Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
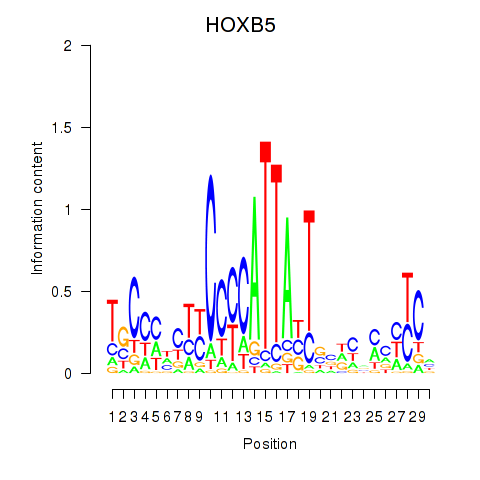
Results for HOXB5
Z-value: 1.38
Transcription factors associated with HOXB5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXB5
|
ENSG00000120075.5 | homeobox B5 |
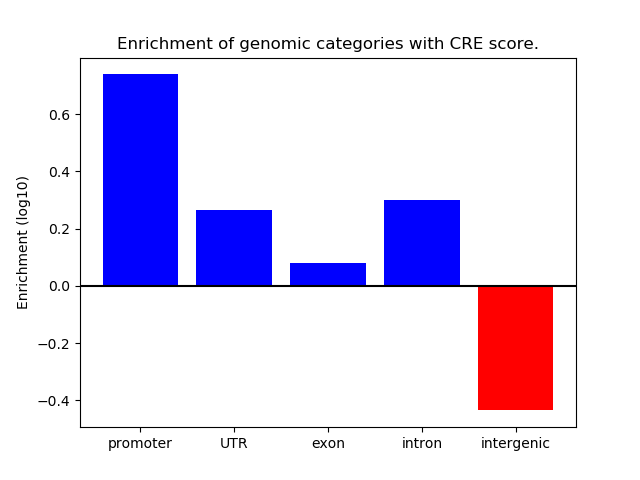
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr17_46670430_46670597 | HOXB5 | 810 | 0.263584 | 0.35 | 3.5e-01 | Click! |
| chr17_46670674_46671049 | HOXB5 | 462 | 0.525865 | -0.21 | 5.9e-01 | Click! |
| chr17_46671392_46671683 | HOXB5 | 214 | 0.805106 | 0.12 | 7.6e-01 | Click! |
Activity of the HOXB5 motif across conditions
Conditions sorted by the z-value of the HOXB5 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
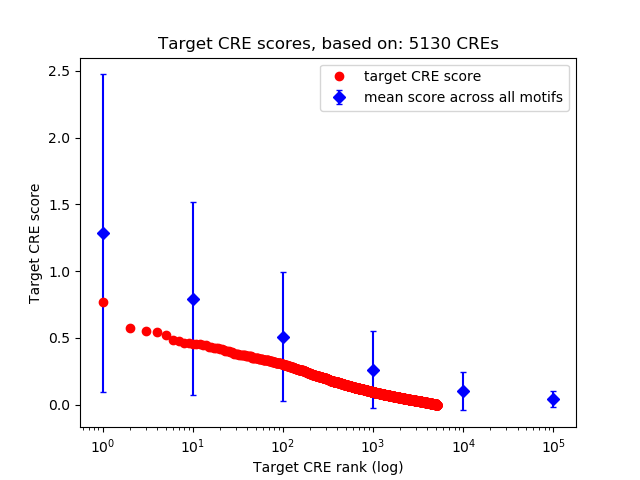
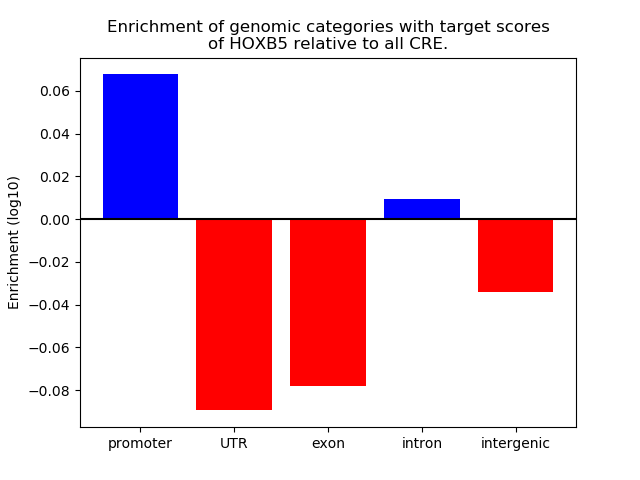
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr3_13457216_13457610 | 0.77 |
NUP210 |
nucleoporin 210kDa |
4396 |
0.28 |
| chr16_50720309_50720716 | 0.57 |
SNX20 |
sorting nexin 20 |
5248 |
0.13 |
| chr2_143883953_143884119 | 0.55 |
ARHGAP15 |
Rho GTPase activating protein 15 |
2847 |
0.35 |
| chr11_85903802_85903953 | 0.55 |
ENSG00000200877 |
. |
39608 |
0.15 |
| chr2_149304182_149304365 | 0.52 |
MBD5 |
methyl-CpG binding domain protein 5 |
77979 |
0.11 |
| chr1_160544322_160544473 | 0.49 |
CD84 |
CD84 molecule |
4866 |
0.16 |
| chr14_22957256_22957407 | 0.48 |
TRAJ51 |
T cell receptor alpha joining 51 (pseudogene) |
1160 |
0.31 |
| chr16_53544923_53545074 | 0.46 |
AKTIP |
AKT interacting protein |
6675 |
0.23 |
| chr6_108278816_108278967 | 0.46 |
RP1-191J18.66 |
|
274 |
0.56 |
| chr5_150623515_150623785 | 0.46 |
GM2A |
GM2 ganglioside activator |
8803 |
0.19 |
| chr18_56324663_56325184 | 0.46 |
MALT1 |
mucosa associated lymphoid tissue lymphoma translocation gene 1 |
13695 |
0.14 |
| chr2_103034112_103034476 | 0.46 |
IL18RAP |
interleukin 18 receptor accessory protein |
855 |
0.53 |
| chr5_175456482_175456633 | 0.45 |
THOC3 |
THO complex 3 |
5126 |
0.16 |
| chr9_20997618_20997943 | 0.44 |
PTPLAD2 |
protein tyrosine phosphatase-like A domain containing 2 |
33828 |
0.16 |
| chr14_52374460_52374611 | 0.44 |
GNG2 |
guanine nucleotide binding protein (G protein), gamma 2 |
5295 |
0.19 |
| chr22_37620188_37620478 | 0.43 |
SSTR3 |
somatostatin receptor 3 |
11971 |
0.13 |
| chr7_151046492_151046873 | 0.43 |
NUB1 |
negative regulator of ubiquitin-like proteins 1 |
4251 |
0.18 |
| chr7_130784590_130784741 | 0.43 |
LINC-PINT |
long intergenic non-protein coding RNA, p53 induced transcript |
7124 |
0.22 |
| chr1_32127594_32127745 | 0.42 |
COL16A1 |
collagen, type XVI, alpha 1 |
4013 |
0.15 |
| chr17_7238561_7238964 | 0.42 |
ACAP1 |
ArfGAP with coiled-coil, ankyrin repeat and PH domains 1 |
1086 |
0.22 |
| chr17_33612463_33612749 | 0.42 |
SLFN5 |
schlafen family member 5 |
42498 |
0.11 |
| chr17_66346042_66346193 | 0.41 |
ARSG |
arylsulfatase G |
58458 |
0.1 |
| chr10_127515808_127515959 | 0.41 |
BCCIP |
BRCA2 and CDKN1A interacting protein |
3768 |
0.14 |
| chr17_4404014_4404514 | 0.40 |
AC118754.4 |
|
1295 |
0.31 |
| chr7_8056755_8056906 | 0.40 |
AC006042.7 |
|
47296 |
0.13 |
| chr6_167534795_167535192 | 0.40 |
CCR6 |
chemokine (C-C motif) receptor 6 |
1264 |
0.46 |
| chr16_28212646_28212858 | 0.40 |
XPO6 |
exportin 6 |
10045 |
0.17 |
| chr20_48843581_48843732 | 0.39 |
CEBPB |
CCAAT/enhancer binding protein (C/EBP), beta |
36280 |
0.16 |
| chr7_12770799_12771102 | 0.38 |
ENSG00000199470 |
. |
30436 |
0.2 |
| chr9_132629831_132629982 | 0.38 |
USP20 |
ubiquitin specific peptidase 20 |
32149 |
0.12 |
| chr13_33296824_33296975 | 0.38 |
PDS5B |
PDS5, regulator of cohesion maintenance, homolog B (S. cerevisiae) |
35408 |
0.19 |
| chr6_110500985_110501198 | 0.38 |
WASF1 |
WAS protein family, member 1 |
35 |
0.77 |
| chr6_106095597_106095936 | 0.38 |
PREP |
prolyl endopeptidase |
244807 |
0.02 |
| chr12_83108332_83108483 | 0.37 |
TMTC2 |
transmembrane and tetratricopeptide repeat containing 2 |
27627 |
0.27 |
| chr5_95161582_95161941 | 0.37 |
GLRX |
glutaredoxin (thioltransferase) |
3052 |
0.2 |
| chrX_118781177_118781328 | 0.37 |
ENSG00000211578 |
. |
441 |
0.81 |
| chr22_23486285_23486436 | 0.37 |
RTDR1 |
rhabdoid tumor deletion region gene 1 |
848 |
0.39 |
| chr2_43355583_43355909 | 0.37 |
ENSG00000207087 |
. |
37114 |
0.2 |
| chr7_102069746_102070456 | 0.37 |
ORAI2 |
ORAI calcium release-activated calcium modulator 2 |
3452 |
0.11 |
| chr5_75709889_75710222 | 0.37 |
IQGAP2 |
IQ motif containing GTPase activating protein 2 |
9806 |
0.28 |
| chr1_157963439_157963975 | 0.37 |
KIRREL |
kin of IRRE like (Drosophila) |
272 |
0.93 |
| chr2_153031608_153031759 | 0.36 |
STAM2 |
signal transducing adaptor molecule (SH3 domain and ITAM motif) 2 |
823 |
0.72 |
| chr14_24021277_24021443 | 0.36 |
ZFHX2 |
zinc finger homeobox 2 |
502 |
0.64 |
| chr18_56382154_56382522 | 0.36 |
RP11-126O1.4 |
|
37492 |
0.12 |
| chr2_99068894_99069462 | 0.36 |
INPP4A |
inositol polyphosphate-4-phosphatase, type I, 107kDa |
7765 |
0.25 |
| chr15_81587451_81588017 | 0.35 |
IL16 |
interleukin 16 |
1520 |
0.42 |
| chr13_100012790_100013013 | 0.35 |
ENSG00000207719 |
. |
4516 |
0.25 |
| chr12_6892610_6892761 | 0.35 |
ENSG00000244532 |
. |
642 |
0.46 |
| chrX_64945443_64945693 | 0.35 |
MSN |
moesin |
58031 |
0.16 |
| chr6_143861183_143861457 | 0.35 |
PHACTR2 |
phosphatase and actin regulator 2 |
3338 |
0.21 |
| chr8_129694153_129694304 | 0.35 |
ENSG00000221351 |
. |
137812 |
0.05 |
| chrX_101857495_101857646 | 0.35 |
ARMCX5 |
armadillo repeat containing, X-linked 5 |
1505 |
0.37 |
| chrX_70324332_70324541 | 0.35 |
CXorf65 |
chromosome X open reading frame 65 |
2019 |
0.18 |
| chr17_72752966_72753168 | 0.35 |
SLC9A3R1 |
solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3 regulator 1 |
5075 |
0.11 |
| chr15_91081084_91081235 | 0.34 |
CRTC3 |
CREB regulated transcription coactivator 3 |
7859 |
0.21 |
| chr1_160537117_160537336 | 0.34 |
CD84 |
CD84 molecule |
12037 |
0.14 |
| chr1_202597717_202597987 | 0.34 |
SYT2 |
synaptotagmin II |
14729 |
0.19 |
| chr14_93105749_93105900 | 0.34 |
RIN3 |
Ras and Rab interactor 3 |
13022 |
0.27 |
| chr11_44630544_44630700 | 0.34 |
RP11-58K22.4 |
|
971 |
0.55 |
| chr3_47025143_47025429 | 0.34 |
CCDC12 |
coiled-coil domain containing 12 |
1814 |
0.32 |
| chr13_52393868_52394096 | 0.34 |
RP11-327P2.5 |
|
15549 |
0.18 |
| chr6_52262511_52262820 | 0.34 |
EFHC1 |
EF-hand domain (C-terminal) containing 1 |
22441 |
0.2 |
| chr6_53199538_53199739 | 0.34 |
ELOVL5 |
ELOVL fatty acid elongase 5 |
13949 |
0.2 |
| chr5_39189552_39189909 | 0.34 |
FYB |
FYN binding protein |
13399 |
0.27 |
| chr6_45563562_45563713 | 0.33 |
ENSG00000252738 |
. |
50204 |
0.18 |
| chr5_1314270_1314421 | 0.33 |
ENSG00000263670 |
. |
4853 |
0.19 |
| chr1_158255508_158255815 | 0.33 |
CD1C |
CD1c molecule |
3915 |
0.22 |
| chr1_226863115_226863281 | 0.33 |
ITPKB-IT1 |
ITPKB intronic transcript 1 (non-protein coding) |
430 |
0.85 |
| chr5_176172799_176173168 | 0.33 |
UNC5A |
unc-5 homolog A (C. elegans) |
64495 |
0.09 |
| chr16_88759405_88759694 | 0.33 |
RP5-1142A6.5 |
|
3360 |
0.09 |
| chr19_41813754_41814061 | 0.33 |
CCDC97 |
coiled-coil domain containing 97 |
2187 |
0.19 |
| chr2_71562064_71562215 | 0.33 |
ZNF638 |
zinc finger protein 638 |
3227 |
0.29 |
| chr14_61977064_61977273 | 0.33 |
RP11-47I22.4 |
|
18678 |
0.19 |
| chrX_135702154_135702305 | 0.33 |
ENSG00000233093 |
. |
19473 |
0.14 |
| chr4_109031937_109032201 | 0.32 |
LEF1 |
lymphoid enhancer-binding factor 1 |
55388 |
0.13 |
| chr3_53063759_53063910 | 0.32 |
SFMBT1 |
Scm-like with four mbt domains 1 |
15447 |
0.17 |
| chr1_36678399_36678593 | 0.32 |
THRAP3 |
thyroid hormone receptor associated protein 3 |
11521 |
0.15 |
| chr15_63783402_63783553 | 0.32 |
USP3 |
ubiquitin specific peptidase 3 |
13316 |
0.23 |
| chr5_66483406_66483586 | 0.32 |
CD180 |
CD180 molecule |
9131 |
0.28 |
| chr10_115967390_115967541 | 0.32 |
TDRD1 |
tudor domain containing 1 |
28116 |
0.14 |
| chr1_184944092_184944619 | 0.32 |
FAM129A |
family with sequence similarity 129, member A |
673 |
0.75 |
| chr1_89486291_89486615 | 0.32 |
ENSG00000202385 |
. |
405 |
0.78 |
| chrX_123353114_123353327 | 0.31 |
ENSG00000252693 |
. |
21627 |
0.25 |
| chr17_15905285_15905436 | 0.31 |
ZSWIM7 |
zinc finger, SWIM-type containing 7 |
2354 |
0.22 |
| chr13_99916204_99916355 | 0.31 |
GPR18 |
G protein-coupled receptor 18 |
2281 |
0.31 |
| chr16_30379858_30380009 | 0.31 |
TBC1D10B |
TBC1 domain family, member 10B |
1652 |
0.17 |
| chr6_112311050_112311201 | 0.31 |
WISP3 |
WNT1 inducible signaling pathway protein 3 |
64150 |
0.11 |
| chr15_45024051_45024202 | 0.31 |
TRIM69 |
tripartite motif containing 69 |
2940 |
0.21 |
| chr11_128382093_128382244 | 0.31 |
ETS1 |
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
6879 |
0.25 |
| chr12_58027267_58027418 | 0.31 |
B4GALNT1 |
beta-1,4-N-acetyl-galactosaminyl transferase 1 |
204 |
0.86 |
| chr3_45430424_45430606 | 0.31 |
LARS2 |
leucyl-tRNA synthetase 2, mitochondrial |
404 |
0.9 |
| chr1_38316786_38316995 | 0.31 |
MTF1 |
metal-regulatory transcription factor 1 |
8402 |
0.1 |
| chr6_155070964_155071137 | 0.31 |
SCAF8 |
SR-related CTD-associated factor 8 |
16500 |
0.23 |
| chr1_158984706_158984928 | 0.31 |
IFI16 |
interferon, gamma-inducible protein 16 |
346 |
0.89 |
| chr1_28453002_28453170 | 0.31 |
ENSG00000253005 |
. |
16266 |
0.12 |
| chr3_15597201_15597352 | 0.31 |
COLQ |
collagen-like tail subunit (single strand of homotrimer) of asymmetric acetylcholinesterase |
34018 |
0.12 |
| chr11_123552269_123552420 | 0.31 |
SCN3B |
sodium channel, voltage-gated, type III, beta subunit |
26392 |
0.14 |
| chr17_8858889_8859261 | 0.31 |
PIK3R5 |
phosphoinositide-3-kinase, regulatory subunit 5 |
9949 |
0.24 |
| chr6_43299390_43299611 | 0.30 |
ZNF318 |
zinc finger protein 318 |
11074 |
0.14 |
| chr17_72984009_72984428 | 0.30 |
CDR2L |
cerebellar degeneration-related protein 2-like |
491 |
0.63 |
| chr5_66809962_66810113 | 0.30 |
ENSG00000222939 |
. |
49601 |
0.2 |
| chr3_10280429_10280763 | 0.30 |
TATDN2 |
TatD DNase domain containing 2 |
9111 |
0.13 |
| chr7_142378423_142378715 | 0.30 |
MTRNR2L6 |
MT-RNR2-like 6 |
4465 |
0.28 |
| chr5_142737307_142737458 | 0.30 |
NR3C1 |
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
43035 |
0.2 |
| chr21_34606770_34607111 | 0.30 |
IFNAR2 |
interferon (alpha, beta and omega) receptor 2 |
4226 |
0.17 |
| chr6_118002568_118002719 | 0.30 |
NUS1 |
nuclear undecaprenyl pyrophosphate synthase 1 homolog (S. cerevisiae) |
5978 |
0.3 |
| chr10_13148496_13148807 | 0.30 |
OPTN |
optineurin |
6426 |
0.2 |
| chr17_62010060_62010211 | 0.30 |
CD79B |
CD79b molecule, immunoglobulin-associated beta |
421 |
0.74 |
| chr8_146018396_146018558 | 0.30 |
RPL8 |
ribosomal protein L8 |
505 |
0.65 |
| chr22_17570345_17570505 | 0.30 |
IL17RA |
interleukin 17 receptor A |
4576 |
0.19 |
| chr11_117884308_117884597 | 0.29 |
IL10RA |
interleukin 10 receptor, alpha |
27343 |
0.14 |
| chr6_34152409_34152560 | 0.29 |
GRM4 |
glutamate receptor, metabotropic 4 |
29085 |
0.17 |
| chrX_118789581_118789748 | 0.29 |
ENSG00000211578 |
. |
8853 |
0.17 |
| chr3_130613754_130614033 | 0.29 |
ATP2C1 |
ATPase, Ca++ transporting, type 2C, member 1 |
408 |
0.84 |
| chr8_2077178_2077450 | 0.29 |
MYOM2 |
myomesin 2 |
84130 |
0.11 |
| chr13_99931416_99931591 | 0.29 |
GPR18 |
G protein-coupled receptor 18 |
17505 |
0.18 |
| chr1_9477618_9477769 | 0.29 |
ENSG00000252956 |
. |
20144 |
0.21 |
| chr1_158218506_158218726 | 0.29 |
CD1A |
CD1a molecule |
5311 |
0.22 |
| chr18_43268903_43269487 | 0.29 |
SLC14A2 |
solute carrier family 14 (urea transporter), member 2 |
23088 |
0.16 |
| chr15_86249746_86250319 | 0.29 |
RP11-815J21.1 |
|
5692 |
0.13 |
| chr9_140195094_140195255 | 0.29 |
NRARP |
NOTCH-regulated ankyrin repeat protein |
1529 |
0.21 |
| chr1_1712444_1712922 | 0.29 |
NADK |
NAD kinase |
1175 |
0.4 |
| chr13_74591900_74592051 | 0.28 |
KLF12 |
Kruppel-like factor 12 |
22789 |
0.29 |
| chr3_186776851_186777002 | 0.28 |
ST6GAL1 |
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
15395 |
0.23 |
| chr5_169744528_169744722 | 0.28 |
LCP2 |
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa) |
19394 |
0.19 |
| chr12_56733182_56733440 | 0.28 |
IL23A |
interleukin 23, alpha subunit p19 |
648 |
0.46 |
| chrX_100662156_100662307 | 0.28 |
GLA |
galactosidase, alpha |
682 |
0.45 |
| chrX_65225891_65226383 | 0.28 |
RP6-159A1.3 |
|
6544 |
0.22 |
| chr4_89199715_89199866 | 0.28 |
ENSG00000200469 |
. |
3330 |
0.19 |
| chr12_113621285_113621493 | 0.28 |
DDX54 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 54 |
218 |
0.86 |
| chr4_103546216_103546367 | 0.28 |
NFKB1 |
nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 |
47270 |
0.15 |
| chr1_110331899_110332050 | 0.28 |
EPS8L3 |
EPS8-like 3 |
25325 |
0.13 |
| chr15_30468678_30468972 | 0.27 |
ENSG00000221785 |
. |
31745 |
0.11 |
| chr20_48329492_48329643 | 0.27 |
B4GALT5 |
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 5 |
848 |
0.61 |
| chr2_27071961_27072112 | 0.27 |
DPYSL5 |
dihydropyrimidinase-like 5 |
660 |
0.72 |
| chr15_86325217_86325368 | 0.27 |
ENSG00000221634 |
. |
11483 |
0.11 |
| chr3_17621324_17621533 | 0.27 |
TBC1D5 |
TBC1 domain family, member 5 |
43980 |
0.2 |
| chr9_15300217_15300592 | 0.27 |
TTC39B |
tetratricopeptide repeat domain 39B |
6803 |
0.25 |
| chr1_36634160_36634311 | 0.27 |
MAP7D1 |
MAP7 domain containing 1 |
8083 |
0.14 |
| chr20_3725478_3725931 | 0.27 |
HSPA12B |
heat shock 70kD protein 12B |
12346 |
0.12 |
| chr20_62579033_62579428 | 0.27 |
UCKL1 |
uridine-cytidine kinase 1-like 1 |
3249 |
0.09 |
| chrY_7188749_7188900 | 0.27 |
ENSG00000252633 |
. |
3812 |
0.24 |
| chr18_21514931_21515082 | 0.27 |
LAMA3 |
laminin, alpha 3 |
6803 |
0.22 |
| chr11_113948126_113948329 | 0.27 |
ENSG00000221112 |
. |
9424 |
0.21 |
| chr1_156457116_156457326 | 0.27 |
MEF2D |
myocyte enhancer factor 2D |
3170 |
0.16 |
| chr1_182599288_182599608 | 0.27 |
RGS16 |
regulator of G-protein signaling 16 |
25905 |
0.17 |
| chrX_135747491_135747642 | 0.27 |
CD40LG |
CD40 ligand |
17180 |
0.17 |
| chr9_316050_316201 | 0.26 |
DOCK8 |
dedicator of cytokinesis 8 |
43055 |
0.14 |
| chr15_32654105_32654382 | 0.26 |
ENSG00000221444 |
. |
31430 |
0.1 |
| chr11_121521536_121521776 | 0.26 |
SORL1 |
sortilin-related receptor, L(DLR class) A repeats containing |
60528 |
0.16 |
| chr18_74176432_74176583 | 0.26 |
ZNF516 |
zinc finger protein 516 |
26228 |
0.16 |
| chr10_17548800_17549076 | 0.26 |
ST8SIA6 |
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 6 |
52609 |
0.13 |
| chr16_3122157_3122311 | 0.26 |
ENSG00000252561 |
. |
2710 |
0.09 |
| chr14_68951873_68952122 | 0.26 |
RAD51B |
RAD51 paralog B |
73770 |
0.11 |
| chr2_198075933_198076206 | 0.26 |
ANKRD44 |
ankyrin repeat domain 44 |
13307 |
0.2 |
| chr12_64978366_64978542 | 0.26 |
ENSG00000223294 |
. |
9009 |
0.14 |
| chr12_58175242_58176026 | 0.26 |
TSFM |
Ts translation elongation factor, mitochondrial |
738 |
0.36 |
| chr3_31592528_31592679 | 0.26 |
STT3B |
STT3B, subunit of the oligosaccharyltransferase complex (catalytic) |
18321 |
0.28 |
| chr8_129556248_129556417 | 0.26 |
ENSG00000221351 |
. |
275708 |
0.01 |
| chr1_154312907_154313058 | 0.26 |
ENSG00000238365 |
. |
1763 |
0.21 |
| chr15_70849476_70849729 | 0.26 |
UACA |
uveal autoantigen with coiled-coil domains and ankyrin repeats |
145018 |
0.05 |
| chr17_26876127_26876878 | 0.26 |
UNC119 |
unc-119 homolog (C. elegans) |
85 |
0.93 |
| chr1_248905757_248905908 | 0.26 |
LYPD8 |
LY6/PLAUR domain containing 8 |
2682 |
0.28 |
| chr14_100568921_100569190 | 0.26 |
EVL |
Enah/Vasp-like |
5771 |
0.14 |
| chr15_92401400_92401706 | 0.26 |
SLCO3A1 |
solute carrier organic anion transporter family, member 3A1 |
4202 |
0.3 |
| chr2_198294526_198294745 | 0.26 |
SF3B1 |
splicing factor 3b, subunit 1, 155kDa |
5116 |
0.15 |
| chr3_15319690_15320104 | 0.26 |
SH3BP5 |
SH3-domain binding protein 5 (BTK-associated) |
2345 |
0.23 |
| chr3_122295994_122296195 | 0.26 |
PARP15 |
poly (ADP-ribose) polymerase family, member 15 |
355 |
0.86 |
| chr1_55070670_55070938 | 0.26 |
FAM151A |
family with sequence similarity 151, member A |
18425 |
0.14 |
| chr2_69006771_69007110 | 0.25 |
ARHGAP25 |
Rho GTPase activating protein 25 |
4868 |
0.27 |
| chr10_17703984_17704256 | 0.25 |
ENSG00000251959 |
. |
17067 |
0.14 |
| chr2_65172918_65173113 | 0.25 |
ENSG00000238696 |
. |
37195 |
0.12 |
| chr1_76751688_76751938 | 0.25 |
ST6GALNAC3 |
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 3 |
211409 |
0.02 |
| chr10_97415695_97415896 | 0.25 |
ALDH18A1 |
aldehyde dehydrogenase 18 family, member A1 |
668 |
0.74 |
| chr11_118109252_118109403 | 0.25 |
AMICA1 |
adhesion molecule, interacts with CXADR antigen 1 |
13518 |
0.13 |
| chr7_150267291_150267625 | 0.25 |
GIMAP4 |
GTPase, IMAP family member 4 |
2934 |
0.27 |
| chr3_186657505_186657656 | 0.25 |
ST6GAL1 |
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
8439 |
0.21 |
| chr7_77317684_77317835 | 0.25 |
RSBN1L |
round spermatid basic protein 1-like |
8001 |
0.26 |
| chr2_99312607_99313038 | 0.25 |
MGAT4A |
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A |
30337 |
0.18 |
| chr8_126661069_126661220 | 0.25 |
ENSG00000266452 |
. |
204337 |
0.03 |
| chr18_60881389_60881563 | 0.25 |
ENSG00000238988 |
. |
19578 |
0.21 |
| chr15_75642153_75642345 | 0.25 |
NEIL1 |
nei endonuclease VIII-like 1 (E. coli) |
1509 |
0.26 |
| chr11_121331911_121332238 | 0.25 |
RP11-730K11.1 |
|
8352 |
0.25 |
| chr1_156183961_156184321 | 0.25 |
PMF1-BGLAP |
PMF1-BGLAP readthrough |
1341 |
0.22 |
| chr1_111436803_111436998 | 0.24 |
CD53 |
CD53 molecule |
21124 |
0.16 |
| chr9_6422321_6422472 | 0.24 |
UHRF2 |
ubiquitin-like with PHD and ring finger domains 2, E3 ubiquitin protein ligase |
1365 |
0.45 |
| chr19_30172794_30172945 | 0.24 |
PLEKHF1 |
pleckstrin homology domain containing, family F (with FYVE domain) member 1 |
15088 |
0.21 |
| chrX_19795655_19795879 | 0.24 |
SH3KBP1 |
SH3-domain kinase binding protein 1 |
22102 |
0.26 |
| chr1_111180656_111180873 | 0.24 |
KCNA2 |
potassium voltage-gated channel, shaker-related subfamily, member 2 |
6668 |
0.19 |
| chr3_42390480_42390631 | 0.24 |
LYZL4 |
lysozyme-like 4 |
61495 |
0.1 |
| chr9_130532441_130532713 | 0.24 |
SH2D3C |
SH2 domain containing 3C |
1140 |
0.29 |
| chr8_126661631_126661782 | 0.24 |
ENSG00000266452 |
. |
204899 |
0.02 |
| chr8_95229930_95230081 | 0.24 |
CDH17 |
cadherin 17, LI cadherin (liver-intestine) |
474 |
0.86 |
| chr6_53202509_53202860 | 0.24 |
ELOVL5 |
ELOVL fatty acid elongase 5 |
10903 |
0.21 |
| chr3_133300803_133300954 | 0.24 |
CDV3 |
CDV3 homolog (mouse) |
7386 |
0.21 |
| chr6_125390047_125390198 | 0.24 |
RNF217 |
ring finger protein 217 |
20764 |
0.24 |
| chr3_50657101_50657507 | 0.24 |
MAPKAPK3 |
mitogen-activated protein kinase-activated protein kinase 3 |
2483 |
0.23 |
| chr20_30973001_30973152 | 0.24 |
ASXL1 |
additional sex combs like 1 (Drosophila) |
25526 |
0.17 |
| chr2_198192116_198192267 | 0.24 |
AC010746.3 |
|
16074 |
0.15 |
| chr14_67983023_67983217 | 0.23 |
TMEM229B |
transmembrane protein 229B |
932 |
0.54 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.1 | 0.1 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.1 | 0.2 | GO:0009301 | snRNA transcription(GO:0009301) |
| 0.1 | 0.3 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.1 | GO:0034625 | fatty acid elongation, monounsaturated fatty acid(GO:0034625) |
| 0.0 | 0.3 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 0.1 | GO:0016137 | glycoside metabolic process(GO:0016137) glycoside catabolic process(GO:0016139) |
| 0.0 | 0.1 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.1 | GO:0045074 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.0 | 0.1 | GO:0031946 | regulation of glucocorticoid biosynthetic process(GO:0031946) |
| 0.0 | 0.1 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.1 | GO:0051140 | regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) |
| 0.0 | 0.2 | GO:0002335 | mature B cell differentiation(GO:0002335) |
| 0.0 | 0.2 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.0 | 0.2 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.0 | 0.1 | GO:0070561 | vitamin D receptor signaling pathway(GO:0070561) |
| 0.0 | 0.1 | GO:0090116 | DNA methylation on cytosine(GO:0032776) C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 0.1 | GO:0002606 | positive regulation of dendritic cell antigen processing and presentation(GO:0002606) |
| 0.0 | 0.1 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.0 | 0.1 | GO:0034122 | negative regulation of toll-like receptor signaling pathway(GO:0034122) |
| 0.0 | 0.1 | GO:0002713 | negative regulation of B cell mediated immunity(GO:0002713) negative regulation of immunoglobulin mediated immune response(GO:0002890) |
| 0.0 | 0.1 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.1 | GO:2000041 | regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 0.0 | 0.1 | GO:0008049 | male courtship behavior(GO:0008049) |
| 0.0 | 0.1 | GO:0060129 | thyroid-stimulating hormone-secreting cell differentiation(GO:0060129) |
| 0.0 | 0.3 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.0 | 0.1 | GO:0070254 | mucus secretion(GO:0070254) |
| 0.0 | 0.1 | GO:1904035 | endothelial cell apoptotic process(GO:0072577) epithelial cell apoptotic process(GO:1904019) regulation of epithelial cell apoptotic process(GO:1904035) regulation of endothelial cell apoptotic process(GO:2000351) |
| 0.0 | 0.1 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.1 | GO:0045341 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) |
| 0.0 | 0.1 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 0.0 | GO:0003402 | planar cell polarity pathway involved in axis elongation(GO:0003402) |
| 0.0 | 0.1 | GO:0006210 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.0 | 0.1 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.0 | 0.1 | GO:0046599 | regulation of centriole replication(GO:0046599) |
| 0.0 | 0.0 | GO:0046137 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.0 | 0.2 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.1 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.0 | 0.1 | GO:0000042 | protein targeting to Golgi(GO:0000042) establishment of protein localization to Golgi(GO:0072600) |
| 0.0 | 0.0 | GO:0002579 | positive regulation of antigen processing and presentation(GO:0002579) |
| 0.0 | 0.1 | GO:0021780 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) oligodendrocyte cell fate specification(GO:0021778) glial cell fate specification(GO:0021780) |
| 0.0 | 0.0 | GO:0010882 | regulation of cardiac muscle contraction by calcium ion signaling(GO:0010882) |
| 0.0 | 0.1 | GO:0031061 | negative regulation of histone methylation(GO:0031061) |
| 0.0 | 0.2 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.0 | GO:2001012 | mesenchymal cell differentiation involved in kidney development(GO:0072161) metanephric mesenchymal cell differentiation(GO:0072162) mesenchymal cell differentiation involved in renal system development(GO:2001012) |
| 0.0 | 0.1 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.2 | GO:0001782 | B cell homeostasis(GO:0001782) |
| 0.0 | 0.2 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 0.1 | GO:0016093 | polyprenol metabolic process(GO:0016093) dolichol metabolic process(GO:0019348) |
| 0.0 | 0.1 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.0 | 0.1 | GO:0052167 | pathogen-associated molecular pattern dependent induction by symbiont of host innate immune response(GO:0052033) positive regulation by symbiont of host innate immune response(GO:0052166) modulation by symbiont of host innate immune response(GO:0052167) pathogen-associated molecular pattern dependent modulation by symbiont of host innate immune response(GO:0052169) pathogen-associated molecular pattern dependent induction by organism of innate immune response of other organism involved in symbiotic interaction(GO:0052257) positive regulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052305) modulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052306) pathogen-associated molecular pattern dependent modulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052308) |
| 0.0 | 0.0 | GO:0002874 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) |
| 0.0 | 0.1 | GO:0031342 | negative regulation of leukocyte mediated cytotoxicity(GO:0001911) negative regulation of cell killing(GO:0031342) |
| 0.0 | 0.1 | GO:0046502 | uroporphyrinogen III metabolic process(GO:0046502) |
| 0.0 | 0.1 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 | 0.1 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.2 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.1 | GO:0071364 | cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.0 | 0.0 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.0 | 0.0 | GO:0033091 | positive regulation of immature T cell proliferation(GO:0033091) |
| 0.0 | 0.0 | GO:0021801 | cerebral cortex radial glia guided migration(GO:0021801) telencephalon glial cell migration(GO:0022030) |
| 0.0 | 0.1 | GO:0010310 | regulation of hydrogen peroxide metabolic process(GO:0010310) |
| 0.0 | 0.1 | GO:0045906 | negative regulation of vasoconstriction(GO:0045906) |
| 0.0 | 0.1 | GO:0042996 | regulation of Golgi to plasma membrane protein transport(GO:0042996) |
| 0.0 | 0.1 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.0 | GO:0002513 | tolerance induction to self antigen(GO:0002513) |
| 0.0 | 0.1 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.0 | 0.1 | GO:0032328 | alanine transport(GO:0032328) |
| 0.0 | 0.1 | GO:0046880 | regulation of follicle-stimulating hormone secretion(GO:0046880) negative regulation of follicle-stimulating hormone secretion(GO:0046882) |
| 0.0 | 0.1 | GO:0019883 | antigen processing and presentation of endogenous antigen(GO:0019883) |
| 0.0 | 0.1 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 0.1 | GO:0008356 | asymmetric cell division(GO:0008356) |
| 0.0 | 0.1 | GO:0022038 | corpus callosum development(GO:0022038) |
| 0.0 | 0.1 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.1 | GO:0032203 | telomere formation via telomerase(GO:0032203) |
| 0.0 | 0.1 | GO:0032784 | regulation of DNA-templated transcription, elongation(GO:0032784) |
| 0.0 | 0.1 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.0 | 0.1 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.0 | 0.0 | GO:0007100 | mitotic centrosome separation(GO:0007100) |
| 0.0 | 0.1 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.0 | 0.1 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.0 | 0.1 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.0 | GO:0002678 | positive regulation of chronic inflammatory response(GO:0002678) |
| 0.0 | 0.0 | GO:0009296 | obsolete flagellum assembly(GO:0009296) |
| 0.0 | 0.0 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.0 | 0.1 | GO:0032627 | interleukin-23 production(GO:0032627) regulation of interleukin-23 production(GO:0032667) positive regulation of interleukin-23 production(GO:0032747) |
| 0.0 | 0.0 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.1 | GO:0030889 | negative regulation of B cell proliferation(GO:0030889) |
| 0.0 | 0.0 | GO:0002361 | CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) |
| 0.0 | 0.0 | GO:0045002 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) double-strand break repair via single-strand annealing(GO:0045002) |
| 0.0 | 0.0 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.0 | 0.1 | GO:0003341 | cilium movement(GO:0003341) |
| 0.0 | 0.1 | GO:0008291 | acetylcholine metabolic process(GO:0008291) acetate ester metabolic process(GO:1900619) |
| 0.0 | 0.0 | GO:0050849 | negative regulation of calcium-mediated signaling(GO:0050849) |
| 0.0 | 0.1 | GO:0009812 | flavonoid metabolic process(GO:0009812) |
| 0.0 | 0.2 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.0 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.1 | GO:0009169 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.0 | 0.1 | GO:0032703 | negative regulation of interleukin-2 production(GO:0032703) |
| 0.0 | 0.1 | GO:0006528 | asparagine metabolic process(GO:0006528) |
| 0.0 | 0.1 | GO:0035066 | positive regulation of histone acetylation(GO:0035066) positive regulation of protein acetylation(GO:1901985) positive regulation of peptidyl-lysine acetylation(GO:2000758) |
| 0.0 | 0.0 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.0 | GO:1901072 | amino sugar catabolic process(GO:0046348) glucosamine-containing compound catabolic process(GO:1901072) |
| 0.0 | 0.0 | GO:0002475 | antigen processing and presentation via MHC class Ib(GO:0002475) |
| 0.0 | 0.0 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.0 | 0.0 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.1 | GO:0045022 | early endosome to late endosome transport(GO:0045022) vesicle-mediated transport between endosomal compartments(GO:0098927) |
| 0.0 | 0.1 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.0 | 0.0 | GO:0090322 | regulation of superoxide metabolic process(GO:0090322) |
| 0.0 | 0.0 | GO:0002125 | maternal aggressive behavior(GO:0002125) |
| 0.0 | 0.1 | GO:0008628 | hormone-mediated apoptotic signaling pathway(GO:0008628) |
| 0.0 | 0.0 | GO:0042109 | lymphotoxin A production(GO:0032641) lymphotoxin A biosynthetic process(GO:0042109) |
| 0.0 | 0.1 | GO:0046666 | retinal cell programmed cell death(GO:0046666) |
| 0.0 | 0.0 | GO:0015816 | glycine transport(GO:0015816) |
| 0.0 | 0.2 | GO:0050672 | negative regulation of mononuclear cell proliferation(GO:0032945) negative regulation of lymphocyte proliferation(GO:0050672) negative regulation of leukocyte proliferation(GO:0070664) |
| 0.0 | 0.1 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.0 | 0.2 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.0 | 0.0 | GO:0046958 | nonassociative learning(GO:0046958) |
| 0.0 | 0.0 | GO:0015959 | diadenosine polyphosphate metabolic process(GO:0015959) |
| 0.0 | 0.0 | GO:0060437 | lung growth(GO:0060437) |
| 0.0 | 0.0 | GO:0009414 | response to water deprivation(GO:0009414) response to water(GO:0009415) |
| 0.0 | 0.3 | GO:0006612 | protein targeting to membrane(GO:0006612) |
| 0.0 | 0.3 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.0 | GO:0036037 | CD8-positive, alpha-beta T cell activation(GO:0036037) CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.1 | GO:0032099 | negative regulation of appetite(GO:0032099) |
| 0.0 | 0.0 | GO:0016102 | retinoic acid biosynthetic process(GO:0002138) diterpenoid biosynthetic process(GO:0016102) |
| 0.0 | 0.0 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.1 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.0 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.1 | GO:0032069 | regulation of nuclease activity(GO:0032069) |
| 0.0 | 0.1 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 0.0 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.1 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.0 | GO:0006154 | adenosine catabolic process(GO:0006154) |
| 0.0 | 0.0 | GO:0002883 | hypersensitivity(GO:0002524) regulation of hypersensitivity(GO:0002883) positive regulation of hypersensitivity(GO:0002885) |
| 0.0 | 0.3 | GO:0050772 | positive regulation of axonogenesis(GO:0050772) |
| 0.0 | 0.0 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.0 | GO:0043628 | ncRNA 3'-end processing(GO:0043628) |
| 0.0 | 0.0 | GO:0016241 | regulation of macroautophagy(GO:0016241) |
| 0.0 | 0.1 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.1 | GO:0043584 | nose development(GO:0043584) |
| 0.0 | 0.0 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.0 | GO:0045056 | transcytosis(GO:0045056) |
| 0.0 | 0.0 | GO:0048537 | mucosal-associated lymphoid tissue development(GO:0048537) |
| 0.0 | 0.0 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.0 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.1 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 | 0.4 | GO:0042059 | negative regulation of epidermal growth factor receptor signaling pathway(GO:0042059) negative regulation of ERBB signaling pathway(GO:1901185) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.1 | 0.2 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.0 | 0.1 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.1 | GO:0032449 | CBM complex(GO:0032449) |
| 0.0 | 0.1 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.0 | 0.1 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.1 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.4 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.1 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.1 | GO:0071778 | obsolete WINAC complex(GO:0071778) |
| 0.0 | 0.1 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.0 | 0.0 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.1 | GO:0042583 | chromaffin granule(GO:0042583) chromaffin granule membrane(GO:0042584) |
| 0.0 | 0.1 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.1 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.3 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 0.0 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.1 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.0 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.2 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.1 | GO:0090533 | cation-transporting ATPase complex(GO:0090533) |
| 0.0 | 0.2 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.1 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.1 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.1 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.1 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.1 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.1 | GO:0000791 | euchromatin(GO:0000791) |
| 0.0 | 0.0 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 0.0 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.1 | GO:0048500 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.0 | 0.0 | GO:0005638 | lamin filament(GO:0005638) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.1 | 0.1 | GO:0043175 | RNA polymerase core enzyme binding(GO:0043175) |
| 0.1 | 0.3 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 0.1 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.2 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.2 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.2 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.1 | GO:0016362 | activin receptor activity, type II(GO:0016362) |
| 0.0 | 0.3 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.1 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.0 | 0.1 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.1 | GO:0015925 | galactosidase activity(GO:0015925) |
| 0.0 | 0.1 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.1 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.0 | 0.1 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.1 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.1 | GO:0055106 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.1 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.1 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.0 | 0.2 | GO:0009922 | fatty acid elongase activity(GO:0009922) |
| 0.0 | 0.5 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.0 | 0.1 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.0 | 0.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.2 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.1 | GO:0001099 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.0 | 0.1 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.2 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.1 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.1 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 0.2 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.1 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.1 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.0 | 0.2 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.1 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.4 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.1 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.1 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.2 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.0 | 0.1 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.1 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.1 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.0 | 0.1 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.1 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.0 | 0.4 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.1 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.1 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.1 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.0 | GO:0042156 | obsolete zinc-mediated transcriptional activator activity(GO:0042156) |
| 0.0 | 0.1 | GO:0034593 | phosphatidylinositol bisphosphate phosphatase activity(GO:0034593) |
| 0.0 | 0.2 | GO:0017136 | NAD-dependent histone deacetylase activity(GO:0017136) |
| 0.0 | 0.1 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.1 | GO:0061630 | ubiquitin-ubiquitin ligase activity(GO:0034450) ubiquitin protein ligase activity(GO:0061630) ubiquitin-like protein ligase activity(GO:0061659) |
| 0.0 | 0.0 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.1 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.1 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.0 | 0.1 | GO:0004904 | interferon receptor activity(GO:0004904) |
| 0.0 | 0.1 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.0 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.0 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.3 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.0 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0003711 | obsolete transcription elongation regulator activity(GO:0003711) |
| 0.0 | 0.1 | GO:0003964 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.0 | 0.1 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.1 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.0 | 0.1 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.1 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 0.0 | 0.2 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.3 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.0 | 0.1 | GO:0003709 | obsolete RNA polymerase III transcription factor activity(GO:0003709) |
| 0.0 | 0.0 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.0 | 0.0 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.0 | 0.2 | GO:0016505 | peptidase activator activity involved in apoptotic process(GO:0016505) |
| 0.0 | 0.3 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.0 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.1 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.0 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.0 | 0.1 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 0.1 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 0.0 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.0 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.1 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.0 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.0 | 0.1 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.0 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.0 | 0.1 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.0 | 0.1 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 0.5 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.1 | GO:0008312 | 7S RNA binding(GO:0008312) |
| 0.0 | 0.0 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.0 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.0 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.3 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.1 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.1 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 0.7 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.5 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.5 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.1 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.2 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.3 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.4 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 0.2 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.5 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.5 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.3 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 0.1 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.1 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.2 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.1 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.0 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 0.3 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.3 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.2 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.3 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.8 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.2 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.6 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.0 | 0.4 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.2 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.1 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.6 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.2 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.0 | 0.3 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 0.2 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.2 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.2 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.1 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.1 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 0.2 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.2 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.0 | 0.0 | REACTOME SCF BETA TRCP MEDIATED DEGRADATION OF EMI1 | Genes involved in SCF-beta-TrCP mediated degradation of Emi1 |
| 0.0 | 0.0 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.0 | 0.1 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.0 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.0 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.0 | 0.2 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.3 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |