Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
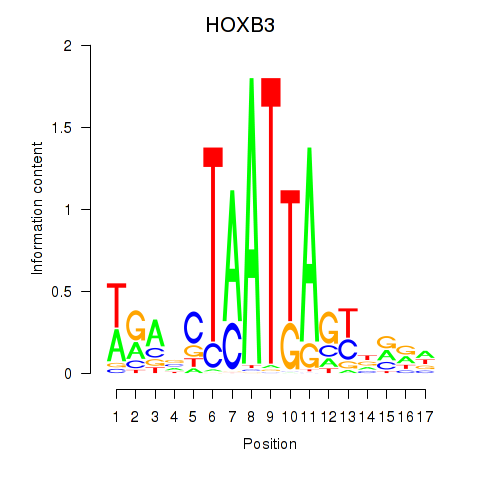
Results for HOXB3
Z-value: 1.50
Transcription factors associated with HOXB3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXB3
|
ENSG00000120093.7 | homeobox B3 |
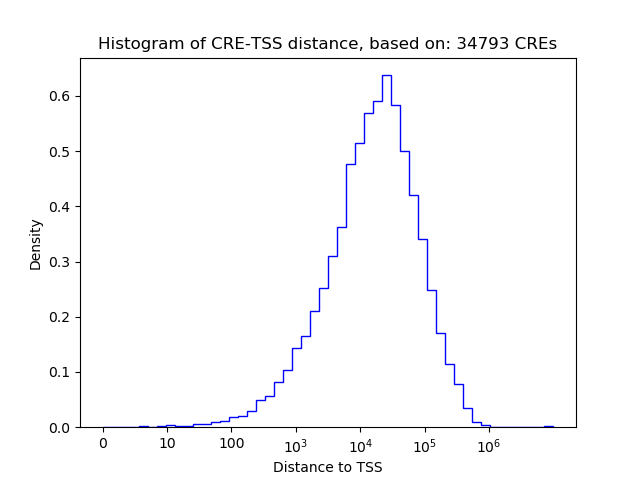
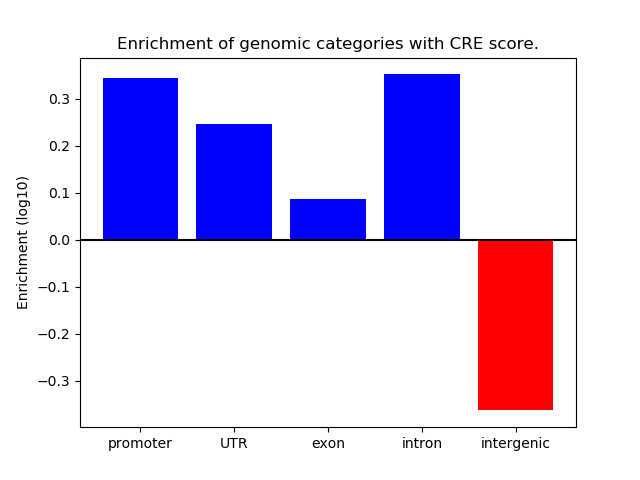
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr17_46654451_46654696 | HOXB3 | 2135 | 0.116487 | -0.84 | 4.5e-03 | Click! |
| chr17_46632798_46632949 | HOXB3 | 1010 | 0.266210 | -0.84 | 4.9e-03 | Click! |
| chr17_46655643_46655794 | HOXB3 | 1001 | 0.238854 | -0.83 | 5.8e-03 | Click! |
| chr17_46662373_46662833 | HOXB3 | 2619 | 0.098821 | -0.82 | 7.1e-03 | Click! |
| chr17_46654100_46654251 | HOXB3 | 1737 | 0.143918 | -0.80 | 9.0e-03 | Click! |
Activity of the HOXB3 motif across conditions
Conditions sorted by the z-value of the HOXB3 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
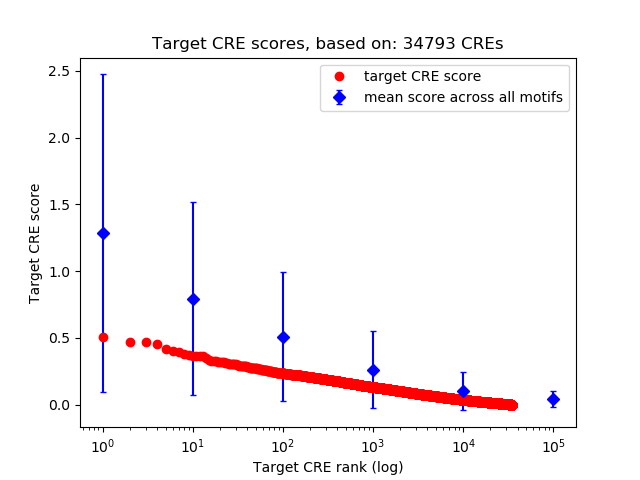
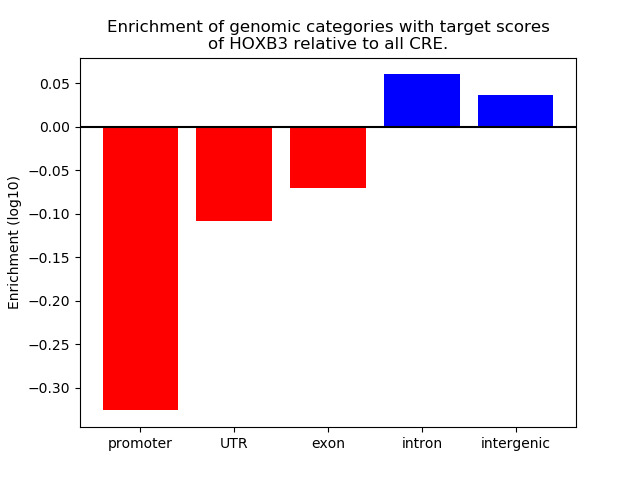
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr13_51051372_51051556 | 0.51 |
ENSG00000221198 |
. |
114091 |
0.07 |
| chr17_66379311_66379462 | 0.47 |
ENSG00000207561 |
. |
41303 |
0.14 |
| chr15_39652479_39652630 | 0.47 |
RP11-624L4.1 |
|
60356 |
0.14 |
| chr10_89436229_89436444 | 0.45 |
PAPSS2 |
3'-phosphoadenosine 5'-phosphosulfate synthase 2 |
15628 |
0.19 |
| chr7_150784838_150785537 | 0.42 |
AGAP3 |
ArfGAP with GTPase domain, ankyrin repeat and PH domain 3 |
1033 |
0.32 |
| chr9_3360340_3360491 | 0.40 |
RFX3 |
regulatory factor X, 3 (influences HLA class II expression) |
13651 |
0.3 |
| chr15_50194108_50194259 | 0.40 |
ATP8B4 |
ATPase, class I, type 8B, member 4 |
6107 |
0.24 |
| chr10_30347671_30348341 | 0.38 |
KIAA1462 |
KIAA1462 |
447 |
0.9 |
| chr7_133267178_133267329 | 0.37 |
EXOC4 |
exocyst complex component 4 |
6037 |
0.35 |
| chr4_89647157_89647308 | 0.37 |
FAM13A-AS1 |
FAM13A antisense RNA 1 |
4366 |
0.21 |
| chrX_39832066_39832217 | 0.37 |
BCOR |
BCL6 corepressor |
90049 |
0.09 |
| chr8_29057628_29057779 | 0.36 |
ENSG00000264328 |
. |
57493 |
0.09 |
| chr12_62931646_62931797 | 0.36 |
ENSG00000202034 |
. |
7738 |
0.19 |
| chr12_12016346_12017014 | 0.35 |
ETV6 |
ets variant 6 |
22191 |
0.26 |
| chr1_64549059_64549210 | 0.34 |
ENSG00000200508 |
. |
16721 |
0.18 |
| chr6_97629282_97629433 | 0.33 |
MMS22L |
MMS22-like, DNA repair protein |
81871 |
0.11 |
| chr9_88955484_88955635 | 0.33 |
ZCCHC6 |
zinc finger, CCHC domain containing 6 |
2479 |
0.34 |
| chr12_81338456_81338607 | 0.32 |
LIN7A |
lin-7 homolog A (C. elegans) |
6827 |
0.19 |
| chr2_33662966_33663243 | 0.32 |
RASGRP3 |
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
1713 |
0.48 |
| chr20_43124845_43125142 | 0.32 |
SERINC3 |
serine incorporator 3 |
8539 |
0.13 |
| chrX_37702580_37702731 | 0.32 |
DYNLT3 |
dynein, light chain, Tctex-type 3 |
4006 |
0.26 |
| chr1_97899901_97900052 | 0.32 |
DPYD-IT1 |
DPYD intronic transcript 1 (non-protein coding) |
14279 |
0.31 |
| chr11_71524282_71524863 | 0.32 |
ZNF705E |
zinc finger protein 705E |
6261 |
0.1 |
| chr17_66380501_66380857 | 0.31 |
ENSG00000207561 |
. |
40010 |
0.14 |
| chr2_28344167_28344318 | 0.31 |
ENSG00000265321 |
. |
125008 |
0.05 |
| chr6_134871939_134872090 | 0.31 |
SGK1 |
serum/glucocorticoid regulated kinase 1 |
232764 |
0.02 |
| chr16_87840292_87840843 | 0.31 |
RP4-536B24.2 |
|
29571 |
0.14 |
| chr6_39787204_39787470 | 0.30 |
DAAM2 |
dishevelled associated activator of morphogenesis 2 |
26543 |
0.21 |
| chr8_80226395_80226546 | 0.30 |
ENSG00000264969 |
. |
216872 |
0.02 |
| chr2_118804547_118804712 | 0.30 |
ENSG00000243510 |
. |
30671 |
0.13 |
| chr2_223875941_223876092 | 0.30 |
KCNE4 |
potassium voltage-gated channel, Isk-related family, member 4 |
40516 |
0.17 |
| chr11_6376113_6376264 | 0.30 |
PRKCDBP |
protein kinase C, delta binding protein |
34311 |
0.11 |
| chr8_101294374_101294525 | 0.29 |
RNF19A |
ring finger protein 19A, RBR E3 ubiquitin protein ligase |
21009 |
0.2 |
| chr1_36908138_36908289 | 0.29 |
OSCP1 |
organic solute carrier partner 1 |
1637 |
0.28 |
| chr5_5374817_5374968 | 0.29 |
KIAA0947 |
KIAA0947 |
47915 |
0.19 |
| chr7_39480916_39481067 | 0.29 |
POU6F2-AS1 |
POU6F2 antisense RNA 1 |
35046 |
0.19 |
| chr2_153267355_153267720 | 0.29 |
FMNL2 |
formin-like 2 |
75786 |
0.12 |
| chr2_145225492_145225643 | 0.29 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
37430 |
0.19 |
| chr2_33501178_33501329 | 0.29 |
LTBP1 |
latent transforming growth factor beta binding protein 1 |
277 |
0.93 |
| chr2_189847314_189847465 | 0.28 |
ENSG00000221502 |
. |
4571 |
0.25 |
| chr16_17931428_17931579 | 0.28 |
XYLT1 |
xylosyltransferase I |
366765 |
0.01 |
| chr1_92207941_92208092 | 0.28 |
ENSG00000239794 |
. |
87615 |
0.09 |
| chr7_15498785_15499062 | 0.28 |
AGMO |
alkylglycerol monooxygenase |
93076 |
0.1 |
| chr1_44412862_44413792 | 0.28 |
IPO13 |
importin 13 |
716 |
0.44 |
| chr15_79204771_79204922 | 0.28 |
ENSG00000252061 |
. |
13568 |
0.17 |
| chr10_94452668_94452819 | 0.28 |
HHEX |
hematopoietically expressed homeobox |
1131 |
0.51 |
| chr6_107645107_107645353 | 0.27 |
PDSS2 |
prenyl (decaprenyl) diphosphate synthase, subunit 2 |
111867 |
0.06 |
| chr15_71824033_71824261 | 0.27 |
THSD4 |
thrombospondin, type I, domain containing 4 |
15428 |
0.25 |
| chr4_25407599_25407888 | 0.27 |
ANAPC4 |
anaphase promoting complex subunit 4 |
28801 |
0.19 |
| chr15_93579557_93579854 | 0.27 |
CHD2 |
chromodomain helicase DNA binding protein 2 |
27609 |
0.2 |
| chr3_156975213_156975364 | 0.27 |
ENSG00000243176 |
. |
27220 |
0.2 |
| chr3_197784203_197784354 | 0.27 |
ENSG00000212297 |
. |
14010 |
0.14 |
| chr3_14418133_14418284 | 0.27 |
ENSG00000199609 |
. |
17940 |
0.21 |
| chr5_36300489_36300640 | 0.27 |
RANBP3L |
RAN binding protein 3-like |
1438 |
0.48 |
| chr9_3768105_3768256 | 0.27 |
RP11-252M18.3 |
|
107404 |
0.07 |
| chr5_57006064_57006215 | 0.27 |
ENSG00000221195 |
. |
67326 |
0.13 |
| chr1_185390714_185391145 | 0.26 |
ENSG00000252407 |
. |
12447 |
0.26 |
| chr9_128640336_128640592 | 0.26 |
PBX3 |
pre-B-cell leukemia homeobox 3 |
12914 |
0.3 |
| chr2_119427478_119427629 | 0.26 |
EN1 |
engrailed homeobox 1 |
177701 |
0.03 |
| chr7_116218439_116218590 | 0.26 |
AC006159.4 |
|
7226 |
0.19 |
| chr2_190018216_190018510 | 0.26 |
ENSG00000264725 |
. |
20526 |
0.19 |
| chr18_56240273_56240424 | 0.26 |
RP11-126O1.2 |
|
16046 |
0.15 |
| chr13_115076948_115077099 | 0.26 |
CHAMP1 |
chromosome alignment maintaining phosphoprotein 1 |
2965 |
0.23 |
| chr5_38924606_38924757 | 0.26 |
OSMR |
oncostatin M receptor |
20181 |
0.26 |
| chr8_69903972_69904123 | 0.26 |
ENSG00000238808 |
. |
118762 |
0.07 |
| chr8_19516504_19516655 | 0.26 |
CSGALNACT1 |
chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
23515 |
0.27 |
| chr1_90290727_90290878 | 0.25 |
LRRC8D |
leucine rich repeat containing 8 family, member D |
3322 |
0.28 |
| chr3_29228707_29228858 | 0.25 |
RBMS3 |
RNA binding motif, single stranded interacting protein 3 |
93691 |
0.09 |
| chr14_82456075_82456226 | 0.25 |
SEL1L |
sel-1 suppressor of lin-12-like (C. elegans) |
455945 |
0.01 |
| chr22_46426709_46426860 | 0.25 |
RP6-109B7.5 |
|
22189 |
0.1 |
| chr4_119732850_119733001 | 0.25 |
SEC24D |
SEC24 family member D |
24368 |
0.24 |
| chr9_80524258_80524409 | 0.25 |
GNAQ |
guanine nucleotide binding protein (G protein), q polypeptide |
86418 |
0.1 |
| chr12_110388287_110388838 | 0.25 |
GIT2 |
G protein-coupled receptor kinase interacting ArfGAP 2 |
45467 |
0.11 |
| chr10_74248398_74248549 | 0.25 |
RP11-167P22.3 |
|
13031 |
0.19 |
| chr12_7596689_7596840 | 0.25 |
CD163L1 |
CD163 molecule-like 1 |
15 |
0.98 |
| chr4_40375661_40375812 | 0.25 |
ENSG00000239010 |
. |
3794 |
0.24 |
| chr14_101308246_101309004 | 0.25 |
ENSG00000214548 |
. |
7020 |
0.04 |
| chr17_66379496_66379718 | 0.25 |
ENSG00000207561 |
. |
41082 |
0.14 |
| chr5_36928360_36928511 | 0.24 |
NIPBL |
Nipped-B homolog (Drosophila) |
51543 |
0.18 |
| chr1_52043843_52044143 | 0.24 |
OSBPL9 |
oxysterol binding protein-like 9 |
1142 |
0.5 |
| chr14_100240950_100241315 | 0.24 |
EML1 |
echinoderm microtubule associated protein like 1 |
1091 |
0.59 |
| chr5_169308984_169309135 | 0.24 |
FAM196B |
family with sequence similarity 196, member B |
98685 |
0.07 |
| chr12_1727335_1727720 | 0.24 |
WNT5B |
wingless-type MMTV integration site family, member 5B |
1305 |
0.46 |
| chr5_138288736_138289663 | 0.24 |
CTNNA1 |
catenin (cadherin-associated protein), alpha 1, 102kDa |
22872 |
0.17 |
| chr10_75798433_75798584 | 0.24 |
RP11-417O11.5 |
|
33613 |
0.15 |
| chr11_19619185_19619336 | 0.24 |
ENSG00000207407 |
. |
6422 |
0.22 |
| chr3_173610727_173610878 | 0.24 |
NLGN1-AS1 |
NLGN1 antisense RNA 1 |
27646 |
0.27 |
| chr5_90592330_90592999 | 0.24 |
ENSG00000199643 |
. |
26120 |
0.23 |
| chr9_113926997_113927193 | 0.24 |
ENSG00000212409 |
. |
67402 |
0.12 |
| chr1_56098686_56098837 | 0.24 |
ENSG00000272051 |
. |
148116 |
0.04 |
| chr2_118817741_118818188 | 0.24 |
INSIG2 |
insulin induced gene 2 |
28086 |
0.15 |
| chr11_63278500_63278680 | 0.24 |
LGALS12 |
lectin, galactoside-binding, soluble, 12 |
3131 |
0.21 |
| chr12_105617824_105617982 | 0.24 |
APPL2 |
adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 2 |
5735 |
0.25 |
| chr11_34338329_34338672 | 0.24 |
ABTB2 |
ankyrin repeat and BTB (POZ) domain containing 2 |
40302 |
0.19 |
| chr14_74724538_74725207 | 0.24 |
VSX2 |
visual system homeobox 2 |
18697 |
0.14 |
| chr7_101457741_101458764 | 0.24 |
CUX1 |
cut-like homeobox 1 |
707 |
0.73 |
| chr2_8012045_8012196 | 0.24 |
ENSG00000221255 |
. |
295148 |
0.01 |
| chr15_70023529_70023680 | 0.24 |
ENSG00000238870 |
. |
443 |
0.88 |
| chr2_158326449_158327167 | 0.24 |
CYTIP |
cytohesin 1 interacting protein |
18533 |
0.22 |
| chr8_13078515_13078765 | 0.23 |
DLC1 |
deleted in liver cancer 1 |
55415 |
0.14 |
| chr12_43709526_43709677 | 0.23 |
ENSG00000215993 |
. |
24218 |
0.27 |
| chr11_109453346_109453497 | 0.23 |
RP11-708B6.2 |
|
948 |
0.73 |
| chr15_57527692_57527843 | 0.23 |
TCF12 |
transcription factor 12 |
12447 |
0.25 |
| chr1_109740426_109740857 | 0.23 |
ENSG00000238310 |
. |
9829 |
0.15 |
| chr5_52192649_52192800 | 0.23 |
ITGA2 |
integrin, alpha 2 (CD49B, alpha 2 subunit of VLA-2 receptor) |
92432 |
0.07 |
| chr11_12436390_12436541 | 0.23 |
PARVA |
parvin, alpha |
16677 |
0.24 |
| chr11_121979710_121979861 | 0.23 |
RP11-166D19.1 |
|
3135 |
0.2 |
| chr11_103924198_103924349 | 0.23 |
PDGFD |
platelet derived growth factor D |
16519 |
0.22 |
| chr2_238221351_238221502 | 0.23 |
AC112715.2 |
Uncharacterized protein |
55692 |
0.13 |
| chr4_86718935_86719260 | 0.23 |
ARHGAP24 |
Rho GTPase activating protein 24 |
19238 |
0.25 |
| chr4_26346811_26346962 | 0.23 |
RBPJ |
recombination signal binding protein for immunoglobulin kappa J region |
2124 |
0.47 |
| chr4_169768965_169769116 | 0.23 |
RP11-635L1.3 |
|
14952 |
0.19 |
| chr15_65851048_65851199 | 0.23 |
ENSG00000207449 |
. |
5598 |
0.14 |
| chr4_77680501_77680652 | 0.23 |
RP11-359D14.2 |
|
949 |
0.62 |
| chr17_3705503_3705718 | 0.23 |
ITGAE |
integrin, alpha E (antigen CD103, human mucosal lymphocyte antigen 1; alpha polypeptide) |
1073 |
0.45 |
| chr9_90131509_90131738 | 0.23 |
DAPK1 |
death-associated protein kinase 1 |
17703 |
0.23 |
| chr2_36720605_36720756 | 0.23 |
CRIM1 |
cysteine rich transmembrane BMP regulator 1 (chordin-like) |
20153 |
0.19 |
| chr2_147081299_147081450 | 0.23 |
ENSG00000251905 |
. |
178650 |
0.04 |
| chr12_52585492_52585885 | 0.23 |
KRT80 |
keratin 80 |
96 |
0.95 |
| chr7_37011204_37011355 | 0.23 |
ELMO1 |
engulfment and cell motility 1 |
13386 |
0.2 |
| chr9_112955106_112955494 | 0.23 |
C9orf152 |
chromosome 9 open reading frame 152 |
15169 |
0.23 |
| chr1_161691025_161691176 | 0.23 |
FCRLB |
Fc receptor-like B |
253 |
0.86 |
| chr13_24862558_24862709 | 0.23 |
SPATA13 |
spermatogenesis associated 13 |
15806 |
0.14 |
| chr2_62987338_62987489 | 0.23 |
ENSG00000252436 |
. |
33538 |
0.18 |
| chr11_71778137_71778288 | 0.23 |
NUMA1 |
nuclear mitotic apparatus protein 1 |
2921 |
0.12 |
| chr2_145785636_145785787 | 0.23 |
ENSG00000253036 |
. |
306927 |
0.01 |
| chr7_28104329_28104480 | 0.23 |
JAZF1 |
JAZF zinc finger 1 |
6899 |
0.27 |
| chr2_841280_841431 | 0.23 |
AC113607.3 |
|
75455 |
0.1 |
| chr1_184026988_184027139 | 0.23 |
TSEN15 |
TSEN15 tRNA splicing endonuclease subunit |
6183 |
0.25 |
| chr2_145135123_145135336 | 0.23 |
GTDC1 |
glycosyltransferase-like domain containing 1 |
45146 |
0.17 |
| chr12_10194519_10194670 | 0.23 |
CLEC9A |
C-type lectin domain family 9, member A |
11318 |
0.12 |
| chr13_114871936_114872087 | 0.23 |
RASA3-IT1 |
RASA3 intronic transcript 1 (non-protein coding) |
2284 |
0.35 |
| chr15_50148578_50148980 | 0.22 |
ATP8B4 |
ATPase, class I, type 8B, member 4 |
20113 |
0.21 |
| chrX_121759874_121760025 | 0.22 |
ENSG00000221317 |
. |
154278 |
0.04 |
| chr2_52146184_52146335 | 0.22 |
ENSG00000202195 |
. |
378874 |
0.01 |
| chr13_21289490_21289641 | 0.22 |
ENSG00000265710 |
. |
11723 |
0.19 |
| chr4_122569979_122570130 | 0.22 |
ANXA5 |
annexin A5 |
48063 |
0.16 |
| chr13_77971472_77971623 | 0.22 |
MYCBP2 |
MYC binding protein 2, E3 ubiquitin protein ligase |
70368 |
0.12 |
| chr6_11590032_11590183 | 0.22 |
TMEM170B |
transmembrane protein 170B |
51596 |
0.15 |
| chr11_101316311_101316462 | 0.22 |
ENSG00000263885 |
. |
74250 |
0.12 |
| chr12_89453867_89454269 | 0.22 |
ENSG00000238302 |
. |
221994 |
0.02 |
| chr21_40856650_40856801 | 0.22 |
SH3BGR |
SH3 domain binding glutamic acid-rich protein |
15024 |
0.18 |
| chr8_57425756_57425907 | 0.22 |
RP11-17A4.2 |
|
24174 |
0.24 |
| chr15_29022183_29022334 | 0.22 |
RP11-578F21.10 |
|
11490 |
0.15 |
| chr3_151082583_151082734 | 0.22 |
P2RY12 |
purinergic receptor P2Y, G-protein coupled, 12 |
19942 |
0.17 |
| chr20_48295346_48295512 | 0.22 |
B4GALT5 |
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 5 |
34986 |
0.15 |
| chr3_159740833_159741097 | 0.22 |
LINC01100 |
long intergenic non-protein coding RNA 1100 |
7154 |
0.2 |
| chr2_238442752_238442903 | 0.22 |
MLPH |
melanophilin |
6681 |
0.17 |
| chr4_148710085_148710236 | 0.22 |
ENSG00000264274 |
. |
6414 |
0.2 |
| chr8_134591977_134592128 | 0.22 |
ST3GAL1 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
7869 |
0.3 |
| chr5_31990290_31990441 | 0.22 |
ENSG00000266243 |
. |
54100 |
0.13 |
| chr2_36731284_36731435 | 0.22 |
CRIM1 |
cysteine rich transmembrane BMP regulator 1 (chordin-like) |
9474 |
0.2 |
| chr9_138952025_138952443 | 0.22 |
NACC2 |
NACC family member 2, BEN and BTB (POZ) domain containing |
9808 |
0.22 |
| chr2_200180531_200180682 | 0.22 |
RP11-486F17.1 |
|
9427 |
0.3 |
| chr5_90603533_90603789 | 0.22 |
ENSG00000199643 |
. |
37117 |
0.18 |
| chr3_148722415_148722820 | 0.22 |
GYG1 |
glycogenin 1 |
12459 |
0.19 |
| chr11_92117975_92118126 | 0.22 |
RP11-675M1.2 |
|
23292 |
0.23 |
| chr8_16535817_16535968 | 0.22 |
MSR1 |
macrophage scavenger receptor 1 |
111021 |
0.07 |
| chr16_68100077_68100483 | 0.22 |
ENSG00000221789 |
. |
10493 |
0.1 |
| chr14_32977387_32977538 | 0.22 |
AKAP6 |
A kinase (PRKA) anchor protein 6 |
13184 |
0.26 |
| chr15_56269998_56270149 | 0.22 |
NEDD4 |
neural precursor cell expressed, developmentally down-regulated 4, E3 ubiquitin protein ligase |
15871 |
0.2 |
| chr1_10303434_10303585 | 0.22 |
KIF1B |
kinesin family member 1B |
11201 |
0.16 |
| chr10_4426416_4426567 | 0.22 |
ENSG00000207124 |
. |
130653 |
0.06 |
| chr10_31272970_31273270 | 0.22 |
ZNF438 |
zinc finger protein 438 |
235 |
0.96 |
| chr4_54870791_54871088 | 0.22 |
AC110792.1 |
HCG2027126; Uncharacterized protein |
56274 |
0.12 |
| chr15_63739639_63739912 | 0.22 |
USP3 |
ubiquitin specific peptidase 3 |
57018 |
0.12 |
| chr4_81228983_81229134 | 0.22 |
C4orf22 |
chromosome 4 open reading frame 22 |
27816 |
0.2 |
| chr2_158113855_158114082 | 0.22 |
GALNT5 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 5 (GalNAc-T5) |
142 |
0.97 |
| chr11_122129149_122129397 | 0.22 |
RP11-716H6.2 |
|
36625 |
0.18 |
| chr3_153331537_153331688 | 0.22 |
ENSG00000200162 |
. |
6446 |
0.31 |
| chr2_198094184_198094335 | 0.22 |
ANKRD44 |
ankyrin repeat domain 44 |
31497 |
0.16 |
| chr4_138463404_138463555 | 0.22 |
PCDH18 |
protocadherin 18 |
9831 |
0.33 |
| chr2_64515406_64515557 | 0.21 |
ENSG00000264297 |
. |
52412 |
0.14 |
| chr6_125541820_125541971 | 0.21 |
TPD52L1 |
tumor protein D52-like 1 |
904 |
0.71 |
| chr3_81913343_81913495 | 0.21 |
GBE1 |
glucan (1,4-alpha-), branching enzyme 1 |
102107 |
0.09 |
| chr10_17151320_17151471 | 0.21 |
CUBN |
cubilin (intrinsic factor-cobalamin receptor) |
18441 |
0.23 |
| chr5_71501439_71501590 | 0.21 |
MAP1B |
microtubule-associated protein 1B |
26059 |
0.19 |
| chr16_70057514_70057665 | 0.21 |
ENSG00000239118 |
. |
6660 |
0.16 |
| chr7_55491154_55491305 | 0.21 |
LANCL2 |
LanC lantibiotic synthetase component C-like 2 (bacterial) |
58088 |
0.14 |
| chr3_108988791_108989046 | 0.21 |
ENSG00000252889 |
. |
44942 |
0.15 |
| chr6_52430093_52430244 | 0.21 |
TRAM2 |
translocation associated membrane protein 2 |
11545 |
0.25 |
| chr1_78841651_78841802 | 0.21 |
ENSG00000212308 |
. |
773 |
0.75 |
| chr3_145791643_145791794 | 0.21 |
RP11-274H2.3 |
|
4746 |
0.16 |
| chr13_73379676_73380042 | 0.21 |
DIS3 |
DIS3 mitotic control homolog (S. cerevisiae) |
23625 |
0.15 |
| chr6_64389523_64389709 | 0.21 |
PHF3 |
PHD finger protein 3 |
30 |
0.99 |
| chr4_178223159_178223383 | 0.21 |
NEIL3 |
nei endonuclease VIII-like 3 (E. coli) |
7719 |
0.23 |
| chr6_13437908_13438059 | 0.21 |
GFOD1 |
glucose-fructose oxidoreductase domain containing 1 |
29614 |
0.15 |
| chr3_112329096_112329326 | 0.21 |
CCDC80 |
coiled-coil domain containing 80 |
84 |
0.98 |
| chr15_38437640_38437791 | 0.21 |
SPRED1 |
sprouty-related, EVH1 domain containing 1 |
106812 |
0.08 |
| chr2_169772940_169773098 | 0.21 |
SPC25 |
SPC25, NDC80 kinetochore complex component |
3232 |
0.28 |
| chr18_21470429_21470580 | 0.21 |
ENSG00000221389 |
. |
3666 |
0.25 |
| chr17_68163620_68163771 | 0.21 |
KCNJ2 |
potassium inwardly-rectifying channel, subfamily J, member 2 |
1119 |
0.45 |
| chr3_183173438_183173589 | 0.21 |
ENSG00000251730 |
. |
1597 |
0.22 |
| chr10_100288310_100288461 | 0.21 |
HPS1 |
Hermansky-Pudlak syndrome 1 |
81702 |
0.09 |
| chr2_9292545_9292696 | 0.21 |
ASAP2 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2 |
54274 |
0.16 |
| chr4_16771566_16771717 | 0.21 |
LDB2 |
LIM domain binding 2 |
95705 |
0.09 |
| chr21_35908494_35908645 | 0.21 |
RCAN1 |
regulator of calcineurin 1 |
9308 |
0.2 |
| chr5_35771841_35771992 | 0.21 |
SPEF2 |
sperm flagellar 2 |
7354 |
0.25 |
| chr7_81405925_81406300 | 0.21 |
HGF |
hepatocyte growth factor (hepapoietin A; scatter factor) |
6358 |
0.33 |
| chr8_90731390_90731592 | 0.21 |
RIPK2 |
receptor-interacting serine-threonine kinase 2 |
38484 |
0.22 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:1900115 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.1 | 0.3 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 0.2 | GO:0007132 | meiotic metaphase I(GO:0007132) |
| 0.1 | 0.2 | GO:0043578 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.1 | 0.2 | GO:0071436 | sodium ion export(GO:0071436) |
| 0.1 | 0.1 | GO:0021825 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) substrate-dependent cerebral cortex tangential migration(GO:0021825) postnatal olfactory bulb interneuron migration(GO:0021827) |
| 0.1 | 0.3 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.1 | 0.2 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.1 | 0.2 | GO:0002328 | pro-B cell differentiation(GO:0002328) |
| 0.1 | 0.2 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.1 | 0.2 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.1 | 0.3 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.1 | 0.2 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
| 0.1 | 0.3 | GO:0070498 | interleukin-1-mediated signaling pathway(GO:0070498) |
| 0.1 | 0.3 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.1 | 0.2 | GO:0033603 | positive regulation of dopamine secretion(GO:0033603) |
| 0.0 | 0.1 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.1 | GO:0045627 | positive regulation of T-helper 1 cell differentiation(GO:0045627) |
| 0.0 | 0.1 | GO:0070828 | heterochromatin assembly(GO:0031507) heterochromatin organization(GO:0070828) |
| 0.0 | 0.2 | GO:0061001 | dendritic spine morphogenesis(GO:0060997) regulation of dendritic spine morphogenesis(GO:0061001) dendritic spine organization(GO:0097061) |
| 0.0 | 0.2 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.0 | 0.2 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.0 | 1.1 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.3 | GO:0006896 | Golgi to endosome transport(GO:0006895) Golgi to vacuole transport(GO:0006896) |
| 0.0 | 0.2 | GO:0031641 | regulation of myelination(GO:0031641) |
| 0.0 | 0.2 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 | 0.1 | GO:0003078 | obsolete regulation of natriuresis(GO:0003078) |
| 0.0 | 0.1 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.1 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.1 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.0 | 0.2 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 0.1 | GO:0022030 | cerebral cortex radial glia guided migration(GO:0021801) telencephalon glial cell migration(GO:0022030) |
| 0.0 | 0.1 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.1 | GO:0002023 | reduction of food intake in response to dietary excess(GO:0002023) |
| 0.0 | 0.2 | GO:0033137 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) |
| 0.0 | 0.1 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.1 | GO:0034616 | response to laminar fluid shear stress(GO:0034616) |
| 0.0 | 0.0 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.0 | 0.8 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.1 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) negative regulation of neural precursor cell proliferation(GO:2000178) |
| 0.0 | 0.1 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.0 | 0.1 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.1 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.0 | 0.1 | GO:0046476 | glycosylceramide biosynthetic process(GO:0046476) |
| 0.0 | 0.1 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 0.1 | GO:0002870 | lymphocyte anergy(GO:0002249) regulation of T cell anergy(GO:0002667) positive regulation of T cell anergy(GO:0002669) T cell anergy(GO:0002870) regulation of lymphocyte anergy(GO:0002911) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.0 | 0.1 | GO:2001279 | regulation of prostaglandin biosynthetic process(GO:0031392) positive regulation of prostaglandin biosynthetic process(GO:0031394) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.0 | 0.1 | GO:0045989 | positive regulation of striated muscle contraction(GO:0045989) |
| 0.0 | 0.1 | GO:0060437 | lung growth(GO:0060437) |
| 0.0 | 0.1 | GO:0032506 | cytokinetic process(GO:0032506) |
| 0.0 | 0.0 | GO:0002679 | respiratory burst involved in defense response(GO:0002679) |
| 0.0 | 0.1 | GO:0003323 | type B pancreatic cell development(GO:0003323) |
| 0.0 | 0.1 | GO:0090030 | regulation of steroid hormone biosynthetic process(GO:0090030) |
| 0.0 | 0.1 | GO:0009414 | response to water deprivation(GO:0009414) |
| 0.0 | 0.1 | GO:0046607 | positive regulation of centrosome cycle(GO:0046607) |
| 0.0 | 0.0 | GO:0003283 | atrial septum development(GO:0003283) atrial septum morphogenesis(GO:0060413) |
| 0.0 | 0.1 | GO:2000095 | regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.0 | 0.1 | GO:0060760 | positive regulation of response to cytokine stimulus(GO:0060760) |
| 0.0 | 0.1 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.0 | 0.1 | GO:0046882 | negative regulation of follicle-stimulating hormone secretion(GO:0046882) |
| 0.0 | 0.1 | GO:0001779 | natural killer cell differentiation(GO:0001779) |
| 0.0 | 0.1 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.0 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.0 | 0.1 | GO:0001547 | antral ovarian follicle growth(GO:0001547) |
| 0.0 | 0.1 | GO:0070874 | negative regulation of glycogen metabolic process(GO:0070874) |
| 0.0 | 0.1 | GO:0032431 | activation of phospholipase A2 activity(GO:0032431) |
| 0.0 | 0.1 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.1 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.0 | 0.1 | GO:0009169 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.0 | 0.1 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) |
| 0.0 | 0.1 | GO:0060129 | thyroid-stimulating hormone-secreting cell differentiation(GO:0060129) |
| 0.0 | 0.1 | GO:0045091 | regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045091) |
| 0.0 | 0.1 | GO:0000492 | small nucleolar ribonucleoprotein complex assembly(GO:0000491) box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.0 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.0 | 0.1 | GO:0016115 | diterpenoid catabolic process(GO:0016103) terpenoid catabolic process(GO:0016115) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.1 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 0.0 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.0 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.0 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.0 | 0.1 | GO:0060754 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 0.1 | GO:0032933 | SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.1 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.0 | 0.1 | GO:0060398 | regulation of growth hormone receptor signaling pathway(GO:0060398) |
| 0.0 | 0.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.3 | GO:0021846 | cell proliferation in forebrain(GO:0021846) |
| 0.0 | 0.1 | GO:0021800 | cerebral cortex tangential migration(GO:0021800) |
| 0.0 | 0.1 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.0 | GO:0010766 | negative regulation of sodium ion transport(GO:0010766) |
| 0.0 | 0.0 | GO:0002885 | positive regulation of hypersensitivity(GO:0002885) |
| 0.0 | 0.1 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.1 | GO:0042504 | tyrosine phosphorylation of Stat4 protein(GO:0042504) regulation of tyrosine phosphorylation of Stat4 protein(GO:0042519) positive regulation of tyrosine phosphorylation of Stat4 protein(GO:0042520) |
| 0.0 | 0.0 | GO:0044321 | cellular response to leptin stimulus(GO:0044320) response to leptin(GO:0044321) |
| 0.0 | 0.0 | GO:0009125 | nucleoside monophosphate catabolic process(GO:0009125) |
| 0.0 | 0.1 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.3 | GO:0006195 | purine nucleotide catabolic process(GO:0006195) |
| 0.0 | 0.2 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.1 | GO:0071347 | cellular response to interleukin-1(GO:0071347) |
| 0.0 | 0.1 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 | 0.1 | GO:0021508 | floor plate formation(GO:0021508) |
| 0.0 | 0.0 | GO:0002540 | arachidonic acid metabolite production involved in inflammatory response(GO:0002538) leukotriene production involved in inflammatory response(GO:0002540) |
| 0.0 | 0.1 | GO:0070528 | protein kinase C signaling(GO:0070528) |
| 0.0 | 0.0 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) |
| 0.0 | 0.0 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.0 | 0.0 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.0 | 0.0 | GO:0015864 | pyrimidine nucleoside transport(GO:0015864) |
| 0.0 | 0.0 | GO:0048631 | regulation of skeletal muscle tissue growth(GO:0048631) |
| 0.0 | 0.1 | GO:0031033 | myosin filament organization(GO:0031033) |
| 0.0 | 0.1 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.0 | 0.2 | GO:0010458 | exit from mitosis(GO:0010458) |
| 0.0 | 0.1 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.0 | GO:0042339 | keratan sulfate metabolic process(GO:0042339) |
| 0.0 | 0.0 | GO:0035385 | Roundabout signaling pathway(GO:0035385) |
| 0.0 | 0.1 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.0 | 0.1 | GO:0032354 | response to follicle-stimulating hormone(GO:0032354) |
| 0.0 | 0.0 | GO:0010837 | regulation of keratinocyte proliferation(GO:0010837) |
| 0.0 | 0.0 | GO:0060638 | mesenchymal-epithelial cell signaling(GO:0060638) |
| 0.0 | 0.1 | GO:0010886 | positive regulation of cholesterol storage(GO:0010886) |
| 0.0 | 0.0 | GO:0051459 | corticotropin secretion(GO:0051458) regulation of corticotropin secretion(GO:0051459) positive regulation of corticotropin secretion(GO:0051461) |
| 0.0 | 0.0 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.0 | 0.0 | GO:0032817 | regulation of natural killer cell proliferation(GO:0032817) |
| 0.0 | 0.1 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.0 | GO:0002866 | positive regulation of acute inflammatory response to antigenic stimulus(GO:0002866) |
| 0.0 | 0.0 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.0 | 0.0 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) |
| 0.0 | 0.1 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.0 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.0 | 0.0 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.1 | GO:0032776 | DNA methylation on cytosine(GO:0032776) C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 0.0 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.0 | 0.0 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.0 | 0.1 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.0 | GO:0032236 | obsolete positive regulation of calcium ion transport via store-operated calcium channel activity(GO:0032236) |
| 0.0 | 0.0 | GO:0009261 | ribonucleotide catabolic process(GO:0009261) |
| 0.0 | 0.0 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.0 | 0.0 | GO:0060014 | granulosa cell differentiation(GO:0060014) |
| 0.0 | 0.0 | GO:0045915 | positive regulation of catecholamine metabolic process(GO:0045915) positive regulation of dopamine metabolic process(GO:0045964) |
| 0.0 | 0.1 | GO:0043268 | positive regulation of potassium ion transport(GO:0043268) |
| 0.0 | 0.0 | GO:0030540 | female genitalia development(GO:0030540) |
| 0.0 | 0.0 | GO:0014834 | skeletal muscle satellite cell maintenance involved in skeletal muscle regeneration(GO:0014834) |
| 0.0 | 0.1 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.0 | 0.1 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.0 | 0.0 | GO:0043312 | neutrophil activation involved in immune response(GO:0002283) neutrophil degranulation(GO:0043312) |
| 0.0 | 0.0 | GO:0021591 | ventricular system development(GO:0021591) |
| 0.0 | 0.0 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 0.0 | GO:2000189 | regulation of cholesterol homeostasis(GO:2000188) positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.0 | GO:0072526 | pyridine-containing compound catabolic process(GO:0072526) |
| 0.0 | 0.0 | GO:0034371 | chylomicron remodeling(GO:0034371) |
| 0.0 | 0.1 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.0 | 0.0 | GO:0071731 | response to nitric oxide(GO:0071731) cellular response to nitric oxide(GO:0071732) cellular response to reactive nitrogen species(GO:1902170) |
| 0.0 | 0.1 | GO:0042693 | muscle cell fate commitment(GO:0042693) |
| 0.0 | 0.0 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.0 | 0.0 | GO:0000019 | regulation of mitotic recombination(GO:0000019) |
| 0.0 | 0.1 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.0 | 0.2 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.0 | 0.0 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.0 | 0.1 | GO:0090231 | regulation of spindle checkpoint(GO:0090231) |
| 0.0 | 0.2 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.1 | GO:0019673 | GDP-mannose metabolic process(GO:0019673) |
| 0.0 | 0.1 | GO:0009629 | response to gravity(GO:0009629) |
| 0.0 | 0.0 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 | 0.1 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.1 | GO:0043537 | negative regulation of blood vessel endothelial cell migration(GO:0043537) |
| 0.0 | 0.0 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) platelet activating factor metabolic process(GO:0046469) |
| 0.0 | 0.0 | GO:0006824 | cobalt ion transport(GO:0006824) |
| 0.0 | 0.1 | GO:0044872 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.0 | 0.1 | GO:0009435 | NAD biosynthetic process(GO:0009435) |
| 0.0 | 0.1 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.0 | GO:0071034 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.0 | 0.0 | GO:1901797 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) negative regulation of signal transduction by p53 class mediator(GO:1901797) |
| 0.0 | 0.0 | GO:0097576 | vacuole fusion(GO:0097576) |
| 0.0 | 0.0 | GO:0031297 | replication fork processing(GO:0031297) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.1 | 0.2 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.1 | 0.2 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 0.3 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 0.4 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.2 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.1 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.1 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.2 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.2 | GO:0098642 | network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.0 | 0.1 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.2 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.2 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.0 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.1 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.0 | 0.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.2 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.1 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.2 | GO:0001527 | microfibril(GO:0001527) |
| 0.0 | 0.1 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.3 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.2 | GO:0098644 | complex of collagen trimers(GO:0098644) |
| 0.0 | 0.1 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.2 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.2 | GO:0043256 | laminin complex(GO:0043256) |
| 0.0 | 0.1 | GO:0060199 | clathrin-sculpted glutamate transport vesicle(GO:0060199) clathrin-sculpted glutamate transport vesicle membrane(GO:0060203) |
| 0.0 | 0.1 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.1 | GO:0045277 | mitochondrial respiratory chain complex IV(GO:0005751) respiratory chain complex IV(GO:0045277) |
| 0.0 | 0.1 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.1 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.0 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.1 | GO:0044291 | intercalated disc(GO:0014704) cell-cell contact zone(GO:0044291) |
| 0.0 | 0.1 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.0 | 0.1 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.0 | GO:0001652 | granular component(GO:0001652) |
| 0.0 | 0.1 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.0 | 0.1 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.2 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.0 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.0 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.1 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 0.0 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.0 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.1 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.0 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.1 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 1.0 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.0 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.1 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.1 | 0.3 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.1 | 0.4 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.1 | 0.2 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.1 | 0.2 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.1 | 0.1 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.4 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.0 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.1 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.0 | 0.3 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.3 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.3 | GO:0005536 | glucose binding(GO:0005536) |
| 0.0 | 0.1 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.0 | 0.3 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.1 | GO:0000987 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.0 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.1 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.1 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.1 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.0 | 0.4 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.1 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.1 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.4 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.3 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.1 | GO:0030354 | melanin-concentrating hormone activity(GO:0030354) |
| 0.0 | 0.1 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.2 | GO:0022821 | potassium ion antiporter activity(GO:0022821) |
| 0.0 | 0.1 | GO:0005350 | pyrimidine nucleobase transmembrane transporter activity(GO:0005350) |
| 0.0 | 0.1 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.2 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.1 | GO:0004516 | nicotinate phosphoribosyltransferase activity(GO:0004516) |
| 0.0 | 0.1 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.0 | 0.1 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.0 | 0.3 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.1 | GO:0071553 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.0 | 0.1 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.1 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.1 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.1 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.1 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.1 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.2 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.1 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.0 | 0.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.1 | GO:0042808 | obsolete neuronal Cdc2-like kinase binding(GO:0042808) |
| 0.0 | 0.0 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.0 | 0.5 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.5 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.1 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.2 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.0 | 0.3 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 0.2 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.1 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.0 | 0.1 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.1 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.2 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.2 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
| 0.0 | 0.1 | GO:0004525 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.0 | 0.3 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.2 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.0 | 0.1 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.1 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.1 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.0 | 0.0 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.0 | 0.1 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.0 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 0.1 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.0 | 0.1 | GO:0045569 | TRAIL binding(GO:0045569) |
| 0.0 | 0.1 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 0.0 | 0.1 | GO:0016520 | growth hormone-releasing hormone receptor activity(GO:0016520) |
| 0.0 | 0.0 | GO:0050543 | icosatetraenoic acid binding(GO:0050543) |
| 0.0 | 0.3 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0099602 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.0 | 0.1 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.1 | GO:0032450 | maltose alpha-glucosidase activity(GO:0032450) |
| 0.0 | 0.1 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.1 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.0 | GO:0004340 | glucokinase activity(GO:0004340) |
| 0.0 | 0.1 | GO:0004738 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.1 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.1 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.0 | 0.0 | GO:0004075 | biotin carboxylase activity(GO:0004075) |
| 0.0 | 0.2 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.2 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.1 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.0 | 0.2 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.5 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.0 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.1 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.1 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.0 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.1 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.1 | GO:0070696 | transmembrane receptor protein serine/threonine kinase binding(GO:0070696) |
| 0.0 | 0.0 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.1 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.0 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) |
| 0.0 | 0.1 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 0.0 | 0.1 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.0 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.0 | 0.1 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.0 | 0.0 | GO:0052834 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.1 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.0 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.1 | GO:0008518 | reduced folate carrier activity(GO:0008518) |
| 0.0 | 0.0 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.1 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.1 | GO:0055106 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.0 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.1 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.0 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.0 | 0.1 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.1 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.2 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.0 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.1 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.0 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.1 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.2 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.0 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.0 | 0.1 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.0 | 0.0 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.0 | 0.2 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.0 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.0 | 0.1 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.0 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.1 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.0 | 0.2 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.0 | GO:0019958 | C-X-C chemokine binding(GO:0019958) interleukin-8 binding(GO:0019959) |
| 0.0 | 0.2 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.0 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.0 | 0.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.1 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.0 | 0.0 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.0 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.0 | 0.1 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.1 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.0 | 0.0 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.0 | 0.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.0 | GO:0004946 | bombesin receptor activity(GO:0004946) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 0.1 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 1.3 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.0 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.0 | 0.0 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.0 | 0.5 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.1 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 0.3 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.0 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.0 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 0.5 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.2 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 2.0 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.2 | PID ARF 3PATHWAY | Arf1 pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | REACTOME DAG AND IP3 SIGNALING | Genes involved in DAG and IP3 signaling |
| 0.0 | 0.3 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.0 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.0 | 0.2 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.3 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.6 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 1.1 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.3 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.0 | 0.2 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.0 | 0.3 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.2 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.1 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 0.3 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.3 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.1 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.0 | 0.2 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.1 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.0 | REACTOME P75NTR SIGNALS VIA NFKB | Genes involved in p75NTR signals via NF-kB |
| 0.0 | 0.1 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.1 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.1 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.3 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |