Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
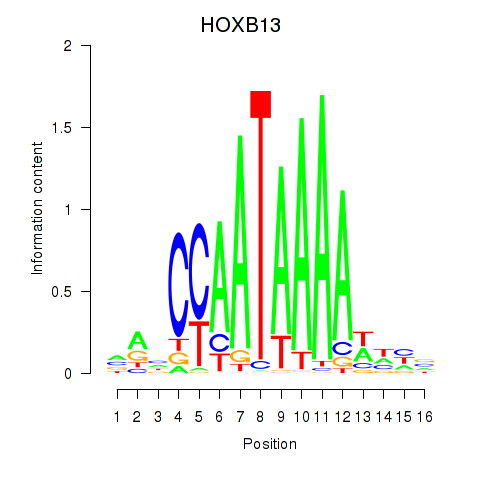
Results for HOXB13
Z-value: 0.77
Transcription factors associated with HOXB13
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXB13
|
ENSG00000159184.7 | homeobox B13 |
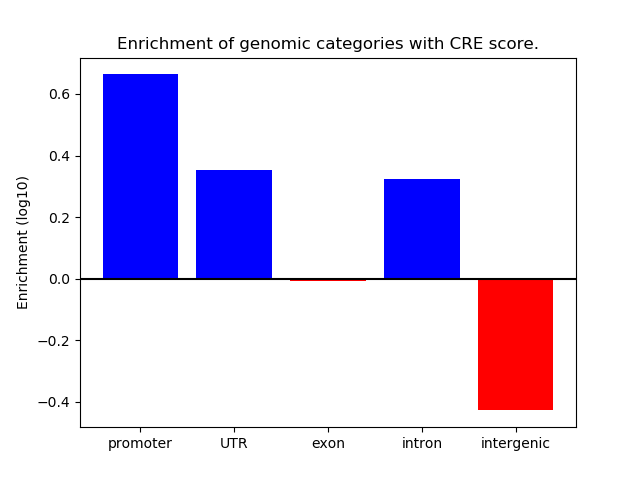
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr17_46806332_46806483 | HOXB13 | 133 | 0.921349 | 0.41 | 2.7e-01 | Click! |
| chr17_46816297_46816448 | HOXB13 | 9832 | 0.100685 | 0.32 | 4.0e-01 | Click! |
| chr17_46810841_46811006 | HOXB13 | 4383 | 0.117601 | 0.22 | 5.7e-01 | Click! |
| chr17_46805969_46806120 | HOXB13 | 496 | 0.646259 | 0.18 | 6.5e-01 | Click! |
| chr17_46811332_46811483 | HOXB13 | 4867 | 0.113937 | 0.11 | 7.7e-01 | Click! |
Activity of the HOXB13 motif across conditions
Conditions sorted by the z-value of the HOXB13 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
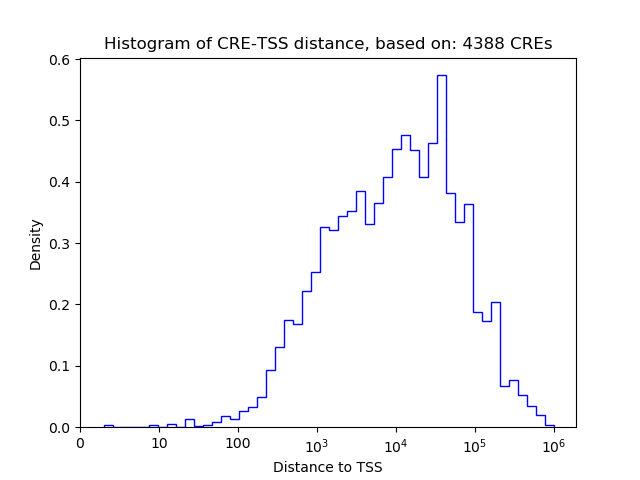
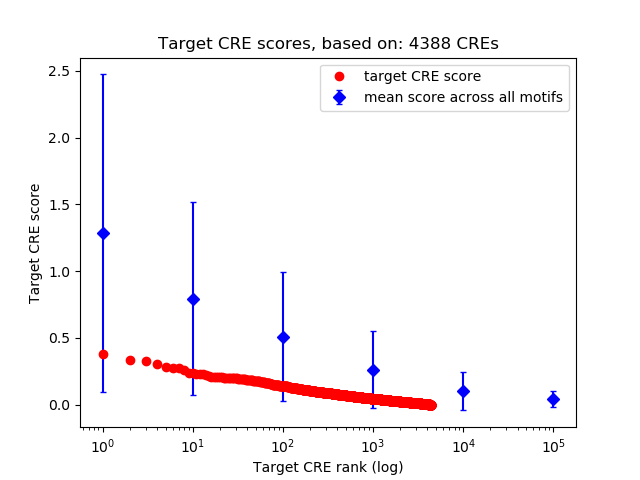
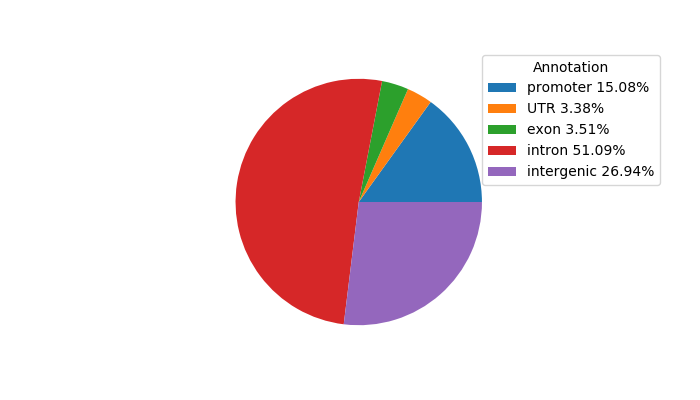
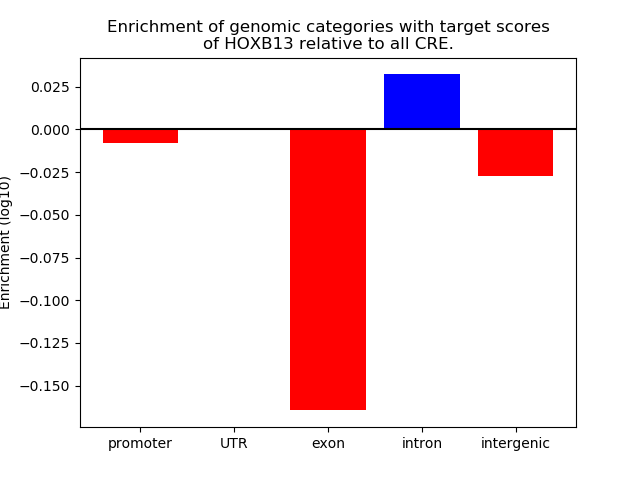
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr2_216484078_216484386 | 0.38 |
AC012462.1 |
|
183256 |
0.03 |
| chr20_10600399_10600550 | 0.33 |
JAG1 |
jagged 1 |
42680 |
0.18 |
| chr2_135814073_135814224 | 0.33 |
RAB3GAP1 |
RAB3 GTPase activating protein subunit 1 (catalytic) |
4279 |
0.2 |
| chr8_122360906_122361134 | 0.31 |
ENSG00000221644 |
. |
161888 |
0.04 |
| chr6_161527421_161527572 | 0.28 |
AGPAT4 |
1-acylglycerol-3-phosphate O-acyltransferase 4 |
42825 |
0.18 |
| chr15_60370131_60370351 | 0.28 |
FOXB1 |
forkhead box B1 |
73820 |
0.12 |
| chr12_23131728_23131879 | 0.28 |
ETNK1 |
ethanolamine kinase 1 |
353512 |
0.01 |
| chr8_27154876_27155049 | 0.26 |
TRIM35 |
tripartite motif containing 35 |
3310 |
0.25 |
| chr11_111990686_111990837 | 0.24 |
SDHD |
succinate dehydrogenase complex, subunit D, integral membrane protein |
33138 |
0.08 |
| chr8_19069882_19070033 | 0.24 |
SH2D4A |
SH2 domain containing 4A |
101171 |
0.08 |
| chr16_50236477_50236828 | 0.23 |
ADCY7 |
adenylate cyclase 7 |
43396 |
0.12 |
| chr2_148804260_148804811 | 0.23 |
ORC4 |
origin recognition complex, subunit 4 |
25388 |
0.19 |
| chr1_198498527_198498869 | 0.23 |
ATP6V1G3 |
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G3 |
11106 |
0.29 |
| chr3_159173828_159173979 | 0.22 |
ENSG00000263634 |
. |
173468 |
0.03 |
| chr10_101692086_101692460 | 0.21 |
DNMBP |
dynamin binding protein |
1520 |
0.41 |
| chr5_36879254_36879912 | 0.21 |
NIPBL |
Nipped-B homolog (Drosophila) |
2691 |
0.41 |
| chr5_86422918_86423069 | 0.21 |
ENSG00000265919 |
. |
12222 |
0.22 |
| chr7_17234854_17235017 | 0.21 |
AC003075.4 |
|
85992 |
0.1 |
| chr2_176030685_176030836 | 0.21 |
ENSG00000215973 |
. |
1677 |
0.22 |
| chr20_36618887_36619038 | 0.20 |
ENSG00000199683 |
. |
15079 |
0.17 |
| chr17_7653466_7653617 | 0.20 |
DNAH2 |
dynein, axonemal, heavy chain 2 |
30502 |
0.08 |
| chr15_59598180_59598331 | 0.20 |
RP11-429D19.1 |
|
34894 |
0.11 |
| chr12_25487343_25487617 | 0.20 |
ENSG00000201439 |
. |
69882 |
0.1 |
| chr17_70388485_70388636 | 0.20 |
SOX9 |
SRY (sex determining region Y)-box 9 |
271399 |
0.02 |
| chr21_36251212_36251413 | 0.20 |
RUNX1 |
runt-related transcription factor 1 |
8168 |
0.31 |
| chr5_38872858_38873009 | 0.20 |
OSMR |
oncostatin M receptor |
26832 |
0.24 |
| chr19_38700842_38701258 | 0.20 |
ENSG00000266428 |
. |
10176 |
0.1 |
| chr6_10416011_10416162 | 0.20 |
TFAP2A |
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha) |
337 |
0.82 |
| chr7_56130478_56130743 | 0.20 |
SUMF2 |
sulfatase modifying factor 2 |
1085 |
0.32 |
| chr17_15941252_15941403 | 0.20 |
NCOR1 |
nuclear receptor corepressor 1 |
1453 |
0.3 |
| chr20_10963665_10963956 | 0.20 |
RP11-103J8.1 |
|
3513 |
0.34 |
| chr10_101723594_101723781 | 0.20 |
DNMBP |
dynamin binding protein |
32934 |
0.15 |
| chr1_199582244_199582395 | 0.19 |
ENSG00000263805 |
. |
2089 |
0.49 |
| chr10_29916759_29916910 | 0.19 |
SVIL |
supervillin |
7067 |
0.21 |
| chr5_167126219_167126370 | 0.19 |
CTB-78F1.2 |
|
22074 |
0.2 |
| chr22_38244115_38244422 | 0.19 |
ENSG00000207696 |
. |
487 |
0.41 |
| chr5_37841386_37841537 | 0.19 |
GDNF |
glial cell derived neurotrophic factor |
1673 |
0.47 |
| chr10_72593489_72593705 | 0.19 |
SGPL1 |
sphingosine-1-phosphate lyase 1 |
10657 |
0.2 |
| chr7_14020149_14020300 | 0.19 |
ETV1 |
ets variant 1 |
5842 |
0.29 |
| chr17_69436901_69437052 | 0.19 |
ENSG00000222563 |
. |
129667 |
0.06 |
| chr21_29020378_29020529 | 0.19 |
ADAMTS5 |
ADAM metallopeptidase with thrombospondin type 1 motif, 5 |
681621 |
0.0 |
| chr11_13964368_13964519 | 0.19 |
ENSG00000201856 |
. |
35306 |
0.23 |
| chr2_227625941_227626092 | 0.18 |
IRS1 |
insulin receptor substrate 1 |
38459 |
0.16 |
| chr13_49005626_49005777 | 0.18 |
LPAR6 |
lysophosphatidic acid receptor 6 |
4658 |
0.32 |
| chr6_33548164_33548316 | 0.18 |
BAK1 |
BCL2-antagonist/killer 1 |
221 |
0.9 |
| chr11_67354003_67354154 | 0.18 |
GSTP1 |
glutathione S-transferase pi 1 |
2865 |
0.13 |
| chr2_75847358_75847509 | 0.18 |
ENSG00000221638 |
. |
7646 |
0.14 |
| chr1_12678723_12678924 | 0.18 |
DHRS3 |
dehydrogenase/reductase (SDR family) member 3 |
1086 |
0.5 |
| chr14_71956022_71956173 | 0.18 |
SIPA1L1 |
signal-induced proliferation-associated 1 like 1 |
39945 |
0.17 |
| chr4_151074226_151074377 | 0.18 |
ENSG00000238721 |
. |
56557 |
0.15 |
| chr1_31889861_31890012 | 0.18 |
SERINC2 |
serine incorporator 2 |
3276 |
0.21 |
| chr5_77268268_77268488 | 0.18 |
AP3B1 |
adaptor-related protein complex 3, beta 1 subunit |
66596 |
0.12 |
| chr7_7196352_7196827 | 0.18 |
C1GALT1 |
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1 |
24 |
0.98 |
| chr3_145902395_145902546 | 0.18 |
PLOD2 |
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2 |
21030 |
0.2 |
| chr2_40079335_40079486 | 0.18 |
SLC8A1-AS1 |
SLC8A1 antisense RNA 1 |
65817 |
0.13 |
| chr2_99342877_99343146 | 0.17 |
MGAT4A |
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A |
148 |
0.97 |
| chr9_36194329_36194526 | 0.17 |
CLTA |
clathrin, light chain A |
3467 |
0.23 |
| chr7_156792904_156793239 | 0.17 |
MNX1-AS2 |
MNX1 antisense RNA 2 |
5930 |
0.17 |
| chr1_149816060_149816683 | 0.17 |
HIST2H2AA3 |
histone cluster 2, H2aa3 |
1893 |
0.12 |
| chr20_46815691_46815842 | 0.17 |
ENSG00000252227 |
. |
51000 |
0.18 |
| chr22_47212286_47212467 | 0.17 |
ENSG00000221672 |
. |
31427 |
0.17 |
| chr18_53145450_53145601 | 0.17 |
ENSG00000264571 |
. |
927 |
0.57 |
| chrX_24166137_24167088 | 0.17 |
ZFX |
zinc finger protein, X-linked |
1134 |
0.35 |
| chr5_172328463_172328614 | 0.17 |
ERGIC1 |
endoplasmic reticulum-golgi intermediate compartment (ERGIC) 1 |
3682 |
0.23 |
| chr5_153826475_153827364 | 0.16 |
SAP30L |
SAP30-like |
814 |
0.51 |
| chr5_179770020_179770171 | 0.16 |
GFPT2 |
glutamine-fructose-6-phosphate transaminase 2 |
9805 |
0.21 |
| chr1_43420334_43420485 | 0.16 |
SLC2A1 |
solute carrier family 2 (facilitated glucose transporter), member 1 |
4091 |
0.22 |
| chr7_76213485_76213833 | 0.16 |
POMZP3 |
POM121 and ZP3 fusion |
33958 |
0.13 |
| chr3_57958497_57958723 | 0.16 |
FLNB |
filamin B, beta |
35517 |
0.18 |
| chr14_31641329_31641480 | 0.16 |
HECTD1 |
HECT domain containing E3 ubiquitin protein ligase 1 |
3857 |
0.22 |
| chr15_102156555_102156706 | 0.16 |
ENSG00000252614 |
. |
31885 |
0.15 |
| chr2_44798144_44798295 | 0.16 |
CAMKMT |
calmodulin-lysine N-methyltransferase |
198338 |
0.03 |
| chr12_58781035_58781186 | 0.16 |
RP11-362K2.2 |
Protein LOC100506869 |
156797 |
0.04 |
| chr6_110508362_110508550 | 0.16 |
CDC40 |
cell division cycle 40 |
6832 |
0.24 |
| chr6_122793405_122793976 | 0.16 |
PKIB |
protein kinase (cAMP-dependent, catalytic) inhibitor beta |
614 |
0.51 |
| chr17_40949013_40949515 | 0.15 |
COA3 |
cytochrome c oxidase assembly factor 3 |
1458 |
0.18 |
| chr2_12618919_12619301 | 0.15 |
ENSG00000207183 |
. |
67483 |
0.14 |
| chr11_33049563_33049714 | 0.15 |
TCP11L1 |
t-complex 11, testis-specific-like 1 |
11325 |
0.17 |
| chr12_24753774_24754027 | 0.15 |
ENSG00000240481 |
. |
105822 |
0.08 |
| chr6_140784427_140784578 | 0.15 |
ENSG00000221336 |
. |
197348 |
0.03 |
| chr2_46303608_46303759 | 0.15 |
AC017006.2 |
|
2284 |
0.4 |
| chrY_14665326_14665477 | 0.15 |
GYG2P1 |
glycogenin 2 pseudogene 1 |
133280 |
0.05 |
| chr6_38229324_38229475 | 0.15 |
ENSG00000200706 |
. |
54349 |
0.16 |
| chr12_68374810_68375098 | 0.15 |
IFNG-AS1 |
IFNG antisense RNA 1 |
8271 |
0.31 |
| chr14_52077289_52077440 | 0.15 |
ENSG00000251771 |
. |
5746 |
0.23 |
| chr3_127454839_127454990 | 0.15 |
MGLL |
monoglyceride lipase |
286 |
0.93 |
| chr22_38898988_38899381 | 0.15 |
DDX17 |
DEAD (Asp-Glu-Ala-Asp) box helicase 17 |
3133 |
0.2 |
| chr12_26899168_26899462 | 0.15 |
ITPR2 |
inositol 1,4,5-trisphosphate receptor, type 2 |
86399 |
0.09 |
| chr5_133328742_133328893 | 0.14 |
VDAC1 |
voltage-dependent anion channel 1 |
11509 |
0.22 |
| chr4_169691508_169691659 | 0.14 |
PALLD |
palladin, cytoskeletal associated protein |
58273 |
0.12 |
| chr5_107009300_107009451 | 0.14 |
EFNA5 |
ephrin-A5 |
2779 |
0.38 |
| chr5_136483987_136484138 | 0.14 |
SPOCK1 |
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 1 |
165165 |
0.04 |
| chr1_64449955_64450106 | 0.14 |
ENSG00000207190 |
. |
44642 |
0.17 |
| chr6_126362121_126362442 | 0.14 |
TRMT11 |
tRNA methyltransferase 11 homolog (S. cerevisiae) |
34293 |
0.18 |
| chr17_64345059_64345333 | 0.14 |
ENSG00000244044 |
. |
14951 |
0.2 |
| chr4_100368387_100368679 | 0.14 |
ADH7 |
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
11639 |
0.22 |
| chr6_128833498_128833649 | 0.14 |
RP1-86D1.4 |
|
7303 |
0.17 |
| chr2_105460725_105461021 | 0.14 |
LINC01158 |
long intergenic non-protein coding RNA 1158 |
7038 |
0.15 |
| chr16_84766620_84766890 | 0.14 |
USP10 |
ubiquitin specific peptidase 10 |
33109 |
0.16 |
| chr5_141707709_141707943 | 0.14 |
AC005592.2 |
|
2963 |
0.28 |
| chr3_65982631_65982782 | 0.14 |
ENSG00000202071 |
. |
6909 |
0.21 |
| chr10_73600697_73600848 | 0.14 |
ENSG00000238446 |
. |
3779 |
0.22 |
| chr5_108992233_108992456 | 0.14 |
ENSG00000266090 |
. |
28937 |
0.18 |
| chr12_79960162_79960313 | 0.14 |
PAWR |
PRKC, apoptosis, WT1, regulator |
30077 |
0.18 |
| chr5_150160797_150161161 | 0.14 |
AC010441.1 |
|
3112 |
0.19 |
| chr3_59761256_59761407 | 0.14 |
NPCDR1 |
nasopharyngeal carcinoma, down-regulated 1 |
196252 |
0.03 |
| chr2_128615982_128616171 | 0.14 |
POLR2D |
polymerase (RNA) II (DNA directed) polypeptide D |
345 |
0.84 |
| chr5_83222299_83222450 | 0.14 |
ENSG00000216125 |
. |
47331 |
0.18 |
| chr1_174131198_174131349 | 0.14 |
RABGAP1L |
RAB GTPase activating protein 1-like |
2545 |
0.33 |
| chr3_42861494_42861645 | 0.14 |
ACKR2 |
atypical chemokine receptor 2 |
458 |
0.72 |
| chr6_126305019_126305170 | 0.14 |
TRMT11 |
tRNA methyltransferase 11 homolog (S. cerevisiae) |
2482 |
0.26 |
| chr6_80853031_80853182 | 0.14 |
BCKDHB |
branched chain keto acid dehydrogenase E1, beta polypeptide |
36723 |
0.23 |
| chr18_55853503_55853654 | 0.14 |
NEDD4L |
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
9044 |
0.23 |
| chr9_116641852_116642003 | 0.14 |
ZNF618 |
zinc finger protein 618 |
3297 |
0.35 |
| chr4_82390108_82390350 | 0.13 |
RASGEF1B |
RasGEF domain family, member 1B |
2161 |
0.47 |
| chr12_80950525_80950676 | 0.13 |
RP11-272K23.3 |
|
36880 |
0.19 |
| chr21_39992446_39992744 | 0.13 |
ERG |
v-ets avian erythroblastosis virus E26 oncogene homolog |
35726 |
0.23 |
| chrX_44730783_44731431 | 0.13 |
KDM6A |
lysine (K)-specific demethylase 6A |
1650 |
0.39 |
| chr4_123461472_123461766 | 0.13 |
IL21-AS1 |
IL21 antisense RNA 1 |
78519 |
0.09 |
| chr18_19971465_19971616 | 0.13 |
CTAGE1 |
cutaneous T-cell lymphoma-associated antigen 1 |
23290 |
0.24 |
| chr14_69535729_69536049 | 0.13 |
ENSG00000206768 |
. |
74781 |
0.09 |
| chr6_26049516_26049667 | 0.13 |
HIST1H3C |
histone cluster 1, H3c |
3952 |
0.08 |
| chr13_51104174_51104472 | 0.13 |
ENSG00000221198 |
. |
166950 |
0.04 |
| chr8_87535059_87535465 | 0.13 |
CPNE3 |
copine III |
6029 |
0.21 |
| chr2_233453893_233454044 | 0.13 |
AC073254.1 |
|
2564 |
0.17 |
| chr13_76867098_76867386 | 0.13 |
ENSG00000243274 |
. |
158175 |
0.04 |
| chr12_106630836_106630987 | 0.13 |
RP11-651L5.2 |
|
9823 |
0.18 |
| chr7_151434053_151434204 | 0.13 |
PRKAG2 |
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
728 |
0.74 |
| chr11_130036128_130036279 | 0.13 |
ST14 |
suppression of tumorigenicity 14 (colon carcinoma) |
6746 |
0.27 |
| chr17_60074998_60075962 | 0.13 |
ENSG00000242398 |
. |
7868 |
0.17 |
| chr14_103859829_103860122 | 0.13 |
MARK3 |
MAP/microtubule affinity-regulating kinase 3 |
7399 |
0.18 |
| chr2_70142925_70143800 | 0.13 |
MXD1 |
MAX dimerization protein 1 |
1042 |
0.4 |
| chr6_27833560_27833860 | 0.13 |
HIST1H2AL |
histone cluster 1, H2al |
676 |
0.39 |
| chr13_74271902_74272053 | 0.12 |
KLF12 |
Kruppel-like factor 12 |
297209 |
0.01 |
| chr12_10876643_10876807 | 0.12 |
YBX3 |
Y box binding protein 3 |
814 |
0.61 |
| chr1_198143089_198143887 | 0.12 |
NEK7 |
NIMA-related kinase 7 |
17237 |
0.29 |
| chr7_123387049_123387200 | 0.12 |
WASL |
Wiskott-Aldrich syndrome-like |
1997 |
0.27 |
| chr6_14128901_14129195 | 0.12 |
CD83 |
CD83 molecule |
11176 |
0.25 |
| chr2_216529775_216530114 | 0.12 |
ENSG00000212055 |
. |
213698 |
0.02 |
| chr5_83018281_83018896 | 0.12 |
HAPLN1 |
hyaluronan and proteoglycan link protein 1 |
1156 |
0.52 |
| chr6_43894280_43894431 | 0.12 |
C6orf223 |
chromosome 6 open reading frame 223 |
73962 |
0.08 |
| chr3_12879901_12880174 | 0.12 |
ENSG00000207496 |
. |
1912 |
0.22 |
| chr10_35626541_35627695 | 0.12 |
CCNY |
cyclin Y |
1316 |
0.4 |
| chr4_169752762_169753076 | 0.12 |
PALLD |
palladin, cytoskeletal associated protein |
237 |
0.89 |
| chr7_156739962_156740113 | 0.12 |
NOM1 |
nucleolar protein with MIF4G domain 1 |
2380 |
0.29 |
| chr2_65453360_65453632 | 0.12 |
ACTR2 |
ARP2 actin-related protein 2 homolog (yeast) |
1391 |
0.5 |
| chr3_194391812_194392295 | 0.12 |
LSG1 |
large 60S subunit nuclear export GTPase 1 |
1153 |
0.38 |
| chr19_8383856_8384007 | 0.12 |
NDUFA7 |
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 7, 14.5kDa |
2349 |
0.15 |
| chr11_46535797_46536138 | 0.12 |
ENSG00000265014 |
. |
62528 |
0.09 |
| chr12_76414837_76415483 | 0.12 |
RP11-290L1.3 |
|
9114 |
0.19 |
| chr3_177624333_177624484 | 0.12 |
ENSG00000199858 |
. |
101159 |
0.09 |
| chr2_86408072_86408223 | 0.12 |
IMMT |
inner membrane protein, mitochondrial |
7262 |
0.12 |
| chr5_57557351_57557502 | 0.12 |
ENSG00000238899 |
. |
191411 |
0.03 |
| chr2_102681351_102681502 | 0.12 |
IL1R1 |
interleukin 1 receptor, type I |
422 |
0.88 |
| chr1_2205577_2205728 | 0.12 |
SKI |
v-ski avian sarcoma viral oncogene homolog |
45518 |
0.08 |
| chr16_9189167_9189953 | 0.12 |
C16orf72 |
chromosome 16 open reading frame 72 |
4055 |
0.18 |
| chr20_37485148_37485299 | 0.12 |
ENSG00000240474 |
. |
16190 |
0.19 |
| chr9_34252490_34252641 | 0.12 |
ENSG00000222426 |
. |
30103 |
0.1 |
| chrX_13007464_13007615 | 0.12 |
TMSB4X |
thymosin beta 4, X-linked |
13762 |
0.22 |
| chr3_111793003_111793154 | 0.12 |
C3orf52 |
chromosome 3 open reading frame 52 |
12104 |
0.16 |
| chr1_151592190_151592613 | 0.11 |
RP11-404E16.1 |
|
6562 |
0.11 |
| chr1_83030740_83030891 | 0.11 |
LPHN2 |
latrophilin 2 |
585242 |
0.0 |
| chr12_22387753_22387904 | 0.11 |
ST8SIA1 |
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 1 |
14152 |
0.25 |
| chr5_14810981_14811591 | 0.11 |
ENSG00000264792 |
. |
14835 |
0.24 |
| chr3_156510400_156510551 | 0.11 |
LEKR1 |
leucine, glutamate and lysine rich 1 |
33614 |
0.21 |
| chr11_124542402_124542553 | 0.11 |
SPA17 |
sperm autoantigenic protein 17 |
1217 |
0.28 |
| chr17_27897968_27898119 | 0.11 |
TP53I13 |
tumor protein p53 inducible protein 13 |
1405 |
0.19 |
| chr10_131750921_131751072 | 0.11 |
EBF3 |
early B-cell factor 3 |
11109 |
0.3 |
| chr15_52753628_52753923 | 0.11 |
MYO5A |
myosin VA (heavy chain 12, myoxin) |
25908 |
0.21 |
| chr4_119608098_119608296 | 0.11 |
METTL14 |
methyltransferase like 14 |
1627 |
0.48 |
| chr7_104918256_104918407 | 0.11 |
SRPK2 |
SRSF protein kinase 2 |
8869 |
0.22 |
| chr1_43151129_43151715 | 0.11 |
YBX1 |
Y box binding protein 1 |
2764 |
0.23 |
| chrX_22318328_22318479 | 0.11 |
ZNF645 |
zinc finger protein 645 |
27338 |
0.25 |
| chr2_205995260_205995411 | 0.11 |
NRP2 |
neuropilin 2 |
551889 |
0.0 |
| chr11_122603673_122604064 | 0.11 |
ENSG00000239079 |
. |
6849 |
0.26 |
| chr4_151231130_151231281 | 0.11 |
LRBA |
LPS-responsive vesicle trafficking, beach and anchor containing |
7342 |
0.31 |
| chr1_225843948_225844099 | 0.11 |
ENAH |
enabled homolog (Drosophila) |
3179 |
0.27 |
| chr4_54975045_54975230 | 0.11 |
ENSG00000221219 |
. |
5314 |
0.18 |
| chr13_109783595_109783805 | 0.11 |
MYO16-AS2 |
MYO16 antisense RNA 2 |
29433 |
0.24 |
| chr1_229415776_229415934 | 0.11 |
RAB4A |
RAB4A, member RAS oncogene family |
8976 |
0.15 |
| chr5_109029475_109029626 | 0.11 |
MAN2A1 |
mannosidase, alpha, class 2A, member 1 |
4483 |
0.22 |
| chr5_57484730_57484881 | 0.11 |
ENSG00000238899 |
. |
118790 |
0.06 |
| chr20_43590444_43590650 | 0.11 |
TOMM34 |
translocase of outer mitochondrial membrane 34 |
1420 |
0.33 |
| chr11_45870605_45870756 | 0.11 |
CRY2 |
cryptochrome 2 (photolyase-like) |
1723 |
0.3 |
| chr2_134900167_134900318 | 0.11 |
ENSG00000263813 |
. |
15546 |
0.23 |
| chr11_57539746_57540045 | 0.11 |
CTNND1 |
catenin (cadherin-associated protein), delta 1 |
8570 |
0.15 |
| chr4_129733836_129734549 | 0.11 |
JADE1 |
jade family PHD finger 1 |
1193 |
0.62 |
| chr2_203498647_203499096 | 0.11 |
FAM117B |
family with sequence similarity 117, member B |
1030 |
0.67 |
| chr20_48470048_48470199 | 0.11 |
ENSG00000252123 |
. |
25594 |
0.13 |
| chr10_23634401_23634613 | 0.11 |
C10orf67 |
chromosome 10 open reading frame 67 |
733 |
0.58 |
| chr9_89696027_89696178 | 0.11 |
C9orf170 |
chromosome 9 open reading frame 170 |
67457 |
0.13 |
| chr3_48858285_48858436 | 0.11 |
PRKAR2A-AS1 |
PRKAR2A antisense RNA 1 |
26645 |
0.12 |
| chr1_115866355_115866506 | 0.11 |
NGF |
nerve growth factor (beta polypeptide) |
14427 |
0.24 |
| chr11_62439776_62440015 | 0.11 |
C11orf48 |
chromosome 11 open reading frame 48 |
168 |
0.78 |
| chr15_67492948_67493403 | 0.11 |
SMAD3 |
SMAD family member 3 |
16062 |
0.2 |
| chr13_32663117_32663268 | 0.11 |
FRY |
furry homolog (Drosophila) |
28191 |
0.21 |
| chr1_180600327_180600734 | 0.11 |
XPR1 |
xenotropic and polytropic retrovirus receptor 1 |
610 |
0.8 |
| chr12_72240351_72240502 | 0.11 |
TBC1D15 |
TBC1 domain family, member 15 |
6868 |
0.27 |
| chr3_63954596_63954752 | 0.11 |
ATXN7 |
ataxin 7 |
1254 |
0.4 |
| chr12_24714032_24714515 | 0.11 |
ENSG00000240481 |
. |
145449 |
0.05 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0043497 | regulation of protein heterodimerization activity(GO:0043497) |
| 0.1 | 0.2 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 0.1 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.2 | GO:0090312 | positive regulation of protein deacetylation(GO:0090312) |
| 0.0 | 0.1 | GO:0044413 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.0 | 0.1 | GO:0060676 | ureteric bud formation(GO:0060676) |
| 0.0 | 0.1 | GO:0071731 | response to nitric oxide(GO:0071731) |
| 0.0 | 0.1 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.1 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.0 | 0.0 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.0 | 0.0 | GO:0072033 | renal vesicle formation(GO:0072033) |
| 0.0 | 0.0 | GO:0033591 | response to L-ascorbic acid(GO:0033591) |
| 0.0 | 0.2 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.0 | 0.0 | GO:0071436 | sodium ion export(GO:0071436) |
| 0.0 | 0.1 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.0 | 0.0 | GO:0016102 | retinoic acid biosynthetic process(GO:0002138) diterpenoid biosynthetic process(GO:0016102) |
| 0.0 | 0.0 | GO:0010799 | regulation of peptidyl-threonine phosphorylation(GO:0010799) |
| 0.0 | 0.1 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.1 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.0 | GO:0002606 | positive regulation of dendritic cell antigen processing and presentation(GO:0002606) |
| 0.0 | 0.0 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.0 | 0.0 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.0 | GO:0060605 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.0 | 0.1 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.0 | 0.0 | GO:0090273 | regulation of somatostatin secretion(GO:0090273) |
| 0.0 | 0.0 | GO:0000966 | RNA 5'-end processing(GO:0000966) |
| 0.0 | 0.0 | GO:0090206 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.0 | 0.0 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.0 | GO:0043276 | anoikis(GO:0043276) |
| 0.0 | 0.0 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.0 | GO:0030185 | nitric oxide transport(GO:0030185) |
| 0.0 | 0.1 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.1 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.1 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.0 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) |
| 0.0 | 0.0 | GO:0010837 | regulation of keratinocyte proliferation(GO:0010837) |
| 0.0 | 0.0 | GO:0031558 | obsolete induction of apoptosis in response to chemical stimulus(GO:0031558) |
| 0.0 | 0.0 | GO:0033689 | negative regulation of osteoblast proliferation(GO:0033689) |
| 0.0 | 0.1 | GO:0051155 | positive regulation of striated muscle cell differentiation(GO:0051155) |
| 0.0 | 0.0 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.0 | 0.0 | GO:0002072 | optic cup morphogenesis involved in camera-type eye development(GO:0002072) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.0 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.0 | 0.1 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.0 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.2 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 0.0 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.1 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.1 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.1 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.2 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.2 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.1 | GO:0001098 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.1 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.2 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.2 | GO:0048531 | beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.0 | 0.1 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.0 | 0.1 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.0 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.0 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.1 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.0 | GO:0042808 | obsolete neuronal Cdc2-like kinase binding(GO:0042808) |
| 0.0 | 0.0 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.0 | GO:0008934 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.1 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.0 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.0 | 0.1 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.1 | GO:0004661 | protein geranylgeranyltransferase activity(GO:0004661) |
| 0.0 | 0.0 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.0 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.0 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.0 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.1 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.0 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.0 | 0.1 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.1 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |