54760868_54761170 | 0.42 |
RP5-997D24.3 |
|
9941 |
0.19 |
| chr10_81937813_81938167 | 0.41 |
ANXA11 |
annexin A11 |
5213 |
0.23 |
| chr14_89713300_89713451 | 0.41 |
FOXN3 |
forkhead box N3 |
66287 |
0.12 |
| chr9_120470562_120470843 | 0.41 |
ENSG00000201444 |
. |
322 |
0.9 |
| chr5_150623862_150624013 | 0.41 |
GM2A |
GM2 ganglioside activator |
8516 |
0.19 |
| chr19_50061032_50061281 | 0.40 |
NOSIP |
nitric oxide synthase interacting protein |
2731 |
0.1 |
| chr13_32700633_32700915 | 0.40 |
FRY |
furry homolog (Drosophila) |
65773 |
0.12 |
| chr5_114843276_114843427 | 0.40 |
FEM1C |
fem-1 homolog c (C. elegans) |
37240 |
0.17 |
| chr5_172905193_172905344 | 0.40 |
CTB-33O18.3 |
|
101378 |
0.07 |
| chr8_128520144_128520295 | 0.39 |
CASC8 |
cancer susceptibility candidate 8 (non-protein coding) |
25835 |
0.25 |
| chr9_73021149_73021369 | 0.39 |
KLF9 |
Kruppel-like factor 9 |
8281 |
0.28 |
| chr3_66443269_66443807 | 0.39 |
ENSG00000241587 |
. |
69259 |
0.12 |
| chr19_7423428_7423579 | 0.38 |
CTB-133G6.1 |
|
9655 |
0.15 |
| chr13_24275410_24275561 | 0.38 |
AL139080.1 |
Uncharacterized protein |
46758 |
0.16 |
| chr1_210014679_210014830 | 0.38 |
DIEXF |
digestive organ expansion factor homolog (zebrafish) |
2604 |
0.26 |
| chr4_8305331_8305837 | 0.38 |
HTRA3 |
HtrA serine peptidase 3 |
34080 |
0.17 |
| chr14_106463441_106463592 | 0.38 |
IGHVIII-2-1 |
immunoglobulin heavy variable (III)-2-1 (pseudogene) |
4451 |
0.06 |
| chr17_37922123_37922274 | 0.38 |
IKZF3 |
IKAROS family zinc finger 3 (Aiolos) |
12280 |
0.12 |
| chr1_100742904_100743055 | 0.38 |
ENSG00000207750 |
. |
3818 |
0.17 |
| chr2_85973319_85973470 | 0.37 |
ATOH8 |
atonal homolog 8 (Drosophila) |
7623 |
0.15 |
| chr4_114608486_114608818 | 0.37 |
CAMK2D |
calcium/calmodulin-dependent protein kinase II delta |
73572 |
0.13 |
| chr11_60744102_60744253 | 0.37 |
CD6 |
CD6 molecule |
4839 |
0.13 |
| chr10_63831258_63831409 | 0.37 |
ARID5B |
AT rich interactive domain 5B (MRF1-like) |
22363 |
0.23 |
| chr3_31585090_31585298 | 0.36 |
STT3B |
STT3B, subunit of the oligosaccharyltransferase complex (catalytic) |
10912 |
0.3 |
| chrX_19619969_19620120 | 0.36 |
SH3KBP1 |
SH3-domain kinase binding protein 1 |
68436 |
0.12 |
| chr9_114666357_114666809 | 0.36 |
UGCG |
UDP-glucose ceramide glucosyltransferase |
7537 |
0.2 |
| chr6_112089861_112090078 | 0.35 |
FYN |
FYN oncogene related to SRC, FGR, YES |
8849 |
0.28 |
| chr14_69612705_69612857 | 0.35 |
ENSG00000206768 |
. |
2111 |
0.32 |
| chr20_42143574_42144320 | 0.35 |
L3MBTL1 |
l(3)mbt-like 1 (Drosophila) |
781 |
0.59 |
| chr2_227342979_227343130 | 0.35 |
ENSG00000263363 |
. |
180455 |
0.03 |
| chr12_72260725_72261209 | 0.34 |
TBC1D15 |
TBC1 domain family, member 15 |
27409 |
0.21 |
| chr3_151896872_151897023 | 0.34 |
MBNL1 |
muscleblind-like splicing regulator 1 |
88882 |
0.09 |
| chr13_74438748_74438899 | 0.34 |
KLF12 |
Kruppel-like factor 12 |
130363 |
0.06 |
| chr7_127504383_127504534 | 0.34 |
SND1 |
staphylococcal nuclease and tudor domain containing 1 |
23507 |
0.25 |
| chr16_89373716_89373867 | 0.34 |
AC137932.5 |
|
9527 |
0.13 |
| chr3_106928871_106929022 | 0.34 |
ENSG00000220989 |
. |
155083 |
0.04 |
| chr3_170147264_170147415 | 0.34 |
CLDN11 |
claudin 11 |
10484 |
0.26 |
| chr1_240740736_240741022 | 0.34 |
RP11-467I20.6 |
|
13169 |
0.2 |
| chr14_89722620_89722771 | 0.34 |
FOXN3 |
forkhead box N3 |
75607 |
0.1 |
| chr6_25973027_25973178 | 0.34 |
TRIM38 |
tripartite motif containing 38 |
9827 |
0.1 |
| chr17_64342041_64342192 | 0.33 |
ENSG00000244044 |
. |
18031 |
0.2 |
| chr8_6335605_6335756 | 0.33 |
RP11-115C21.2 |
|
71017 |
0.1 |
| chr22_37475461_37475625 | 0.33 |
RP5-1170K4.7 |
|
565 |
0.62 |
| chr18_20638644_20638795 | 0.33 |
ENSG00000223023 |
. |
34160 |
0.13 |
| chr5_150597516_150598012 | 0.33 |
CCDC69 |
coiled-coil domain containing 69 |
5942 |
0.19 |
| chr2_160058942_160059211 | 0.33 |
WDSUB1 |
WD repeat, sterile alpha motif and U-box domain containing 1 |
83987 |
0.1 |
| chr2_30656489_30656640 | 0.33 |
ENSG00000221377 |
. |
1250 |
0.54 |
| chr3_33087403_33087747 | 0.33 |
GLB1 |
galactosidase, beta 1 |
50709 |
0.11 |
| chr2_69795993_69796144 | 0.33 |
ENSG00000207016 |
. |
48762 |
0.12 |
| chr12_6178946_6179123 | 0.33 |
ENSG00000240533 |
. |
26102 |
0.19 |
| chr1_203606704_203607193 | 0.33 |
ATP2B4 |
ATPase, Ca++ transporting, plasma membrane 4 |
11020 |
0.24 |
| chr3_141133468_141133746 | 0.33 |
ZBTB38 |
zinc finger and BTB domain containing 38 |
11351 |
0.23 |
| chr14_89794652_89794865 | 0.33 |
RP11-356K23.1 |
|
21871 |
0.18 |
| chr7_25891719_25892347 | 0.33 |
ENSG00000199085 |
. |
97573 |
0.09 |
| chr5_177671788_177671939 | 0.32 |
PHYKPL |
5-phosphohydroxy-L-lysine phospho-lyase |
12077 |
0.2 |
| chr7_22707827_22707978 | 0.32 |
AC002480.5 |
|
6491 |
0.2 |
| chr3_53069303_53069506 | 0.32 |
SFMBT1 |
Scm-like with four mbt domains 1 |
9877 |
0.18 |
| chr2_30477316_30477615 | 0.32 |
LBH |
limb bud and heart development |
22419 |
0.22 |
| chr12_111873433_111874308 | 0.32 |
SH2B3 |
SH2B adaptor protein 3 |
1204 |
0.47 |
| chr11_60890001_60890152 | 0.32 |
CD5 |
CD5 molecule |
20095 |
0.15 |
| chr1_67235783_67235934 | 0.32 |
TCTEX1D1 |
Tctex1 domain containing 1 |
17715 |
0.15 |
| chr7_127698825_127699005 | 0.32 |
ENSG00000207588 |
. |
22998 |
0.2 |
| chr22_31848300_31848566 | 0.32 |
RP11-247I13.11 |
|
1804 |
0.22 |
| chr21_34611617_34611944 | 0.32 |
AP000295.9 |
|
7299 |
0.15 |
| chr17_39114507_39114658 | 0.32 |
KRT39 |
keratin 39 |
8562 |
0.08 |
| chr13_30095831_30095982 | 0.32 |
MTUS2-AS1 |
MTUS2 antisense RNA 1 |
31664 |
0.21 |
| chr3_32376656_32376948 | 0.32 |
CMTM7 |
CKLF-like MARVEL transmembrane domain containing 7 |
56361 |
0.12 |
| chr7_130772886_130773250 | 0.32 |
LINC-PINT |
long intergenic non-protein coding RNA, p53 induced transcript |
165 |
0.96 |
| chr1_149984676_149984827 | 0.32 |
OTUD7B |
OTU domain containing 7B |
2126 |
0.26 |
| chr14_98677293_98677464 | 0.32 |
ENSG00000222066 |
. |
120709 |
0.06 |
| chr9_122713224_122713375 | 0.32 |
ENSG00000207814 |
. |
294029 |
0.01 |
| chr18_77265355_77265646 | 0.32 |
AC018445.1 |
Uncharacterized protein |
10557 |
0.27 |
| chr6_112066325_112066681 | 0.32 |
FYN |
FYN oncogene related to SRC, FGR, YES |
13814 |
0.25 |
| chr1_167409803_167409954 | 0.32 |
RP11-104L21.2 |
|
18020 |
0.2 |
| chr17_45358083_45358234 | 0.32 |
ENSG00000238419 |
. |
21372 |
0.12 |
| chr22_43877399_43877550 | 0.32 |
EFCAB6-AS1 |
EFCAB6 antisense RNA 1 |
34523 |
0.19 |
| chr6_112098947_112099288 | 0.31 |
FYN |
FYN oncogene related to SRC, FGR, YES |
3166 |
0.35 |
| chr17_64354339_64354490 | 0.31 |
ENSG00000244044 |
. |
5733 |
0.24 |
| chr11_68023676_68023827 | 0.31 |
C11orf24 |
chromosome 11 open reading frame 24 |
15664 |
0.19 |
| chrX_117647162_117647313 | 0.31 |
DOCK11 |
dedicator of cytokinesis 11 |
17365 |
0.23 |
| chr10_121013984_121014157 | 0.31 |
ENSG00000242853 |
. |
41701 |
0.11 |
| chr14_98689187_98689819 | 0.31 |
ENSG00000222066 |
. |
108584 |
0.07 |
| chr5_106971420_106971571 | 0.31 |
EFNA5 |
ephrin-A5 |
34833 |
0.23 |
| chr11_65876620_65876771 | 0.31 |
PACS1 |
phosphofurin acidic cluster sorting protein 1 |
8461 |
0.11 |
| chr12_57487197_57487835 | 0.31 |
NAB2 |
NGFI-A binding protein 2 (EGR1 binding protein 2) |
4626 |
0.11 |
| chr13_31451769_31451920 | 0.31 |
MEDAG |
mesenteric estrogen-dependent adipogenesis |
28484 |
0.18 |
| chr20_436878_437029 | 0.31 |
ENSG00000206797 |
. |
5646 |
0.17 |
| chr6_24894743_24895047 | 0.31 |
FAM65B |
family with sequence similarity 65, member B |
16300 |
0.2 |
| chr1_55829518_55829669 | 0.31 |
ENSG00000199831 |
. |
12932 |
0.23 |
| chr7_21474691_21474842 | 0.31 |
SP4 |
Sp4 transcription factor |
7105 |
0.2 |
| chr9_125023522_125023809 | 0.31 |
MRRF |
mitochondrial ribosome recycling factor |
3217 |
0.16 |
| chr8_23609447_23609604 | 0.31 |
ENSG00000216123 |
. |
16633 |
0.17 |
| chr11_128227126_128227277 | 0.31 |
ETS1 |
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
148088 |
0.04 |
| chr2_227817637_227817788 | 0.31 |
ENSG00000212391 |
. |
16126 |
0.23 |
| chr12_3808327_3808592 | 0.31 |
ENSG00000222338 |
. |
27034 |
0.21 |
| chr16_25002951_25003162 | 0.31 |
ARHGAP17 |
Rho GTPase activating protein 17 |
23596 |
0.23 |
| chr3_27497662_27497813 | 0.31 |
SLC4A7 |
solute carrier family 4, sodium bicarbonate cotransporter, member 7 |
508 |
0.81 |
| chr2_120578129_120578280 | 0.31 |
PTPN4 |
protein tyrosine phosphatase, non-receptor type 4 (megakaryocyte) |
60404 |
0.13 |
| chr16_64663494_64663645 | 0.31 |
CDH11 |
cadherin 11, type 2, OB-cadherin (osteoblast) |
430012 |
0.01 |
| chr3_152036313_152036556 | 0.31 |
MBNL1 |
muscleblind-like splicing regulator 1 |
18447 |
0.2 |
| chr1_225642878_225643029 | 0.30 |
RP11-496N12.6 |
|
10032 |
0.2 |
| chr1_1659225_1659440 | 0.30 |
CDK11A |
cyclin-dependent kinase 11A |
3566 |
0.15 |
| chr14_90009937_90010173 | 0.30 |
FOXN3-AS2 |
FOXN3 antisense RNA 2 |
32505 |
0.14 |
| chr5_58426674_58426825 | 0.30 |
RP11-266N13.2 |
|
91161 |
0.09 |
| chr1_1593343_1593578 | 0.30 |
RP11-345P4.10 |
|
2674 |
0.13 |
| chr7_36085161_36085312 | 0.30 |
PP13004 |
|
33458 |
0.18 |
| chr21_45512010_45512161 | 0.30 |
PWP2 |
PWP2 periodic tryptophan protein homolog (yeast) |
15086 |
0.16 |
| chr5_156627474_156627850 | 0.30 |
ITK |
IL2-inducible T-cell kinase |
19825 |
0.11 |
| chr3_108566267_108566511 | 0.30 |
TRAT1 |
T cell receptor associated transmembrane adaptor 1 |
24770 |
0.23 |
| chr20_35557608_35557874 | 0.30 |
SAMHD1 |
SAM domain and HD domain 1 |
22370 |
0.17 |
| chr14_99699518_99699950 | 0.30 |
AL109767.1 |
|
29551 |
0.18 |
| chr1_92166568_92166719 | 0.30 |
ENSG00000239794 |
. |
128988 |
0.05 |
| chr1_1362586_1362737 | 0.30 |
TMEM88B |
transmembrane protein 88B |
1153 |
0.23 |
| chr14_91502758_91502909 | 0.30 |
RPS6KA5 |
ribosomal protein S6 kinase, 90kDa, polypeptide 5 |
23645 |
0.17 |
| chr11_95839885_95840036 | 0.30 |
MTMR2 |
myotubularin related protein 2 |
182501 |
0.03 |
| chr7_97958337_97958488 | 0.29 |
RP11-307C18.1 |
|
6247 |
0.2 |
| chr16_78803162_78803313 | 0.29 |
RP11-319G9.3 |
|
127825 |
0.05 |
| chr5_156976034_156976304 | 0.29 |
AC106801.1 |
|
16803 |
0.15 |
| chr11_85761578_85761729 | 0.29 |
PICALM |
phosphatidylinositol binding clathrin assembly protein |
18169 |
0.21 |
| chr9_81181847_81181998 | 0.29 |
PSAT1 |
phosphoserine aminotransferase 1 |
269863 |
0.02 |
| chr12_69034261_69034415 | 0.29 |
ENSG00000206650 |
. |
13183 |
0.19 |
| chr5_543677_544187 | 0.29 |
ENSG00000264233 |
. |
7935 |
0.14 |
| chr10_106739812_106739963 | 0.29 |
SORCS3 |
sortilin-related VPS10 domain containing receptor 3 |
178799 |
0.03 |
| chr6_143255793_143256032 | 0.29 |
HIVEP2 |
human immunodeficiency virus type I enhancer binding protein 2 |
10426 |
0.26 |
| chr10_127859322_127859597 | 0.29 |
ENSG00000222740 |
. |
25308 |
0.25 |
| chr1_21655454_21655664 | 0.29 |
ECE1 |
endothelin converting enzyme 1 |
16438 |
0.2 |
| chr12_57458185_57458336 | 0.29 |
MYO1A |
myosin IA |
13301 |
0.09 |
| chr13_114879199_114879381 | 0.29 |
RASA3-IT1 |
RASA3 intronic transcript 1 (non-protein coding) |
4995 |
0.25 |
| chr6_169958477_169958751 | 0.29 |
WDR27 |
WD repeat domain 27 |
102174 |
0.07 |
| chr1_1951998_1952149 | 0.29 |
GABRD |
gamma-aminobutyric acid (GABA) A receptor, delta |
1293 |
0.33 |
| chr3_112494249_112494410 | 0.29 |
ENSG00000242770 |
. |
26996 |
0.21 |
| chr2_12901157_12901451 | 0.29 |
ENSG00000264370 |
. |
23811 |
0.24 |
| chr12_10874698_10875290 | 0.29 |
YBX3 |
Y box binding protein 3 |
912 |
0.56 |
| chr8_355038_355189 | 0.29 |
FBXO25 |
F-box protein 25 |
1315 |
0.53 |
| chr20_1644883_1645856 | 0.29 |
ENSG00000242348 |
. |
1637 |
0.31 |
| chr5_134545085_134545236 | 0.29 |
C5orf66 |
chromosome 5 open reading frame 66 |
128469 |
0.05 |
| chr17_48104312_48105191 | 0.29 |
ITGA3 |
integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor) |
28588 |
0.11 |
| chr10_3849152_3849639 | 0.29 |
KLF6 |
Kruppel-like factor 6 |
21922 |
0.22 |
| chr7_47366368_47366519 | 0.29 |
TNS3 |
tensin 3 |
46527 |
0.2 |
| chr15_31630334_31631466 | 0.29 |
KLF13 |
Kruppel-like factor 13 |
1236 |
0.62 |
| chr1_240902944_240903095 | 0.29 |
RP11-80B9.1 |
|
25838 |
0.19 |
| chr5_140978009_140978248 | 0.28 |
DIAPH1 |
diaphanous-related formin 1 |
11076 |
0.11 |
| chr12_45678957_45679108 | 0.28 |
ANO6 |
anoctamin 6 |
7434 |
0.32 |
| chr6_41928412_41928563 | 0.28 |
CCND3 |
cyclin D3 |
18901 |
0.13 |
| chr1_183028310_183028461 | 0.28 |
LAMC1 |
laminin, gamma 1 (formerly LAMB2) |
35790 |
0.16 |
| chr1_66818663_66819387 | 0.28 |
PDE4B |
phosphodiesterase 4B, cAMP-specific |
1040 |
0.69 |
| chr15_89203109_89203379 | 0.28 |
ISG20 |
interferon stimulated exonuclease gene 20kDa |
21060 |
0.16 |
| chr10_11360522_11360673 | 0.28 |
CELF2-AS1 |
CELF2 antisense RNA 1 |
1250 |
0.61 |
| chr14_62111525_62111676 | 0.28 |
RP11-47I22.3 |
Uncharacterized protein |
2538 |
0.35 |
| chr9_75731604_75731755 | 0.28 |
ANXA1 |
annexin A1 |
34994 |
0.2 |
| chr17_9336671_9336893 | 0.28 |
AC087501.1 |
|
18732 |
0.19 |
| chr2_19107015_19107166 | 0.28 |
NT5C1B |
5'-nucleotidase, cytosolic IB |
336252 |
0.01 |
| chr1_35448582_35448833 | 0.28 |
ENSG00000241014 |
. |
2207 |
0.22 |
| chr2_98361004_98361155 | 0.28 |
ZAP70 |
zeta-chain (TCR) associated protein kinase 70kDa |
10210 |
0.19 |
| chr2_190126373_190126564 | 0.28 |
ENSG00000266817 |
. |
50019 |
0.15 |
| chr13_99208991_99209442 | 0.28 |
STK24 |
serine/threonine kinase 24 |
19901 |
0.19 |
| chrX_152992299_152993172 | 0.28 |
ABCD1 |
ATP-binding cassette, sub-family D (ALD), member 1 |
1463 |
0.23 |
| chr6_144627394_144627662 | 0.28 |
UTRN |
utrophin |
14563 |
0.24 |
| chr3_58102239_58102390 | 0.28 |
FLNB |
filamin B, beta |
33279 |
0.17 |
| chr3_122807174_122807325 | 0.28 |
PDIA5 |
protein disulfide isomerase family A, member 5 |
21054 |
0.22 |
| chr10_134224918_134226015 | 0.28 |
PWWP2B |
PWWP domain containing 2B |
14764 |
0.17 |
| chr8_61647136_61647287 | 0.28 |
CHD7 |
chromodomain helicase DNA binding protein 7 |
6713 |
0.3 |
| chr2_46997189_46997340 | 0.28 |
LINC01118 |
long intergenic non-protein coding RNA 1118 |
46558 |
0.11 |
| chr3_57018388_57018539 | 0.28 |
ARHGEF3 |
Rho guanine nucleotide exchange factor (GEF) 3 |
7060 |
0.24 |
| chr2_204673809_204673960 | 0.28 |
ENSG00000206970 |
. |
27124 |
0.19 |
| chr11_6424420_6425308 | 0.28 |
APBB1 |
amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65) |
1030 |
0.44 |
| chr17_30902769_30903098 | 0.28 |
RP11-466A19.5 |
|
45866 |
0.1 |
| chr14_68759294_68759585 | 0.28 |
ENSG00000243546 |
. |
56183 |
0.15 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.2 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.2 | 1.1 | GO:0070670 | response to interleukin-4(GO:0070670) |
| 0.2 | 0.8 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 | 0.4 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.1 | 0.4 | GO:0051832 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.1 | 0.5 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.1 | 0.5 | GO:0001911 | negative regulation of leukocyte mediated cytotoxicity(GO:0001911) |
| 0.1 | 0.5 | GO:0045589 | regulation of regulatory T cell differentiation(GO:0045589) |
| 0.1 | 0.3 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 0.3 | GO:0002669 | lymphocyte anergy(GO:0002249) regulation of T cell anergy(GO:0002667) positive regulation of T cell anergy(GO:0002669) T cell anergy(GO:0002870) regulation of lymphocyte anergy(GO:0002911) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.1 | 0.3 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.1 | 0.4 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.1 | 0.3 | GO:0045074 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.1 | 0.3 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.1 | 0.3 | GO:0007132 | meiotic metaphase I(GO:0007132) |
| 0.1 | 0.2 | GO:0045066 | regulatory T cell differentiation(GO:0045066) |
| 0.1 | 0.5 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) negative regulation of signal transduction by p53 class mediator(GO:1901797) |
| 0.1 | 0.2 | GO:0032656 | regulation of interleukin-13 production(GO:0032656) |
| 0.1 | 2.1 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.1 | 0.3 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) chloride ion homeostasis(GO:0055064) |
| 0.1 | 0.1 | GO:0002513 | tolerance induction to self antigen(GO:0002513) |
| 0.1 | 0.2 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.1 | 0.4 | GO:0031646 | positive regulation of neurological system process(GO:0031646) |
| 0.1 | 0.2 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.1 | 0.2 | GO:0034392 | negative regulation of smooth muscle cell apoptotic process(GO:0034392) |
| 0.1 | 0.1 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) |
| 0.1 | 0.6 | GO:0002837 | regulation of response to tumor cell(GO:0002834) positive regulation of response to tumor cell(GO:0002836) regulation of immune response to tumor cell(GO:0002837) positive regulation of immune response to tumor cell(GO:0002839) |
| 0.1 | 0.2 | GO:1903579 | negative regulation of nucleoside metabolic process(GO:0045978) negative regulation of ATP metabolic process(GO:1903579) |
| 0.1 | 0.1 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.1 | 0.3 | GO:0048342 | paraxial mesodermal cell differentiation(GO:0048342) paraxial mesodermal cell fate commitment(GO:0048343) |
| 0.1 | 0.3 | GO:0021831 | embryonic olfactory bulb interneuron precursor migration(GO:0021831) |
| 0.1 | 0.4 | GO:0046325 | negative regulation of glucose import(GO:0046325) |
| 0.1 | 0.2 | GO:0010957 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.1 | 0.5 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.1 | 0.3 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.1 | 0.2 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) positive regulation of lamellipodium organization(GO:1902745) |
| 0.1 | 0.3 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.1 | 0.2 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.1 | 0.2 | GO:0042996 | regulation of Golgi to plasma membrane protein transport(GO:0042996) |
| 0.1 | 0.1 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.1 | 0.1 | GO:0051590 | positive regulation of neurotransmitter transport(GO:0051590) |
| 0.1 | 0.1 | GO:0042940 | D-amino acid transport(GO:0042940) |
| 0.1 | 0.3 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.1 | 0.2 | GO:0010511 | regulation of phosphatidylinositol biosynthetic process(GO:0010511) |
| 0.1 | 0.4 | GO:0097061 | dendritic spine morphogenesis(GO:0060997) regulation of dendritic spine morphogenesis(GO:0061001) dendritic spine organization(GO:0097061) |
| 0.1 | 0.2 | GO:0043383 | negative T cell selection(GO:0043383) |
| 0.1 | 0.1 | GO:0043931 | ossification involved in bone maturation(GO:0043931) |
| 0.1 | 0.2 | GO:0006498 | N-terminal protein lipidation(GO:0006498) |
| 0.1 | 0.3 | GO:0060573 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.1 | 0.1 | GO:0032353 | negative regulation of hormone metabolic process(GO:0032351) negative regulation of hormone biosynthetic process(GO:0032353) |
| 0.1 | 0.1 | GO:0002248 | connective tissue replacement involved in inflammatory response wound healing(GO:0002248) connective tissue replacement(GO:0097709) |
| 0.1 | 0.1 | GO:0048245 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.1 | 0.2 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.1 | 0.2 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.1 | 0.1 | GO:0009214 | cyclic nucleotide catabolic process(GO:0009214) |
| 0.1 | 0.2 | GO:0052169 | pathogen-associated molecular pattern dependent induction by symbiont of host innate immune response(GO:0052033) positive regulation by symbiont of host innate immune response(GO:0052166) modulation by symbiont of host innate immune response(GO:0052167) pathogen-associated molecular pattern dependent modulation by symbiont of host innate immune response(GO:0052169) pathogen-associated molecular pattern dependent induction by organism of innate immune response of other organism involved in symbiotic interaction(GO:0052257) positive regulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052305) modulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052306) pathogen-associated molecular pattern dependent modulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052308) |
| 0.1 | 0.2 | GO:0032431 | activation of phospholipase A2 activity(GO:0032431) |
| 0.1 | 0.2 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.1 | 0.1 | GO:0050955 | thermoception(GO:0050955) |
| 0.1 | 0.2 | GO:0046476 | glycosylceramide biosynthetic process(GO:0046476) |
| 0.1 | 0.6 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 0.2 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.1 | 0.1 | GO:0031065 | positive regulation of histone deacetylation(GO:0031065) |
| 0.0 | 0.1 | GO:0009048 | dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 0.1 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.7 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.1 | GO:0072217 | negative regulation of metanephros development(GO:0072217) |
| 0.0 | 0.2 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.2 | GO:0007195 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.0 | 0.4 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.2 | GO:0060456 | positive regulation of digestive system process(GO:0060456) |
| 0.0 | 0.2 | GO:0042023 | regulation of DNA endoreduplication(GO:0032875) negative regulation of DNA endoreduplication(GO:0032876) DNA endoreduplication(GO:0042023) |
| 0.0 | 0.1 | GO:0019730 | antimicrobial humoral response(GO:0019730) antibacterial humoral response(GO:0019731) |
| 0.0 | 0.0 | GO:0048311 | mitochondrion distribution(GO:0048311) |
| 0.0 | 0.2 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.0 | GO:0060581 | ventral spinal cord interneuron differentiation(GO:0021514) ventral spinal cord interneuron fate commitment(GO:0060579) cell fate commitment involved in pattern specification(GO:0060581) |
| 0.0 | 0.1 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.0 | 0.1 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.0 | 0.1 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.0 | 0.3 | GO:1901976 | regulation of cell cycle checkpoint(GO:1901976) |
| 0.0 | 0.1 | GO:0031282 | regulation of guanylate cyclase activity(GO:0031282) |
| 0.0 | 0.1 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.3 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.6 | GO:0090659 | adult walking behavior(GO:0007628) walking behavior(GO:0090659) |
| 0.0 | 0.1 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 0.1 | GO:0030952 | establishment or maintenance of cytoskeleton polarity(GO:0030952) |
| 0.0 | 0.2 | GO:0071548 | response to dexamethasone(GO:0071548) |
| 0.0 | 0.1 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.0 | 0.2 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 0.0 | GO:0070340 | detection of bacterial lipoprotein(GO:0042494) detection of bacterial lipopeptide(GO:0070340) |
| 0.0 | 0.0 | GO:0032011 | ARF protein signal transduction(GO:0032011) |
| 0.0 | 0.9 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.0 | 0.2 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.0 | 0.1 | GO:0002023 | reduction of food intake in response to dietary excess(GO:0002023) |
| 0.0 | 0.3 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.0 | 0.1 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 0.6 | GO:0032964 | collagen biosynthetic process(GO:0032964) |
| 0.0 | 0.1 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 | 0.0 | GO:0072033 | renal vesicle formation(GO:0072033) |
| 0.0 | 0.4 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.0 | GO:0045636 | positive regulation of melanocyte differentiation(GO:0045636) positive regulation of pigment cell differentiation(GO:0050942) |
| 0.0 | 0.0 | GO:0060129 | thyroid-stimulating hormone-secreting cell differentiation(GO:0060129) |
| 0.0 | 0.0 | GO:0072012 | renal system vasculature development(GO:0061437) kidney vasculature development(GO:0061440) glomerulus vasculature development(GO:0072012) glomerular mesangium development(GO:0072109) |
| 0.0 | 0.2 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.0 | 0.1 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.0 | 0.1 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 0.6 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.2 | GO:0042535 | positive regulation of tumor necrosis factor biosynthetic process(GO:0042535) |
| 0.0 | 0.1 | GO:0042523 | positive regulation of tyrosine phosphorylation of Stat5 protein(GO:0042523) |
| 0.0 | 0.0 | GO:0045354 | interferon-alpha biosynthetic process(GO:0045349) regulation of interferon-alpha biosynthetic process(GO:0045354) positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.0 | 0.2 | GO:0018106 | peptidyl-histidine phosphorylation(GO:0018106) |
| 0.0 | 0.2 | GO:0042977 | activation of JAK2 kinase activity(GO:0042977) |
| 0.0 | 0.1 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.0 | 0.1 | GO:0042661 | regulation of mesodermal cell fate specification(GO:0042661) regulation of mesoderm development(GO:2000380) |
| 0.0 | 0.3 | GO:0042772 | DNA damage response, signal transduction resulting in transcription(GO:0042772) |
| 0.0 | 0.4 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.1 | GO:0034122 | negative regulation of toll-like receptor signaling pathway(GO:0034122) |
| 0.0 | 0.1 | GO:0007008 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.1 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.1 | GO:0001766 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 0.1 | GO:0042117 | monocyte activation(GO:0042117) |
| 0.0 | 0.1 | GO:0060242 | contact inhibition(GO:0060242) |
| 0.0 | 0.1 | GO:0060057 | apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 0.0 | 0.1 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.1 | GO:0031641 | regulation of myelination(GO:0031641) |
| 0.0 | 0.1 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.2 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 0.1 | GO:0002507 | tolerance induction(GO:0002507) |
| 0.0 | 0.1 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.1 | GO:0046639 | negative regulation of alpha-beta T cell differentiation(GO:0046639) |
| 0.0 | 0.1 | GO:0015919 | peroxisomal membrane transport(GO:0015919) |
| 0.0 | 0.0 | GO:0007501 | mesodermal cell fate specification(GO:0007501) |
| 0.0 | 0.1 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.0 | 0.1 | GO:0070091 | glucagon secretion(GO:0070091) |
| 0.0 | 0.2 | GO:0001547 | antral ovarian follicle growth(GO:0001547) |
| 0.0 | 0.5 | GO:0043616 | keratinocyte proliferation(GO:0043616) |
| 0.0 | 0.0 | GO:0071803 | regulation of podosome assembly(GO:0071801) positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.1 | GO:0002677 | negative regulation of chronic inflammatory response(GO:0002677) |
| 0.0 | 0.3 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.2 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.1 | GO:0042518 | negative regulation of tyrosine phosphorylation of Stat3 protein(GO:0042518) |
| 0.0 | 0.2 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.1 | GO:0032825 | regulation of natural killer cell differentiation(GO:0032823) positive regulation of natural killer cell differentiation(GO:0032825) |
| 0.0 | 0.1 | GO:1901419 | regulation of response to alcohol(GO:1901419) |
| 0.0 | 0.1 | GO:0046885 | regulation of hormone biosynthetic process(GO:0046885) |
| 0.0 | 0.1 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.1 | GO:0033087 | immature T cell proliferation in thymus(GO:0033080) negative regulation of immature T cell proliferation(GO:0033087) negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.0 | 0.1 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.0 | 0.1 | GO:0046813 | receptor-mediated virion attachment to host cell(GO:0046813) |
| 0.0 | 0.1 | GO:0061072 | iris morphogenesis(GO:0061072) |
| 0.0 | 0.1 | GO:0045618 | positive regulation of keratinocyte differentiation(GO:0045618) |
| 0.0 | 0.1 | GO:0006210 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.0 | 0.1 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.0 | 0.1 | GO:0072074 | kidney mesenchyme development(GO:0072074) metanephric mesenchyme development(GO:0072075) |
| 0.0 | 0.5 | GO:0032330 | regulation of chondrocyte differentiation(GO:0032330) |
| 0.0 | 0.2 | GO:0006546 | glycine catabolic process(GO:0006546) |
| 0.0 | 0.1 | GO:0032506 | cytokinetic process(GO:0032506) |
| 0.0 | 0.1 | GO:0033962 | cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.0 | 0.3 | GO:0007520 | syncytium formation by plasma membrane fusion(GO:0000768) myoblast fusion(GO:0007520) |
| 0.0 | 0.1 | GO:0032055 | negative regulation of translation in response to stress(GO:0032055) |
| 0.0 | 0.1 | GO:0045542 | positive regulation of cholesterol biosynthetic process(GO:0045542) |
| 0.0 | 0.0 | GO:0042511 | positive regulation of tyrosine phosphorylation of Stat1 protein(GO:0042511) |
| 0.0 | 0.1 | GO:0071354 | cellular response to interleukin-6(GO:0071354) |
| 0.0 | 0.1 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.1 | GO:0046931 | pore complex assembly(GO:0046931) |
| 0.0 | 0.1 | GO:0033688 | regulation of osteoblast proliferation(GO:0033688) |
| 0.0 | 0.0 | GO:0046719 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) regulation by virus of viral protein levels in host cell(GO:0046719) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.0 | 0.1 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) negative regulation of lipase activity(GO:0060192) |
| 0.0 | 0.1 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.1 | GO:0016999 | antibiotic metabolic process(GO:0016999) antibiotic biosynthetic process(GO:0017000) |
| 0.0 | 0.1 | GO:0060605 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.0 | 0.1 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 0.2 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 | 0.3 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.1 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.1 | GO:0071732 | cellular response to nitric oxide(GO:0071732) cellular response to reactive nitrogen species(GO:1902170) |
| 0.0 | 0.2 | GO:0001782 | B cell homeostasis(GO:0001782) |
| 0.0 | 0.1 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.1 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.0 | 0.1 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 0.1 | GO:0033033 | negative regulation of myeloid cell apoptotic process(GO:0033033) |
| 0.0 | 0.1 | GO:0032715 | negative regulation of interleukin-6 production(GO:0032715) |
| 0.0 | 0.1 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.0 | 0.2 | GO:0014037 | Schwann cell differentiation(GO:0014037) |
| 0.0 | 0.3 | GO:0043967 | histone H4 acetylation(GO:0043967) |
| 0.0 | 0.0 | GO:0030011 | maintenance of cell polarity(GO:0030011) |
| 0.0 | 0.0 | GO:0048711 | positive regulation of astrocyte differentiation(GO:0048711) |
| 0.0 | 0.1 | GO:0072048 | pattern specification involved in kidney development(GO:0061004) renal system pattern specification(GO:0072048) pattern specification involved in metanephros development(GO:0072268) |
| 0.0 | 0.0 | GO:0002689 | negative regulation of leukocyte chemotaxis(GO:0002689) |
| 0.0 | 0.0 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.0 | 0.1 | GO:0051024 | positive regulation of immunoglobulin secretion(GO:0051024) |
| 0.0 | 0.0 | GO:0060431 | primary lung bud formation(GO:0060431) |
| 0.0 | 0.0 | GO:0060197 | cloaca development(GO:0035844) cloacal septation(GO:0060197) |
| 0.0 | 0.2 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.0 | 0.1 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.1 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.1 | GO:0010867 | positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.0 | 0.0 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.0 | GO:0000042 | protein targeting to Golgi(GO:0000042) establishment of protein localization to Golgi(GO:0072600) |
| 0.0 | 0.0 | GO:0042362 | fat-soluble vitamin biosynthetic process(GO:0042362) |
| 0.0 | 0.2 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 | 0.1 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.4 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.2 | GO:0034370 | triglyceride-rich lipoprotein particle remodeling(GO:0034370) very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.0 | 0.2 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 0.1 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 | 0.3 | GO:0001937 | negative regulation of endothelial cell proliferation(GO:0001937) |
| 0.0 | 0.1 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.0 | 0.0 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.0 | 0.1 | GO:0034379 | very-low-density lipoprotein particle assembly(GO:0034379) |
| 0.0 | 0.1 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.0 | 0.1 | GO:0010216 | maintenance of DNA methylation(GO:0010216) |
| 0.0 | 0.2 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.0 | 0.0 | GO:0045601 | regulation of endothelial cell differentiation(GO:0045601) |
| 0.0 | 0.1 | GO:0006591 | ornithine metabolic process(GO:0006591) |
| 0.0 | 0.3 | GO:1901224 | activation of NF-kappaB-inducing kinase activity(GO:0007250) NIK/NF-kappaB signaling(GO:0038061) regulation of NIK/NF-kappaB signaling(GO:1901222) positive regulation of NIK/NF-kappaB signaling(GO:1901224) |
| 0.0 | 1.1 | GO:0051291 | protein heterooligomerization(GO:0051291) |
| 0.0 | 0.2 | GO:0090181 | regulation of cholesterol metabolic process(GO:0090181) |
| 0.0 | 0.2 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.0 | 0.2 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.0 | 0.1 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.0 | 0.2 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.0 | 0.3 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.1 | GO:0015816 | glycine transport(GO:0015816) |
| 0.0 | 0.1 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.1 | GO:0042516 | regulation of tyrosine phosphorylation of Stat3 protein(GO:0042516) |
| 0.0 | 0.0 | GO:0046348 | amino sugar catabolic process(GO:0046348) glucosamine-containing compound catabolic process(GO:1901072) |
| 0.0 | 0.0 | GO:0032695 | negative regulation of interleukin-12 production(GO:0032695) |
| 0.0 | 0.0 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.0 | 0.0 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) regulation of gamma-aminobutyric acid secretion(GO:0014052) |
| 0.0 | 0.1 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.0 | 0.1 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.4 | GO:0030838 | positive regulation of actin filament polymerization(GO:0030838) |
| 0.0 | 0.0 | GO:0031943 | regulation of glucocorticoid metabolic process(GO:0031943) |
| 0.0 | 0.1 | GO:0050765 | negative regulation of phagocytosis(GO:0050765) |
| 0.0 | 0.0 | GO:0003079 | obsolete positive regulation of natriuresis(GO:0003079) |
| 0.0 | 0.0 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.0 | GO:0032261 | purine nucleotide salvage(GO:0032261) |
| 0.0 | 0.1 | GO:0006530 | asparagine catabolic process(GO:0006530) asparagine catabolic process via L-aspartate(GO:0033345) |
| 0.0 | 0.1 | GO:0045824 | negative regulation of innate immune response(GO:0045824) |
| 0.0 | 0.1 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.2 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.0 | GO:0006927 | obsolete transformed cell apoptotic process(GO:0006927) |
| 0.0 | 0.1 | GO:0018027 | peptidyl-lysine dimethylation(GO:0018027) |
| 0.0 | 0.0 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 0.0 | GO:0060426 | lung vasculature development(GO:0060426) |
| 0.0 | 0.0 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 0.1 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) double-strand break repair via single-strand annealing(GO:0045002) |
| 0.0 | 0.0 | GO:0043096 | adenine salvage(GO:0006168) purine nucleobase salvage(GO:0043096) adenine metabolic process(GO:0046083) adenine biosynthetic process(GO:0046084) |
| 0.0 | 0.0 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 0.0 | 0.1 | GO:0000132 | establishment of mitotic spindle orientation(GO:0000132) establishment of spindle orientation(GO:0051294) |
| 0.0 | 0.0 | GO:0046958 | nonassociative learning(GO:0046958) |
| 0.0 | 0.1 | GO:0035246 | peptidyl-arginine N-methylation(GO:0035246) |
| 0.0 | 0.0 | GO:0006007 | glucose catabolic process(GO:0006007) |
| 0.0 | 0.1 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.1 | GO:0033194 | response to hydroperoxide(GO:0033194) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.0 | GO:0046668 | retinal cell programmed cell death(GO:0046666) regulation of retinal cell programmed cell death(GO:0046668) |
| 0.0 | 0.1 | GO:0007343 | egg activation(GO:0007343) |
| 0.0 | 0.1 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.0 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.0 | 0.0 | GO:0090311 | regulation of protein deacetylation(GO:0090311) positive regulation of protein deacetylation(GO:0090312) |
| 0.0 | 0.2 | GO:0001662 | behavioral fear response(GO:0001662) |
| 0.0 | 0.1 | GO:0060033 | anatomical structure regression(GO:0060033) |
| 0.0 | 0.1 | GO:0000239 | pachytene(GO:0000239) |
| 0.0 | 0.1 | GO:0046184 | aldehyde biosynthetic process(GO:0046184) |
| 0.0 | 0.1 | GO:0060992 | response to fungicide(GO:0060992) |
| 0.0 | 0.0 | GO:0043497 | regulation of protein heterodimerization activity(GO:0043497) |
| 0.0 | 0.1 | GO:0006998 | nuclear envelope organization(GO:0006998) |
| 0.0 | 0.1 | GO:0010922 | positive regulation of phosphatase activity(GO:0010922) |
| 0.0 | 0.0 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.0 | 0.1 | GO:0043278 | response to isoquinoline alkaloid(GO:0014072) response to morphine(GO:0043278) |
| 0.0 | 0.1 | GO:0046337 | phosphatidylethanolamine biosynthetic process(GO:0006646) phosphatidylethanolamine metabolic process(GO:0046337) |
| 0.0 | 0.1 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.1 | GO:0051788 | response to misfolded protein(GO:0051788) |
| 0.0 | 0.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.1 | GO:0045007 | depurination(GO:0045007) |
| 0.0 | 0.0 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.0 | 0.1 | GO:0048844 | artery morphogenesis(GO:0048844) |
| 0.0 | 0.0 | GO:0060841 | venous blood vessel development(GO:0060841) |
| 0.0 | 0.1 | GO:0032119 | sequestering of zinc ion(GO:0032119) |
| 0.0 | 0.0 | GO:0002890 | negative regulation of B cell mediated immunity(GO:0002713) negative regulation of immunoglobulin mediated immune response(GO:0002890) |
| 0.0 | 0.0 | GO:0036037 | CD8-positive, alpha-beta T cell activation(GO:0036037) CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.0 | GO:0033160 | positive regulation of protein import into nucleus, translocation(GO:0033160) |
| 0.0 | 0.1 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.0 | 0.0 | GO:0060976 | coronary vasculature development(GO:0060976) |
| 0.0 | 0.1 | GO:0000730 | DNA recombinase assembly(GO:0000730) double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.0 | 0.0 | GO:0003211 | cardiac chamber formation(GO:0003207) cardiac ventricle formation(GO:0003211) |
| 0.0 | 0.0 | GO:0022007 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.0 | 0.2 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.1 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.1 | GO:0097191 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) extrinsic apoptotic signaling pathway(GO:0097191) |
| 0.0 | 0.0 | GO:0060664 | epithelial cell proliferation involved in salivary gland morphogenesis(GO:0060664) |
| 0.0 | 0.0 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.0 | 0.1 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.1 | GO:0048679 | regulation of axon regeneration(GO:0048679) regulation of neuron projection regeneration(GO:0070570) |
| 0.0 | 0.0 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.0 | 0.0 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.1 | GO:0007090 | obsolete regulation of S phase of mitotic cell cycle(GO:0007090) |
| 0.0 | 0.0 | GO:0009299 | mRNA transcription(GO:0009299) |
| 0.0 | 0.1 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 0.0 | GO:0003148 | outflow tract septum morphogenesis(GO:0003148) |
| 0.0 | 0.0 | GO:0010940 | positive regulation of necrotic cell death(GO:0010940) |
| 0.0 | 0.0 | GO:0061687 | detoxification of inorganic compound(GO:0061687) stress response to metal ion(GO:0097501) |
| 0.0 | 0.0 | GO:0046022 | regulation of transcription from RNA polymerase II promoter, mitotic(GO:0046021) positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 0.0 | 0.2 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.0 | 0.0 | GO:1904251 | regulation of bile acid biosynthetic process(GO:0070857) regulation of bile acid metabolic process(GO:1904251) |
| 0.0 | 0.0 | GO:0032071 | regulation of endodeoxyribonuclease activity(GO:0032071) |
| 0.0 | 0.0 | GO:0070272 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.0 | 0.0 | GO:0009822 | alkaloid catabolic process(GO:0009822) |
| 0.0 | 0.1 | GO:0042797 | 5S class rRNA transcription from RNA polymerase III type 1 promoter(GO:0042791) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
| 0.0 | 0.0 | GO:0042506 | tyrosine phosphorylation of Stat5 protein(GO:0042506) |
| 0.0 | 0.1 | GO:0071028 | RNA surveillance(GO:0071025) nuclear RNA surveillance(GO:0071027) nuclear mRNA surveillance(GO:0071028) |
| 0.0 | 0.0 | GO:0021860 | pyramidal neuron differentiation(GO:0021859) pyramidal neuron development(GO:0021860) |
| 0.0 | 0.0 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.0 | 0.0 | GO:0006726 | eye pigment biosynthetic process(GO:0006726) eye pigment metabolic process(GO:0042441) pigment metabolic process involved in developmental pigmentation(GO:0043324) pigment metabolic process involved in pigmentation(GO:0043474) |
| 0.0 | 0.1 | GO:0046689 | response to mercury ion(GO:0046689) |
| 0.0 | 0.0 | GO:0032730 | regulation of interleukin-1 alpha production(GO:0032650) positive regulation of interleukin-1 alpha production(GO:0032730) |
| 0.0 | 0.0 | GO:0006772 | thiamine metabolic process(GO:0006772) |
| 0.0 | 0.0 | GO:0060526 | prostate glandular acinus morphogenesis(GO:0060526) prostate epithelial cord arborization involved in prostate glandular acinus morphogenesis(GO:0060527) |
| 0.0 | 0.0 | GO:0042713 | sperm ejaculation(GO:0042713) |
| 0.0 | 0.1 | GO:0000085 | mitotic G2 phase(GO:0000085) G2 phase(GO:0051319) |
| 0.0 | 0.0 | GO:0050482 | arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.0 | 0.1 | GO:0030238 | male sex determination(GO:0030238) |
| 0.0 | 0.1 | GO:0051001 | negative regulation of nitric-oxide synthase activity(GO:0051001) |
| 0.0 | 0.2 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.0 | 0.1 | GO:0010171 | body morphogenesis(GO:0010171) |
| 0.0 | 0.1 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.0 | 0.0 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.1 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.0 | 0.0 | GO:0043537 | negative regulation of blood vessel endothelial cell migration(GO:0043537) |
| 0.0 | 0.0 | GO:0018119 | protein nitrosylation(GO:0017014) peptidyl-cysteine S-nitrosylation(GO:0018119) |
| 0.0 | 0.0 | GO:0010716 | regulation of extracellular matrix disassembly(GO:0010715) negative regulation of extracellular matrix disassembly(GO:0010716) regulation of extracellular matrix organization(GO:1903053) negative regulation of extracellular matrix organization(GO:1903054) |
| 0.0 | 0.1 | GO:0045008 | base-excision repair, AP site formation(GO:0006285) depyrimidination(GO:0045008) |
| 0.0 | 0.0 | GO:0051938 | L-glutamate import(GO:0051938) |
| 0.0 | 0.0 | GO:0005984 | disaccharide metabolic process(GO:0005984) |
| 0.0 | 0.0 | GO:0051894 | positive regulation of focal adhesion assembly(GO:0051894) positive regulation of cell junction assembly(GO:1901890) positive regulation of adherens junction organization(GO:1903393) |
| 0.0 | 0.0 | GO:0060836 | lymphatic endothelial cell differentiation(GO:0060836) |
| 0.0 | 0.4 | GO:0071377 | cellular response to glucagon stimulus(GO:0071377) |
| 0.0 | 0.0 | GO:0010677 | negative regulation of cellular carbohydrate metabolic process(GO:0010677) |
| 0.0 | 0.0 | GO:0072367 | regulation of lipid transport by regulation of transcription from RNA polymerase II promoter(GO:0072367) |
| 0.0 | 0.1 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.1 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.0 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.0 | GO:0090224 | regulation of spindle organization(GO:0090224) |
| 0.0 | 0.1 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.1 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.0 | GO:0035511 | oxidative DNA demethylation(GO:0035511) |
| 0.0 | 0.0 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.0 | 0.0 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.0 | 0.0 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.0 | 0.0 | GO:0007207 | phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.0 | 0.0 | GO:0002829 | negative regulation of type 2 immune response(GO:0002829) |
| 0.0 | 0.2 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.2 | GO:0045103 | intermediate filament-based process(GO:0045103) |
| 0.0 | 0.0 | GO:0071816 | protein insertion into ER membrane(GO:0045048) tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.0 | 0.0 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) |
| 0.0 | 0.1 | GO:0030917 | midbrain-hindbrain boundary development(GO:0030917) |
| 0.0 | 0.0 | GO:0006065 | UDP-glucuronate biosynthetic process(GO:0006065) |
| 0.0 | 0.0 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.0 | 0.0 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.0 | 0.1 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.1 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.0 | 0.0 | GO:0031125 | rRNA 3'-end processing(GO:0031125) |
| 0.0 | 0.3 | GO:0031047 | gene silencing by RNA(GO:0031047) |
| 0.0 | 0.0 | GO:0048024 | regulation of mRNA splicing, via spliceosome(GO:0048024) |
| 0.0 | 0.0 | GO:0016553 | base conversion or substitution editing(GO:0016553) |
| 0.0 | 0.1 | GO:0008089 | anterograde axonal transport(GO:0008089) |
| 0.0 | 0.1 | GO:0045954 | positive regulation of natural killer cell mediated immunity(GO:0002717) positive regulation of natural killer cell mediated cytotoxicity(GO:0045954) |
| 0.0 | 0.0 | GO:0045070 | positive regulation of viral genome replication(GO:0045070) |
| 0.0 | 0.0 | GO:0060789 | hair follicle placode formation(GO:0060789) |
| 0.0 | 0.1 | GO:0051057 | positive regulation of small GTPase mediated signal transduction(GO:0051057) |
| 0.0 | 0.2 | GO:0050772 | positive regulation of axonogenesis(GO:0050772) |
| 0.0 | 0.1 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.0 | 0.0 | GO:0061101 | neuroendocrine cell differentiation(GO:0061101) |
| 0.0 | 0.0 | GO:0040018 | positive regulation of multicellular organism growth(GO:0040018) |
| 0.0 | 0.0 | GO:0051154 | negative regulation of striated muscle cell differentiation(GO:0051154) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.1 | 0.3 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.1 | 0.2 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.1 | 0.2 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 0.2 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 0.2 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.1 | 0.2 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.1 | 0.3 | GO:0071437 | invadopodium(GO:0071437) |
| 0.1 | 0.2 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 0.2 | GO:0000300 | obsolete peripheral to membrane of membrane fraction(GO:0000300) |
| 0.0 | 0.2 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.3 | GO:0031332 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.4 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.7 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.2 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.1 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.2 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.3 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.1 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.0 | 0.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.3 | GO:0005606 | laminin-1 complex(GO:0005606) |
| 0.0 | 0.3 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.2 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.1 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.2 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.1 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.0 | 0.1 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.1 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.2 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.4 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.3 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.2 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.2 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 0.1 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 0.2 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.1 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.1 | GO:0032449 | CBM complex(GO:0032449) |
| 0.0 | 0.1 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.2 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.0 | 0.2 | GO:0005678 | obsolete chromatin assembly complex(GO:0005678) |
| 0.0 | 0.1 | GO:0033011 | perinuclear theca(GO:0033011) |
| 0.0 | 0.2 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.9 | GO:0005626 | obsolete insoluble fraction(GO:0005626) |
| 0.0 | 0.1 | GO:0071942 | XPC complex(GO:0071942) |
| 0.0 | 0.2 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) ribonuclease P complex(GO:0030677) multimeric ribonuclease P complex(GO:0030681) |
| 0.0 | 0.2 | GO:0098651 | basement membrane collagen trimer(GO:0098651) |
| 0.0 | 0.7 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.1 | GO:1903561 | extracellular exosome(GO:0070062) extracellular vesicle(GO:1903561) |
| 0.0 | 0.1 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.1 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.0 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.4 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 0.1 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 0.0 | 0.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.1 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.1 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.1 | GO:0031211 | palmitoyltransferase complex(GO:0002178) serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.4 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.2 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.1 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.5 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.1 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.3 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.2 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.0 | 0.2 | GO:0031105 | septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.0 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.2 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.0 | 0.1 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 0.2 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.1 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.5 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.1 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.0 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.0 | 0.1 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.1 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.0 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.1 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.1 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.1 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.1 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.1 | GO:0016585 | obsolete chromatin remodeling complex(GO:0016585) |
| 0.0 | 0.4 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.1 | GO:0031512 | motile primary cilium(GO:0031512) |
| 0.0 | 0.1 | GO:0071547 | piP-body(GO:0071547) |
| 0.0 | 0.1 | GO:0016942 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.0 | 0.1 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.2 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.1 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.1 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.0 | 0.0 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.1 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 1.6 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.2 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.1 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.2 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.1 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 1.3 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.1 | GO:0005954 | calcium- and calmodulin-dependent protein kinase complex(GO:0005954) |
| 0.0 | 0.0 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 0.1 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.0 | 0.1 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.0 | GO:0070938 | actomyosin contractile ring(GO:0005826) contractile ring(GO:0070938) |
| 0.0 | 0.1 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.6 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.3 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.2 | GO:0098878 | ionotropic glutamate receptor complex(GO:0008328) neurotransmitter receptor complex(GO:0098878) |
| 0.0 | 0.0 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 0.2 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.1 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.2 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.4 | GO:0044309 | neuron spine(GO:0044309) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.0 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.3 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 0.1 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.0 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.0 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.0 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.0 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.1 | GO:0045240 | dihydrolipoyl dehydrogenase complex(GO:0045240) |
| 0.0 | 0.0 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.0 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.1 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.1 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.0 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.0 | GO:0045261 | proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 0.3 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.1 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.1 | GO:0001891 | phagocytic cup(GO:0001891) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.1 | 0.4 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.1 | 0.5 | GO:0052813 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.1 | 0.2 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.1 | 0.2 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.1 | 0.3 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.1 | 0.2 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.1 | 0.1 | GO:0099602 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.1 | 0.3 | GO:0015925 | galactosidase activity(GO:0015925) |
| 0.1 | 0.6 | GO:0034593 | phosphatidylinositol bisphosphate phosphatase activity(GO:0034593) |
| 0.1 | 0.1 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.1 | 0.7 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.1 | 0.2 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.1 | 0.4 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.1 | 0.2 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.1 | 0.2 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.1 | 0.2 | GO:0042808 | obsolete neuronal Cdc2-like kinase binding(GO:0042808) |
| 0.1 | 0.2 | GO:0032296 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.1 | 0.2 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.1 | 0.2 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.1 | 0.3 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.1 | 0.6 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 0.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.1 | 0.3 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.1 | 0.2 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.1 | 0.2 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.1 | 0.2 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 0.2 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.1 | 0.2 | GO:0004607 | phosphatidylcholine-sterol O-acyltransferase activity(GO:0004607) |
| 0.0 | 0.1 | GO:0043175 | RNA polymerase core enzyme binding(GO:0043175) |
| 0.0 | 0.1 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.2 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.2 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.2 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.3 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.2 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.7 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.1 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 2.0 | GO:0001948 | glycoprotein binding(GO:0001948) |
| 0.0 | 0.2 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.2 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.3 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 1.5 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.2 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) bisphosphoglycerate 2-phosphatase activity(GO:0004083) phosphoglycerate mutase activity(GO:0004619) |
| 0.0 | 0.7 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.3 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.0 | 0.1 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.1 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.0 | 0.3 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.2 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.0 | 0.3 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.2 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.8 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.1 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.0 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.0 | 0.0 | GO:0008061 | chitin binding(GO:0008061) |
| 0.0 | 0.7 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.2 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 1.8 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.3 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.2 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.0 | 0.1 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.1 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.0 | GO:0050698 | proteoglycan sulfotransferase activity(GO:0050698) |
| 0.0 | 0.1 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.1 | GO:0004946 | bombesin receptor activity(GO:0004946) |
| 0.0 | 0.1 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 0.1 | GO:0032558 | purine deoxyribonucleotide binding(GO:0032554) adenyl deoxyribonucleotide binding(GO:0032558) |
| 0.0 | 0.2 | GO:0098988 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) G-protein coupled glutamate receptor activity(GO:0098988) |
| 0.0 | 0.1 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.2 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.0 | 0.1 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.1 | GO:0015226 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.0 | 0.2 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
| 0.0 | 0.1 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.0 | 0.1 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.0 | 0.1 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.0 | 0.1 | GO:0034187 | obsolete apolipoprotein E binding(GO:0034187) |
| 0.0 | 0.3 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.1 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.1 | GO:0051998 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 0.0 | 0.0 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.0 | 0.1 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.1 | GO:0055106 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.1 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.1 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.1 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.0 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.2 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.1 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.0 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) |
| 0.0 | 0.1 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.0 | 0.1 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.1 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.1 | GO:0097493 | extracellular matrix constituent conferring elasticity(GO:0030023) structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.1 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.0 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 0.1 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.1 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.8 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 0.1 | GO:0016901 | oxidoreductase activity, acting on the CH-OH group of donors, quinone or similar compound as acceptor(GO:0016901) |
| 0.0 | 0.1 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.1 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.0 | 0.1 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.1 | GO:0016715 | dopamine beta-monooxygenase activity(GO:0004500) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.0 | 0.1 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.0 | 0.1 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.1 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.0 | 0.1 | GO:0008035 | high-density lipoprotein particle binding(GO:0008035) |
| 0.0 | 0.1 | GO:0031545 | procollagen-proline 4-dioxygenase activity(GO:0004656) peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.3 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) |
| 0.0 | 0.2 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.0 | 0.1 | GO:0042910 | xenobiotic-transporting ATPase activity(GO:0008559) xenobiotic transporter activity(GO:0042910) |
| 0.0 | 0.2 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.0 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 0.0 | 0.1 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.0 | GO:0010861 | thyroid hormone receptor activator activity(GO:0010861) |
| 0.0 | 0.0 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.0 | 0.1 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.1 | GO:0005350 | pyrimidine nucleobase transmembrane transporter activity(GO:0005350) |
| 0.0 | 0.1 | GO:0003701 | obsolete RNA polymerase I transcription factor activity(GO:0003701) |
| 0.0 | 0.1 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.1 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.1 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.0 | 0.1 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) |
| 0.0 | 0.4 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.0 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.1 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.1 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.0 | GO:0004952 | dopamine neurotransmitter receptor activity(GO:0004952) |
| 0.0 | 0.1 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.0 | 0.1 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.0 | 0.2 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.1 | GO:0016847 | 1-aminocyclopropane-1-carboxylate synthase activity(GO:0016847) |
| 0.0 | 0.1 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.1 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.1 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 0.0 | 0.1 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.0 | 0.1 | GO:0000832 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) |
| 0.0 | 0.1 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.0 | 0.1 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.0 | 0.2 | GO:0016274 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.0 | 0.1 | GO:0002060 | purine nucleobase binding(GO:0002060) |
| 0.0 | 0.1 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 0.1 | GO:0004470 | malic enzyme activity(GO:0004470) |
| 0.0 | 0.1 | GO:0005338 | nucleotide-sugar transmembrane transporter activity(GO:0005338) |
| 0.0 | 0.1 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.1 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.1 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.1 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.2 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.1 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.1 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.1 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.0 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.0 | 0.3 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.2 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.0 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.0 | 0.2 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.1 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.1 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.1 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.0 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.0 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 0.0 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.0 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.1 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.0 | 0.1 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.1 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.0 | 0.0 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.0 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 0.0 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.1 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.0 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.1 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.1 | GO:0004437 | obsolete inositol or phosphatidylinositol phosphatase activity(GO:0004437) |
| 0.0 | 0.0 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 0.1 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 0.0 | 0.1 | GO:0051636 | obsolete Gram-negative bacterial cell surface binding(GO:0051636) |
| 0.0 | 0.1 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.0 | 0.0 | GO:0005345 | purine nucleobase transmembrane transporter activity(GO:0005345) |
| 0.0 | 0.0 | GO:0015137 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.0 | 0.1 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.1 | GO:0009881 | photoreceptor activity(GO:0009881) |
| 0.0 | 0.0 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.1 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.1 | GO:0032407 | MutSalpha complex binding(GO:0032407) |
| 0.0 | 0.1 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.1 | GO:0008106 | alcohol dehydrogenase (NADP+) activity(GO:0008106) |
| 0.0 | 0.0 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.0 | 1.6 | GO:0003702 | obsolete RNA polymerase II transcription factor activity(GO:0003702) |
| 0.0 | 0.3 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.2 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.2 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.1 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.0 | GO:0016889 | endodeoxyribonuclease activity, producing 3'-phosphomonoesters(GO:0016889) |
| 0.0 | 0.0 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.0 | GO:0034618 | arginine binding(GO:0034618) |
| 0.0 | 0.1 | GO:0005221 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.1 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.1 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.0 | 0.2 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.1 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 0.2 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.0 | 0.2 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.4 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.2 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 0.0 | 0.1 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.0 | 0.0 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.0 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.0 | GO:0035514 | DNA demethylase activity(GO:0035514) DNA-N1-methyladenine dioxygenase activity(GO:0043734) |
| 0.0 | 0.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.2 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.0 | GO:0003720 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.0 | 0.0 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) inositol-1,3,4,5-tetrakisphosphate 5-phosphatase activity(GO:0052659) |
| 0.0 | 0.2 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.0 | GO:0017089 | glycolipid transporter activity(GO:0017089) |
| 0.0 | 0.1 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.0 | 0.1 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.0 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.7 | GO:0004221 | obsolete ubiquitin thiolesterase activity(GO:0004221) |
| 0.0 | 0.1 | GO:0015278 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.0 | 0.0 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.1 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 0.1 | GO:0004945 | angiotensin receptor activity(GO:0001595) angiotensin type II receptor activity(GO:0004945) |
| 0.0 | 0.1 | GO:0016944 | obsolete RNA polymerase II transcription elongation factor activity(GO:0016944) |
| 0.0 | 0.0 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.0 | 0.0 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.0 | GO:0015211 | purine nucleoside transmembrane transporter activity(GO:0015211) |
| 0.0 | 0.1 | GO:0016891 | endoribonuclease activity, producing 5'-phosphomonoesters(GO:0016891) |
| 0.0 | 0.0 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.1 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.0 | 0.0 | GO:0051766 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) inositol trisphosphate kinase activity(GO:0051766) |
| 0.0 | 0.1 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.0 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 0.3 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.0 | 0.2 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.0 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.2 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.2 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.1 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.0 | GO:0005536 | glucose binding(GO:0005536) |
| 0.0 | 0.1 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
| 0.0 | 0.1 | GO:0005110 | frizzled-2 binding(GO:0005110) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.1 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.1 | 0.1 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.1 | 0.1 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.1 | 0.1 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.1 | 0.1 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.1 | 0.1 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.1 | 0.2 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 0.1 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.6 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.9 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 1.0 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.3 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.1 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.1 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 0.1 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 0.3 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 1.4 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.1 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 1.4 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.1 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.8 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.2 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.1 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.6 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.4 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.3 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 0.7 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.8 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 0.7 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 0.0 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 0.3 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.3 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.8 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.3 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.0 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.0 | 0.2 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.0 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.3 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.3 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.2 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.0 | 0.4 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.1 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.4 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.3 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.5 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.0 | 0.1 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 0.1 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.0 | 0.1 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 1.0 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.6 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.0 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 0.5 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.1 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.0 | 0.0 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.1 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.1 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.0 | 0.2 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.1 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.3 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.0 | PID IFNG PATHWAY | IFN-gamma pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.8 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 0.1 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.1 | 0.1 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 0.5 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 0.5 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.5 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.9 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.1 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.0 | 0.1 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.6 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.0 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 0.2 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.3 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.3 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.9 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.0 | 0.5 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.3 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.5 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.6 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 1.0 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.1 | REACTOME RORA ACTIVATES CIRCADIAN EXPRESSION | Genes involved in RORA Activates Circadian Expression |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.2 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.2 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.1 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.0 | 0.4 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.0 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.2 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 1.3 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.5 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.0 | REACTOME BOTULINUM NEUROTOXICITY | Genes involved in Botulinum neurotoxicity |
| 0.0 | 0.3 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 0.0 | REACTOME SIGNALING BY ERBB4 | Genes involved in Signaling by ERBB4 |
| 0.0 | 0.3 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.4 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.2 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.2 | REACTOME CA DEPENDENT EVENTS | Genes involved in Ca-dependent events |
| 0.0 | 0.2 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.2 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.3 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.3 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.1 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.2 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.0 | 0.0 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.0 | 0.1 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.2 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.1 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.2 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.1 | REACTOME CD28 CO STIMULATION | Genes involved in CD28 co-stimulation |
| 0.0 | 0.1 | REACTOME SIGNALING BY TGF BETA RECEPTOR COMPLEX | Genes involved in Signaling by TGF-beta Receptor Complex |
| 0.0 | 0.3 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.2 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.0 | 0.0 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 0.5 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.1 | REACTOME SHC MEDIATED SIGNALLING | Genes involved in SHC-mediated signalling |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.6 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.1 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.0 | 0.3 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.1 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.1 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.3 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.2 | REACTOME GLUCAGON SIGNALING IN METABOLIC REGULATION | Genes involved in Glucagon signaling in metabolic regulation |
| 0.0 | 0.2 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.2 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.0 | 0.1 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.1 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.1 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.0 | 0.2 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.0 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.0 | 0.1 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.2 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.2 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.1 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.1 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |