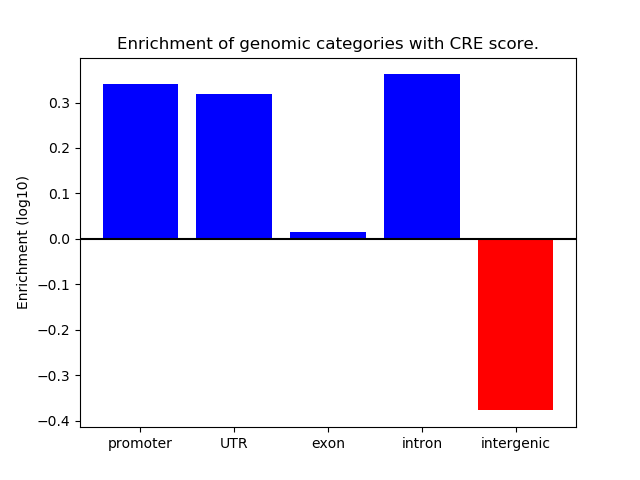
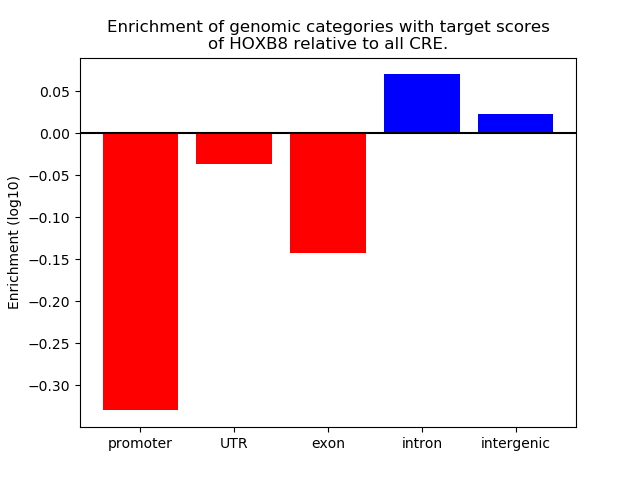
Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
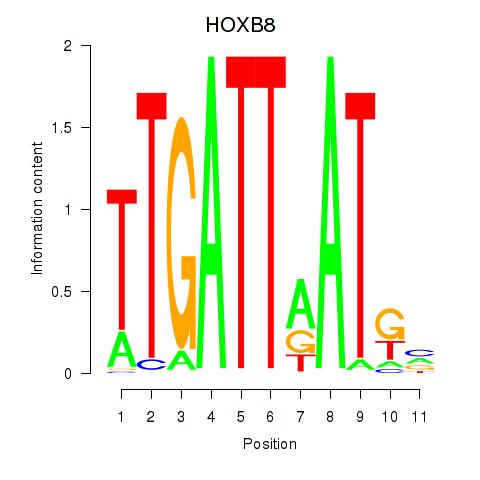
Results for HOXB8
Z-value: 1.30
Transcription factors associated with HOXB8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXB8
|
ENSG00000120068.5 | homeobox B8 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr17_46695333_46695484 | HOXB8 | 2930 | 0.098121 | -0.80 | 1.0e-02 | Click! |
| chr17_46696935_46697184 | HOXB8 | 4581 | 0.080060 | -0.76 | 1.7e-02 | Click! |
| chr17_46690621_46690846 | HOXB8 | 108 | 0.898450 | -0.69 | 3.8e-02 | Click! |
| chr17_46689590_46689741 | HOXB8 | 1176 | 0.198765 | -0.68 | 4.2e-02 | Click! |
| chr17_46695682_46695990 | HOXB8 | 3358 | 0.090943 | -0.67 | 4.7e-02 | Click! |
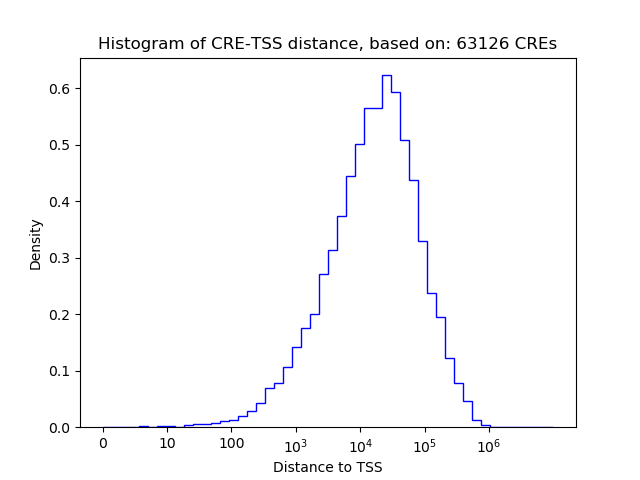
Activity of the HOXB8 motif across conditions
Conditions sorted by the z-value of the HOXB8 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
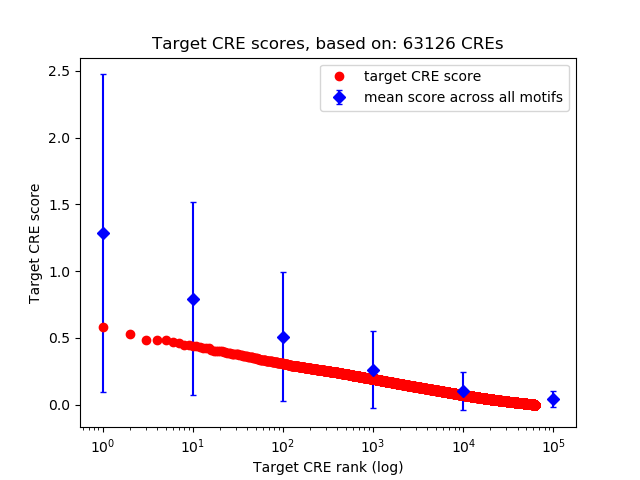
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr7_15498785_15499062 | 0.59 |
AGMO |
alkylglycerol monooxygenase |
93076 |
0.1 |
| chr7_84765663_84765814 | 0.53 |
SEMA3D |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3D |
14487 |
0.32 |
| chr8_118878705_118878856 | 0.49 |
EXT1 |
exostosin glycosyltransferase 1 |
243873 |
0.02 |
| chr4_113437546_113438608 | 0.48 |
NEUROG2 |
neurogenin 2 |
749 |
0.58 |
| chr5_95361917_95362068 | 0.48 |
ENSG00000207578 |
. |
52850 |
0.12 |
| chr1_23609794_23609945 | 0.47 |
HNRNPR |
heterogeneous nuclear ribonucleoprotein R |
60886 |
0.08 |
| chr7_84253629_84253780 | 0.46 |
ENSG00000265050 |
. |
41620 |
0.22 |
| chr2_206185805_206185956 | 0.45 |
NRP2 |
neuropilin 2 |
361344 |
0.01 |
| chr9_128256725_128256876 | 0.45 |
MAPKAP1 |
mitogen-activated protein kinase associated protein 1 |
10020 |
0.22 |
| chr3_134182320_134182615 | 0.44 |
CEP63 |
centrosomal protein 63kDa |
22118 |
0.15 |
| chr5_143293259_143293410 | 0.44 |
HMHB1 |
histocompatibility (minor) HB-1 |
101608 |
0.08 |
| chr3_151581991_151582142 | 0.43 |
SUCNR1 |
succinate receptor 1 |
9365 |
0.23 |
| chr10_69865393_69865544 | 0.42 |
MYPN |
myopalladin |
444 |
0.84 |
| chr8_131417344_131417495 | 0.42 |
ASAP1 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1 |
3202 |
0.4 |
| chr5_167791618_167792032 | 0.42 |
WWC1 |
WW and C2 domain containing 1 |
6601 |
0.29 |
| chr2_69793604_69794058 | 0.41 |
ENSG00000207016 |
. |
46525 |
0.13 |
| chr11_9842791_9843084 | 0.40 |
RP11-1H15.2 |
|
17753 |
0.15 |
| chr12_132488838_132488989 | 0.40 |
ENSG00000271905 |
. |
2593 |
0.25 |
| chr10_29932554_29932705 | 0.40 |
SVIL |
supervillin |
8728 |
0.22 |
| chr4_114305617_114305768 | 0.40 |
ANK2 |
ankyrin 2, neuronal |
11177 |
0.22 |
| chr8_41054271_41054422 | 0.40 |
ENSG00000263372 |
. |
74316 |
0.09 |
| chr7_116418433_116418704 | 0.39 |
MET |
met proto-oncogene |
1339 |
0.49 |
| chr19_381111_381262 | 0.39 |
THEG |
theg spermatid protein |
5173 |
0.15 |
| chr12_93179700_93179851 | 0.39 |
EEA1 |
early endosome antigen 1 |
35100 |
0.18 |
| chr2_216294982_216295133 | 0.39 |
FN1 |
fibronectin 1 |
5733 |
0.24 |
| chr6_17429475_17429626 | 0.38 |
CAP2 |
CAP, adenylate cyclase-associated protein, 2 (yeast) |
35607 |
0.21 |
| chr2_84766841_84767171 | 0.38 |
DNAH6 |
dynein, axonemal, heavy chain 6 |
22063 |
0.25 |
| chr2_27905704_27905855 | 0.38 |
SUPT7L |
suppressor of Ty 7 (S. cerevisiae)-like |
19103 |
0.11 |
| chr13_32331269_32331420 | 0.38 |
RXFP2 |
relaxin/insulin-like family peptide receptor 2 |
17665 |
0.29 |
| chr1_181110732_181111665 | 0.38 |
IER5 |
immediate early response 5 |
53560 |
0.15 |
| chr14_71881120_71881271 | 0.38 |
ENSG00000207444 |
. |
16141 |
0.22 |
| chr11_121336559_121337538 | 0.38 |
RP11-730K11.1 |
|
13326 |
0.24 |
| chr4_154434080_154434421 | 0.37 |
KIAA0922 |
KIAA0922 |
42798 |
0.17 |
| chr11_65045369_65045520 | 0.37 |
POLA2 |
polymerase (DNA directed), alpha 2, accessory subunit |
4534 |
0.15 |
| chr2_174985205_174985356 | 0.37 |
OLA1 |
Obg-like ATPase 1 |
97394 |
0.07 |
| chr18_21207750_21207901 | 0.37 |
ANKRD29 |
ankyrin repeat domain 29 |
21661 |
0.17 |
| chr7_87941676_87941827 | 0.37 |
STEAP4 |
STEAP family member 4 |
5545 |
0.25 |
| chr17_79542900_79543051 | 0.37 |
ENSG00000207021 |
. |
2459 |
0.15 |
| chr14_53342942_53343093 | 0.36 |
FERMT2 |
fermitin family member 2 |
11778 |
0.23 |
| chr11_130717130_130717402 | 0.36 |
SNX19 |
sorting nexin 19 |
46503 |
0.19 |
| chr6_113684543_113684694 | 0.36 |
ENSG00000222677 |
. |
151571 |
0.04 |
| chr7_89900753_89900904 | 0.36 |
C7orf63 |
chromosome 7 open reading frame 63 |
75 |
0.95 |
| chr13_38289097_38289248 | 0.36 |
TRPC4 |
transient receptor potential cation channel, subfamily C, member 4 |
68298 |
0.13 |
| chr3_47844781_47845806 | 0.36 |
DHX30 |
DEAH (Asp-Glu-Ala-His) box helicase 30 |
638 |
0.69 |
| chr8_25771009_25771433 | 0.36 |
EBF2 |
early B-cell factor 2 |
25789 |
0.27 |
| chr7_25891719_25892347 | 0.35 |
ENSG00000199085 |
. |
97573 |
0.09 |
| chr2_28079220_28079371 | 0.35 |
RBKS |
ribokinase |
13267 |
0.18 |
| chr18_6730887_6731038 | 0.35 |
ARHGAP28 |
Rho GTPase activating protein 28 |
920 |
0.46 |
| chr12_96199249_96199495 | 0.35 |
RP11-536G4.1 |
Uncharacterized protein |
2497 |
0.23 |
| chr18_60002993_60003265 | 0.35 |
ENSG00000199867 |
. |
5084 |
0.19 |
| chrX_138971819_138971970 | 0.35 |
ENSG00000221375 |
. |
4254 |
0.21 |
| chr2_217461850_217462001 | 0.34 |
AC073321.3 |
|
9736 |
0.19 |
| chr15_70994047_70994198 | 0.34 |
UACA |
uveal autoantigen with coiled-coil domains and ankyrin repeats |
498 |
0.86 |
| chr14_92620421_92620697 | 0.34 |
CPSF2 |
cleavage and polyadenylation specific factor 2, 100kDa |
964 |
0.57 |
| chr7_41174850_41175131 | 0.34 |
AC005160.3 |
|
359833 |
0.01 |
| chr6_149189778_149189929 | 0.34 |
RP11-162J8.2 |
|
95967 |
0.08 |
| chr14_50691962_50692201 | 0.34 |
SOS2 |
son of sevenless homolog 2 (Drosophila) |
5946 |
0.22 |
| chr4_144318381_144318532 | 0.34 |
GAB1 |
GRB2-associated binding protein 1 |
5793 |
0.26 |
| chr11_12560979_12561130 | 0.34 |
PARVA |
parvin, alpha |
86772 |
0.09 |
| chr5_52785845_52785996 | 0.33 |
FST |
follistatin |
5874 |
0.3 |
| chr5_5340745_5341045 | 0.33 |
KIAA0947 |
KIAA0947 |
81912 |
0.11 |
| chr1_192412658_192412809 | 0.33 |
ENSG00000221145 |
. |
47617 |
0.17 |
| chr15_49231600_49231751 | 0.33 |
RP11-138E16.1 |
|
1242 |
0.44 |
| chr1_205905941_205906092 | 0.33 |
SLC26A9 |
solute carrier family 26 (anion exchanger), member 9 |
1066 |
0.47 |
| chr12_11120355_11120506 | 0.33 |
TAS2R50 |
taste receptor, type 2, member 50 |
19081 |
0.11 |
| chr22_32042185_32042407 | 0.33 |
PISD |
phosphatidylserine decarboxylase |
7865 |
0.21 |
| chr2_201626361_201626512 | 0.33 |
ENSG00000201737 |
. |
13871 |
0.15 |
| chr7_22142934_22143230 | 0.33 |
RAPGEF5 |
Rap guanine nucleotide exchange factor (GEF) 5 |
90445 |
0.09 |
| chr1_185938815_185938966 | 0.33 |
HMCN1 |
hemicentin 1 |
201629 |
0.02 |
| chr10_33304516_33304667 | 0.33 |
ITGB1 |
integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12) |
9871 |
0.23 |
| chr13_80350445_80350596 | 0.33 |
NDFIP2 |
Nedd4 family interacting protein 2 |
294932 |
0.01 |
| chr15_71080718_71080869 | 0.32 |
RP11-138H8.3 |
|
9586 |
0.19 |
| chr8_62634134_62634577 | 0.32 |
ENSG00000264408 |
. |
7008 |
0.24 |
| chr3_8783509_8783660 | 0.32 |
CAV3 |
caveolin 3 |
8032 |
0.14 |
| chr2_242318207_242318358 | 0.32 |
FARP2 |
FERM, RhoGEF and pleckstrin domain protein 2 |
6018 |
0.17 |
| chr3_149867742_149867893 | 0.32 |
RP11-167H9.4 |
|
51998 |
0.14 |
| chr2_214052099_214052250 | 0.32 |
IKZF2 |
IKAROS family zinc finger 2 (Helios) |
35023 |
0.22 |
| chr2_217968796_217968947 | 0.32 |
ENSG00000251849 |
. |
72350 |
0.12 |
| chr4_89647157_89647308 | 0.32 |
FAM13A-AS1 |
FAM13A antisense RNA 1 |
4366 |
0.21 |
| chr2_67628597_67628898 | 0.32 |
ETAA1 |
Ewing tumor-associated antigen 1 |
4296 |
0.38 |
| chr20_33166706_33166857 | 0.32 |
MAP1LC3A |
microtubule-associated protein 1 light chain 3 alpha |
20259 |
0.16 |
| chr11_75798073_75798350 | 0.32 |
UVRAG |
UV radiation resistance associated |
28746 |
0.17 |
| chr2_200307971_200308122 | 0.32 |
SATB2 |
SATB homeobox 2 |
12765 |
0.23 |
| chr2_145700394_145700545 | 0.32 |
ENSG00000253036 |
. |
392169 |
0.01 |
| chr3_176843598_176843767 | 0.32 |
ENSG00000200882 |
. |
226 |
0.94 |
| chr3_101893892_101894043 | 0.32 |
ZPLD1 |
zona pellucida-like domain containing 1 |
75879 |
0.12 |
| chr10_61192443_61192594 | 0.32 |
FAM13C |
family with sequence similarity 13, member C |
69579 |
0.13 |
| chr4_89740050_89740544 | 0.32 |
FAM13A |
family with sequence similarity 13, member A |
4055 |
0.31 |
| chr7_36487948_36488099 | 0.32 |
AC006960.7 |
|
14609 |
0.19 |
| chr13_97959313_97959591 | 0.31 |
MBNL2 |
muscleblind-like splicing regulator 2 |
30994 |
0.21 |
| chr13_74792422_74792795 | 0.31 |
ENSG00000206617 |
. |
70743 |
0.12 |
| chr8_6335605_6335756 | 0.31 |
RP11-115C21.2 |
|
71017 |
0.1 |
| chr1_165860927_165861223 | 0.31 |
UCK2 |
uridine-cytidine kinase 2 |
3732 |
0.24 |
| chr3_8978587_8978738 | 0.31 |
ENSG00000199815 |
. |
5483 |
0.23 |
| chr1_170777177_170777328 | 0.31 |
MROH9 |
maestro heat-like repeat family member 9 |
127360 |
0.05 |
| chr6_113771019_113771170 | 0.31 |
ENSG00000222677 |
. |
65095 |
0.14 |
| chr4_88720136_88720287 | 0.31 |
IBSP |
integrin-binding sialoprotein |
522 |
0.76 |
| chr20_5021981_5022132 | 0.31 |
ENSG00000266199 |
. |
15691 |
0.16 |
| chr22_30014073_30014247 | 0.31 |
NF2 |
neurofibromin 2 (merlin) |
14172 |
0.16 |
| chr2_192121899_192122050 | 0.31 |
MYO1B |
myosin IB |
5751 |
0.29 |
| chr18_46866975_46867126 | 0.31 |
DYM |
dymeclin |
28017 |
0.18 |
| chr17_39415242_39415393 | 0.31 |
KRTAP9-9 |
keratin associated protein 9-9 |
3681 |
0.09 |
| chr3_27565025_27565176 | 0.31 |
ENSG00000201033 |
. |
9738 |
0.2 |
| chr1_92048607_92048758 | 0.31 |
CDC7 |
cell division cycle 7 |
81987 |
0.11 |
| chr1_56098686_56098837 | 0.31 |
ENSG00000272051 |
. |
148116 |
0.04 |
| chr12_90485587_90485738 | 0.31 |
ENSG00000252823 |
. |
337826 |
0.01 |
| chr2_161225682_161226020 | 0.31 |
ENSG00000252465 |
. |
27628 |
0.18 |
| chr13_31784035_31784377 | 0.30 |
B3GALTL |
beta 1,3-galactosyltransferase-like |
10133 |
0.27 |
| chr5_77531337_77531488 | 0.30 |
AP3B1 |
adaptor-related protein complex 3, beta 1 subunit |
59104 |
0.13 |
| chr13_94050738_94050889 | 0.30 |
ENSG00000252365 |
. |
29600 |
0.26 |
| chr15_49967575_49967726 | 0.30 |
RP11-353B9.1 |
|
23314 |
0.2 |
| chr1_88422818_88422969 | 0.30 |
ENSG00000199318 |
. |
503837 |
0.0 |
| chr10_33932286_33932437 | 0.30 |
NRP1 |
neuropilin 1 |
307171 |
0.01 |
| chr6_113011776_113011927 | 0.30 |
ENSG00000252215 |
. |
158467 |
0.04 |
| chr4_188539699_188539850 | 0.30 |
ZFP42 |
ZFP42 zinc finger protein |
377151 |
0.01 |
| chr8_61444637_61444969 | 0.30 |
RAB2A |
RAB2A, member RAS oncogene family |
15063 |
0.21 |
| chr8_119427039_119427190 | 0.30 |
AC023590.1 |
Uncharacterized protein |
132633 |
0.05 |
| chr6_86172282_86172433 | 0.30 |
NT5E |
5'-nucleotidase, ecto (CD73) |
8742 |
0.27 |
| chr15_96817381_96817630 | 0.30 |
NR2F2-AS1 |
NR2F2 antisense RNA 1 |
1720 |
0.37 |
| chr10_112711676_112711827 | 0.30 |
SHOC2 |
soc-2 suppressor of clear homolog (C. elegans) |
12858 |
0.15 |
| chr2_1657675_1657826 | 0.30 |
AC144450.1 |
|
33865 |
0.19 |
| chr8_23268879_23269030 | 0.29 |
LOXL2 |
lysyl oxidase-like 2 |
7201 |
0.19 |
| chr11_12791941_12792092 | 0.29 |
TEAD1 |
TEA domain family member 1 (SV40 transcriptional enhancer factor) |
25428 |
0.2 |
| chr12_118628458_118628609 | 0.29 |
TAOK3 |
TAO kinase 3 |
177 |
0.96 |
| chr4_188524604_188524755 | 0.29 |
ZFP42 |
ZFP42 zinc finger protein |
392246 |
0.01 |
| chr3_40085086_40085237 | 0.29 |
MYRIP |
myosin VIIA and Rab interacting protein |
56341 |
0.14 |
| chr2_68100081_68100232 | 0.29 |
C1D |
C1D nuclear receptor corepressor |
189958 |
0.03 |
| chr6_11973883_11974034 | 0.29 |
RP11-456H18.1 |
|
18287 |
0.24 |
| chr2_86289069_86289220 | 0.29 |
POLR1A |
polymerase (RNA) I polypeptide A, 194kDa |
44134 |
0.12 |
| chr2_26981757_26981908 | 0.29 |
SLC35F6 |
solute carrier family 35, member F6 |
5324 |
0.2 |
| chr12_76345410_76345561 | 0.29 |
ENSG00000243420 |
. |
6425 |
0.27 |
| chr10_5114268_5114419 | 0.29 |
AKR1C3 |
aldo-keto reductase family 1, member C3 |
5487 |
0.16 |
| chr10_33422454_33423119 | 0.29 |
ENSG00000263576 |
. |
35222 |
0.17 |
| chr3_124951031_124951182 | 0.29 |
SLC12A8 |
solute carrier family 12, member 8 |
19497 |
0.22 |
| chr12_91316241_91316392 | 0.29 |
CCER1 |
coiled-coil glutamate-rich protein 1 |
32637 |
0.22 |
| chr17_63660375_63660526 | 0.29 |
CEP112 |
centrosomal protein 112kDa |
23307 |
0.26 |
| chr17_79855827_79855978 | 0.29 |
NPB |
neuropeptide B |
2719 |
0.08 |
| chr10_134224918_134226015 | 0.29 |
PWWP2B |
PWWP domain containing 2B |
14764 |
0.17 |
| chr14_72117760_72117911 | 0.29 |
SIPA1L1 |
signal-induced proliferation-associated 1 like 1 |
32362 |
0.24 |
| chr8_75673011_75673162 | 0.29 |
PI15 |
peptidase inhibitor 15 |
63686 |
0.13 |
| chr1_112106719_112106870 | 0.29 |
ADORA3 |
adenosine A3 receptor |
210 |
0.93 |
| chr2_153226355_153226506 | 0.29 |
FMNL2 |
formin-like 2 |
34679 |
0.22 |
| chr15_37351175_37351326 | 0.29 |
ENSG00000206676 |
. |
8119 |
0.24 |
| chr2_236574553_236574704 | 0.29 |
AGAP1 |
ArfGAP with GTPase domain, ankyrin repeat and PH domain 1 |
3620 |
0.34 |
| chr6_125269183_125269334 | 0.29 |
RNF217 |
ring finger protein 217 |
14433 |
0.24 |
| chr2_206970822_206970973 | 0.29 |
AC007383.4 |
|
9374 |
0.14 |
| chr7_92645118_92645650 | 0.29 |
ENSG00000266794 |
. |
44771 |
0.17 |
| chr21_29050083_29050234 | 0.29 |
ADAMTS5 |
ADAM metallopeptidase with thrombospondin type 1 motif, 5 |
711326 |
0.0 |
| chr3_17082338_17082489 | 0.29 |
PLCL2-AS1 |
PLCL2 antisense RNA 1 |
3271 |
0.3 |
| chr1_180997388_180997539 | 0.29 |
STX6 |
syntaxin 6 |
5416 |
0.18 |
| chr10_10834102_10834253 | 0.29 |
CELF2 |
CUGBP, Elav-like family member 2 |
213082 |
0.02 |
| chr22_36748127_36749442 | 0.29 |
MYH9 |
myosin, heavy chain 9, non-muscle |
12372 |
0.18 |
| chr2_161162069_161162404 | 0.28 |
ENSG00000252465 |
. |
91243 |
0.08 |
| chr8_67443611_67443762 | 0.28 |
ENSG00000206949 |
. |
30140 |
0.15 |
| chr4_151067034_151067185 | 0.28 |
ENSG00000238721 |
. |
63749 |
0.13 |
| chr2_206621541_206621692 | 0.28 |
AC007362.3 |
|
7061 |
0.28 |
| chr2_205416081_205416232 | 0.28 |
PARD3B |
par-3 family cell polarity regulator beta |
5433 |
0.36 |
| chr18_32287372_32287523 | 0.28 |
DTNA |
dystrobrevin, alpha |
2801 |
0.37 |
| chr21_28673144_28673295 | 0.28 |
ADAMTS5 |
ADAM metallopeptidase with thrombospondin type 1 motif, 5 |
334387 |
0.01 |
| chr3_111527873_111528024 | 0.28 |
PHLDB2 |
pleckstrin homology-like domain, family B, member 2 |
50079 |
0.16 |
| chr15_30180552_30180703 | 0.28 |
RP11-680F8.3 |
|
65793 |
0.11 |
| chr10_133761353_133761504 | 0.28 |
PPP2R2D |
protein phosphatase 2, regulatory subunit B, delta |
7895 |
0.25 |
| chr17_48104312_48105191 | 0.28 |
ITGA3 |
integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor) |
28588 |
0.11 |
| chr17_27952739_27952890 | 0.28 |
CORO6 |
coronin 6 |
2892 |
0.11 |
| chrX_39832066_39832217 | 0.28 |
BCOR |
BCL6 corepressor |
90049 |
0.09 |
| chr7_98047722_98048054 | 0.28 |
BAIAP2L1 |
BAI1-associated protein 2-like 1 |
17508 |
0.24 |
| chr3_152049363_152049525 | 0.28 |
TMEM14E |
transmembrane protein 14E |
9335 |
0.23 |
| chr2_225664643_225664794 | 0.28 |
DOCK10 |
dedicator of cytokinesis 10 |
12883 |
0.31 |
| chr14_102537656_102538063 | 0.28 |
RP11-1017G21.4 |
|
5294 |
0.17 |
| chr5_83689497_83689648 | 0.28 |
CTD-2269F5.1 |
|
8275 |
0.25 |
| chr16_69074630_69074781 | 0.28 |
ENSG00000202497 |
. |
33647 |
0.14 |
| chr6_144737070_144737348 | 0.28 |
UTRN |
utrophin |
71972 |
0.12 |
| chr2_56113502_56113653 | 0.28 |
EFEMP1 |
EGF containing fibulin-like extracellular matrix protein 1 |
36779 |
0.17 |
| chr11_101987178_101987329 | 0.28 |
YAP1 |
Yes-associated protein 1 |
4008 |
0.2 |
| chr4_123795223_123795374 | 0.28 |
RP11-170N16.3 |
|
10569 |
0.18 |
| chr3_30323729_30323880 | 0.28 |
ENSG00000199927 |
. |
22223 |
0.27 |
| chr1_85755499_85756121 | 0.28 |
ENSG00000264380 |
. |
5517 |
0.15 |
| chr7_107670861_107671012 | 0.28 |
ENSG00000216085 |
. |
6445 |
0.18 |
| chr18_20898560_20898711 | 0.28 |
RP11-17J14.2 |
|
58317 |
0.13 |
| chr1_100012381_100012668 | 0.28 |
PALMD |
palmdelphin |
98975 |
0.08 |
| chr2_196602136_196602287 | 0.28 |
DNAH7 |
dynein, axonemal, heavy chain 7 |
18711 |
0.24 |
| chr18_20124592_20124743 | 0.28 |
ENSG00000206827 |
. |
102878 |
0.07 |
| chr11_10948283_10948434 | 0.28 |
ZBED5-AS1 |
ZBED5 antisense RNA 1 |
61550 |
0.11 |
| chr8_13130697_13130848 | 0.28 |
DLC1 |
deleted in liver cancer 1 |
3283 |
0.34 |
| chr8_119890733_119890884 | 0.28 |
TNFRSF11B |
tumor necrosis factor receptor superfamily, member 11b |
73631 |
0.11 |
| chr1_41567718_41567869 | 0.28 |
SCMH1 |
sex comb on midleg homolog 1 (Drosophila) |
57812 |
0.13 |
| chr12_76404430_76404581 | 0.28 |
RP11-290L1.3 |
|
19769 |
0.17 |
| chr15_39187982_39188133 | 0.28 |
C15orf53 |
chromosome 15 open reading frame 53 |
199258 |
0.03 |
| chr4_123505443_123505702 | 0.28 |
IL21-AS1 |
IL21 antisense RNA 1 |
34566 |
0.18 |
| chr12_28703830_28703981 | 0.28 |
CCDC91 |
coiled-coil domain containing 91 |
1908 |
0.45 |
| chr17_39114507_39114658 | 0.28 |
KRT39 |
keratin 39 |
8562 |
0.08 |
| chr3_100784256_100784407 | 0.27 |
ABI3BP |
ABI family, member 3 (NESH) binding protein |
71972 |
0.11 |
| chr7_33684637_33684788 | 0.27 |
RP11-89N17.1 |
HCG1643653; Uncharacterized protein |
80881 |
0.1 |
| chr7_65520174_65520325 | 0.27 |
ENSG00000252997 |
. |
1056 |
0.49 |
| chr3_123265492_123265643 | 0.27 |
PTPLB |
protein tyrosine phosphatase-like (proline instead of catalytic arginine), member b |
37835 |
0.15 |
| chr15_48857629_48857780 | 0.27 |
FBN1 |
fibrillin 1 |
80214 |
0.1 |
| chr1_244516671_244516927 | 0.27 |
C1orf100 |
chromosome 1 open reading frame 100 |
862 |
0.69 |
| chr1_184132614_184132871 | 0.27 |
ENSG00000199840 |
. |
8215 |
0.28 |
| chr5_86490215_86490366 | 0.27 |
AC008394.1 |
Uncharacterized protein |
44532 |
0.14 |
| chr18_32287605_32287756 | 0.27 |
DTNA |
dystrobrevin, alpha |
2568 |
0.38 |
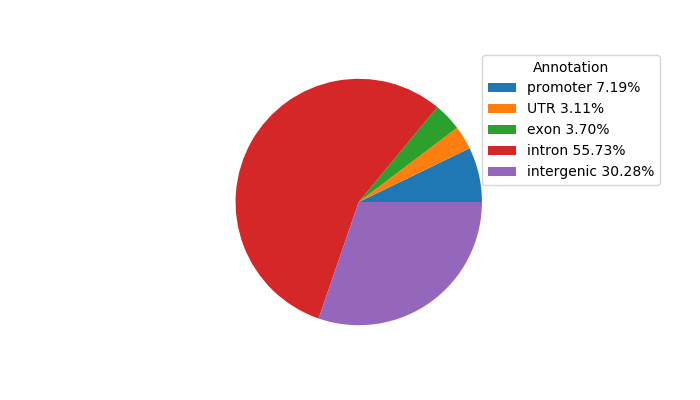
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:1900116 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.3 | 0.3 | GO:0072112 | renal filtration cell differentiation(GO:0061318) glomerular epithelium development(GO:0072010) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.3 | 0.8 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.2 | 0.8 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.2 | 0.6 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.2 | 0.5 | GO:0007132 | meiotic metaphase I(GO:0007132) |
| 0.2 | 1.0 | GO:0061001 | dendritic spine morphogenesis(GO:0060997) regulation of dendritic spine morphogenesis(GO:0061001) dendritic spine organization(GO:0097061) |
| 0.2 | 0.5 | GO:0044413 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.2 | 0.6 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.2 | 0.6 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.1 | 0.6 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.1 | 0.4 | GO:0010766 | negative regulation of sodium ion transport(GO:0010766) |
| 0.1 | 0.4 | GO:0002689 | negative regulation of leukocyte chemotaxis(GO:0002689) |
| 0.1 | 0.4 | GO:0031946 | regulation of glucocorticoid biosynthetic process(GO:0031946) |
| 0.1 | 0.5 | GO:0043497 | regulation of protein heterodimerization activity(GO:0043497) |
| 0.1 | 0.6 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.1 | 0.6 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) negative regulation of signal transduction by p53 class mediator(GO:1901797) |
| 0.1 | 0.4 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.1 | 0.4 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.1 | 0.2 | GO:0050942 | positive regulation of melanocyte differentiation(GO:0045636) positive regulation of pigment cell differentiation(GO:0050942) |
| 0.1 | 0.3 | GO:0060831 | smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:0060831) |
| 0.1 | 0.9 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.1 | 0.4 | GO:0010837 | regulation of keratinocyte proliferation(GO:0010837) |
| 0.1 | 0.4 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.1 | 0.5 | GO:0060638 | mesenchymal-epithelial cell signaling(GO:0060638) |
| 0.1 | 1.1 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.1 | 0.2 | GO:0035511 | oxidative DNA demethylation(GO:0035511) |
| 0.1 | 0.3 | GO:0002551 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.1 | 0.2 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.1 | 0.3 | GO:0009169 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.1 | 0.1 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.1 | 0.3 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.1 | 0.2 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.1 | 0.1 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
| 0.1 | 0.3 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.1 | 0.2 | GO:0060083 | smooth muscle contraction involved in micturition(GO:0060083) |
| 0.1 | 0.2 | GO:0061043 | vascular wound healing(GO:0061042) regulation of vascular wound healing(GO:0061043) |
| 0.1 | 0.2 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.1 | 0.1 | GO:0060129 | thyroid-stimulating hormone-secreting cell differentiation(GO:0060129) |
| 0.1 | 0.1 | GO:0060839 | endothelial cell fate commitment(GO:0060839) |
| 0.1 | 0.2 | GO:0060206 | estrous cycle phase(GO:0060206) |
| 0.1 | 0.1 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.1 | 0.1 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.1 | 0.1 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.1 | 0.2 | GO:0002913 | lymphocyte anergy(GO:0002249) regulation of T cell anergy(GO:0002667) positive regulation of T cell anergy(GO:0002669) T cell anergy(GO:0002870) regulation of lymphocyte anergy(GO:0002911) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.1 | 0.1 | GO:0033091 | positive regulation of immature T cell proliferation(GO:0033091) |
| 0.1 | 0.2 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.1 | 0.2 | GO:0090273 | regulation of somatostatin secretion(GO:0090273) |
| 0.1 | 0.2 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.1 | 1.3 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.1 | 0.1 | GO:0043173 | nucleotide salvage(GO:0043173) |
| 0.1 | 0.2 | GO:0071436 | sodium ion export(GO:0071436) |
| 0.1 | 0.2 | GO:0042339 | keratan sulfate metabolic process(GO:0042339) |
| 0.1 | 0.2 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.1 | 0.4 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.1 | 0.2 | GO:0010834 | obsolete telomere maintenance via telomere shortening(GO:0010834) |
| 0.1 | 0.3 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.1 | 0.1 | GO:0060197 | cloaca development(GO:0035844) cloacal septation(GO:0060197) |
| 0.1 | 0.1 | GO:0033689 | negative regulation of osteoblast proliferation(GO:0033689) |
| 0.1 | 0.2 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.1 | 0.5 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.1 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.3 | GO:0042487 | regulation of odontogenesis of dentin-containing tooth(GO:0042487) |
| 0.0 | 0.0 | GO:0021823 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) substrate-dependent cerebral cortex tangential migration(GO:0021825) postnatal olfactory bulb interneuron migration(GO:0021827) |
| 0.0 | 0.2 | GO:2000400 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.0 | 0.0 | GO:0035385 | Roundabout signaling pathway(GO:0035385) |
| 0.0 | 0.1 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.1 | GO:0006166 | purine ribonucleoside salvage(GO:0006166) |
| 0.0 | 0.1 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.0 | 0.3 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.0 | 0.3 | GO:0008634 | obsolete negative regulation of survival gene product expression(GO:0008634) |
| 0.0 | 0.4 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.2 | GO:0043383 | negative T cell selection(GO:0043383) |
| 0.0 | 0.1 | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.0 | 0.2 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.3 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.0 | 0.2 | GO:0051798 | positive regulation of hair cycle(GO:0042635) positive regulation of hair follicle development(GO:0051798) |
| 0.0 | 0.4 | GO:0048844 | artery morphogenesis(GO:0048844) |
| 0.0 | 0.1 | GO:0010454 | negative regulation of cell fate commitment(GO:0010454) |
| 0.0 | 0.1 | GO:0048245 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.0 | 0.1 | GO:0001743 | optic placode formation(GO:0001743) |
| 0.0 | 0.1 | GO:0014721 | voluntary skeletal muscle contraction(GO:0003010) twitch skeletal muscle contraction(GO:0014721) |
| 0.0 | 0.1 | GO:0071803 | regulation of podosome assembly(GO:0071801) positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.2 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.2 | GO:0051124 | synaptic growth at neuromuscular junction(GO:0051124) |
| 0.0 | 0.0 | GO:0033079 | immature T cell proliferation(GO:0033079) |
| 0.0 | 0.1 | GO:0046322 | negative regulation of fatty acid oxidation(GO:0046322) |
| 0.0 | 1.1 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.1 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.1 | GO:0003308 | negative regulation of Wnt signaling pathway involved in heart development(GO:0003308) |
| 0.0 | 0.2 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.2 | GO:0045989 | positive regulation of striated muscle contraction(GO:0045989) |
| 0.0 | 0.6 | GO:0045104 | intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 | 0.1 | GO:2000095 | regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.0 | 0.1 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.0 | 0.1 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) |
| 0.0 | 0.1 | GO:0060413 | atrial septum development(GO:0003283) atrial septum morphogenesis(GO:0060413) |
| 0.0 | 0.1 | GO:0001302 | replicative cell aging(GO:0001302) |
| 0.0 | 0.0 | GO:0032693 | negative regulation of interleukin-10 production(GO:0032693) |
| 0.0 | 0.2 | GO:0046719 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) regulation by virus of viral protein levels in host cell(GO:0046719) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.0 | 0.0 | GO:0060449 | bud elongation involved in lung branching(GO:0060449) |
| 0.0 | 0.1 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.1 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.1 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.1 | GO:0060340 | positive regulation of type I interferon-mediated signaling pathway(GO:0060340) |
| 0.0 | 0.1 | GO:1903054 | regulation of extracellular matrix disassembly(GO:0010715) negative regulation of extracellular matrix disassembly(GO:0010716) regulation of extracellular matrix organization(GO:1903053) negative regulation of extracellular matrix organization(GO:1903054) |
| 0.0 | 0.0 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.0 | GO:0061046 | regulation of branching involved in lung morphogenesis(GO:0061046) iris morphogenesis(GO:0061072) |
| 0.0 | 0.2 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.1 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.1 | GO:0015868 | purine ribonucleotide transport(GO:0015868) |
| 0.0 | 0.1 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 0.2 | GO:0001547 | antral ovarian follicle growth(GO:0001547) |
| 0.0 | 0.1 | GO:0046476 | glycosylceramide biosynthetic process(GO:0046476) |
| 0.0 | 0.1 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.0 | 0.1 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.5 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.1 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.2 | GO:0021591 | ventricular system development(GO:0021591) |
| 0.0 | 0.2 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 0.1 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.0 | 0.1 | GO:0055089 | fatty acid homeostasis(GO:0055089) |
| 0.0 | 0.1 | GO:0060390 | regulation of SMAD protein import into nucleus(GO:0060390) positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.0 | 0.2 | GO:0070120 | ciliary neurotrophic factor-mediated signaling pathway(GO:0070120) |
| 0.0 | 0.1 | GO:0045657 | positive regulation of monocyte differentiation(GO:0045657) |
| 0.0 | 0.1 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 0.1 | GO:0060317 | cardiac epithelial to mesenchymal transition(GO:0060317) |
| 0.0 | 0.1 | GO:0071043 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.0 | 0.0 | GO:0051305 | chromosome movement towards spindle pole(GO:0051305) |
| 0.0 | 0.1 | GO:0045046 | protein import into peroxisome membrane(GO:0045046) |
| 0.0 | 0.0 | GO:0060435 | bronchiole development(GO:0060435) |
| 0.0 | 0.3 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.1 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.0 | 0.1 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.0 | 0.2 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.0 | 0.2 | GO:0010898 | positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.0 | 0.1 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.0 | 0.1 | GO:0000492 | small nucleolar ribonucleoprotein complex assembly(GO:0000491) box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.0 | GO:0060751 | epithelial cell proliferation involved in mammary gland duct elongation(GO:0060750) branch elongation involved in mammary gland duct branching(GO:0060751) |
| 0.0 | 0.0 | GO:0046877 | regulation of saliva secretion(GO:0046877) |
| 0.0 | 0.1 | GO:0034653 | diterpenoid catabolic process(GO:0016103) terpenoid catabolic process(GO:0016115) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.2 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.1 | GO:0046628 | positive regulation of insulin receptor signaling pathway(GO:0046628) positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.0 | 0.1 | GO:0014912 | negative regulation of smooth muscle cell migration(GO:0014912) |
| 0.0 | 0.2 | GO:0071456 | cellular response to decreased oxygen levels(GO:0036294) cellular response to oxygen levels(GO:0071453) cellular response to hypoxia(GO:0071456) |
| 0.0 | 0.1 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.0 | 0.3 | GO:0046653 | tetrahydrofolate metabolic process(GO:0046653) |
| 0.0 | 0.2 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 0.1 | GO:0072048 | pattern specification involved in kidney development(GO:0061004) renal system pattern specification(GO:0072048) pattern specification involved in metanephros development(GO:0072268) |
| 0.0 | 0.2 | GO:0044091 | membrane biogenesis(GO:0044091) |
| 0.0 | 0.1 | GO:0090504 | wound healing, spreading of epidermal cells(GO:0035313) wound healing, spreading of cells(GO:0044319) epiboly(GO:0090504) epiboly involved in wound healing(GO:0090505) |
| 0.0 | 0.3 | GO:0090114 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) COPII-coated vesicle budding(GO:0090114) |
| 0.0 | 0.1 | GO:0030859 | polarized epithelial cell differentiation(GO:0030859) establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.0 | 0.1 | GO:0015840 | urea transport(GO:0015840) |
| 0.0 | 0.5 | GO:0048146 | positive regulation of fibroblast proliferation(GO:0048146) |
| 0.0 | 0.1 | GO:0009649 | entrainment of circadian clock(GO:0009649) |
| 0.0 | 0.1 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.0 | GO:0070874 | negative regulation of glycogen metabolic process(GO:0070874) |
| 0.0 | 0.1 | GO:0007500 | mesodermal cell fate determination(GO:0007500) |
| 0.0 | 0.1 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.0 | 0.1 | GO:0033603 | positive regulation of dopamine secretion(GO:0033603) |
| 0.0 | 0.0 | GO:0014002 | astrocyte development(GO:0014002) |
| 0.0 | 0.1 | GO:0042984 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.0 | 0.0 | GO:0003148 | outflow tract septum morphogenesis(GO:0003148) |
| 0.0 | 0.2 | GO:0006622 | protein targeting to lysosome(GO:0006622) protein targeting to vacuole(GO:0006623) protein localization to lysosome(GO:0061462) protein localization to vacuole(GO:0072665) establishment of protein localization to vacuole(GO:0072666) |
| 0.0 | 0.2 | GO:0043537 | negative regulation of blood vessel endothelial cell migration(GO:0043537) |
| 0.0 | 0.3 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.0 | GO:0042996 | regulation of Golgi to plasma membrane protein transport(GO:0042996) |
| 0.0 | 0.3 | GO:0007603 | phototransduction, visible light(GO:0007603) |
| 0.0 | 0.1 | GO:0090224 | regulation of spindle organization(GO:0090224) |
| 0.0 | 0.2 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 0.1 | GO:0048319 | axial mesoderm morphogenesis(GO:0048319) |
| 0.0 | 0.4 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 0.2 | GO:0032332 | positive regulation of chondrocyte differentiation(GO:0032332) |
| 0.0 | 0.1 | GO:0009415 | response to water(GO:0009415) |
| 0.0 | 0.1 | GO:0046607 | positive regulation of centrosome cycle(GO:0046607) |
| 0.0 | 0.1 | GO:0061081 | positive regulation of myeloid leukocyte cytokine production involved in immune response(GO:0061081) |
| 0.0 | 0.1 | GO:0033962 | cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.0 | 0.1 | GO:0051459 | corticotropin secretion(GO:0051458) regulation of corticotropin secretion(GO:0051459) positive regulation of corticotropin secretion(GO:0051461) |
| 0.0 | 0.1 | GO:0007619 | courtship behavior(GO:0007619) male courtship behavior(GO:0008049) |
| 0.0 | 0.1 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.1 | GO:0031641 | regulation of myelination(GO:0031641) |
| 0.0 | 0.1 | GO:0071680 | response to indole-3-methanol(GO:0071680) cellular response to indole-3-methanol(GO:0071681) |
| 0.0 | 0.0 | GO:0019471 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 0.0 | GO:0034139 | regulation of toll-like receptor 3 signaling pathway(GO:0034139) |
| 0.0 | 0.1 | GO:0060013 | righting reflex(GO:0060013) |
| 0.0 | 0.0 | GO:0042940 | D-amino acid transport(GO:0042940) |
| 0.0 | 1.2 | GO:0060271 | cilium morphogenesis(GO:0060271) |
| 0.0 | 0.0 | GO:0046639 | negative regulation of alpha-beta T cell differentiation(GO:0046639) |
| 0.0 | 0.0 | GO:0021631 | optic nerve morphogenesis(GO:0021631) |
| 0.0 | 0.1 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.1 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.0 | GO:0014819 | regulation of skeletal muscle contraction(GO:0014819) |
| 0.0 | 0.0 | GO:0016048 | detection of temperature stimulus(GO:0016048) detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.0 | 0.0 | GO:0043129 | surfactant homeostasis(GO:0043129) chemical homeostasis within a tissue(GO:0048875) |
| 0.0 | 0.2 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 0.0 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.0 | GO:0033683 | nucleotide-excision repair, DNA incision(GO:0033683) |
| 0.0 | 0.1 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.0 | 0.0 | GO:0060457 | negative regulation of digestive system process(GO:0060457) |
| 0.0 | 0.0 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 0.0 | 0.1 | GO:0046149 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.0 | 0.0 | GO:0060900 | embryonic camera-type eye formation(GO:0060900) |
| 0.0 | 0.1 | GO:0009820 | alkaloid metabolic process(GO:0009820) |
| 0.0 | 0.2 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.0 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.0 | 0.1 | GO:0046325 | negative regulation of glucose import(GO:0046325) |
| 0.0 | 0.1 | GO:0006534 | cysteine metabolic process(GO:0006534) |
| 0.0 | 0.0 | GO:0072033 | renal vesicle formation(GO:0072033) |
| 0.0 | 0.1 | GO:0042977 | activation of JAK2 kinase activity(GO:0042977) |
| 0.0 | 0.0 | GO:0034244 | negative regulation of DNA-templated transcription, elongation(GO:0032785) negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 0.1 | GO:0009200 | deoxyribonucleoside triphosphate metabolic process(GO:0009200) |
| 0.0 | 0.0 | GO:0046349 | amino sugar biosynthetic process(GO:0046349) |
| 0.0 | 0.1 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.1 | GO:0010224 | response to UV-B(GO:0010224) |
| 0.0 | 0.1 | GO:0010744 | positive regulation of macrophage derived foam cell differentiation(GO:0010744) |
| 0.0 | 0.1 | GO:0006734 | NADH metabolic process(GO:0006734) |
| 0.0 | 0.1 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.1 | GO:0021854 | hypothalamus development(GO:0021854) |
| 0.0 | 0.1 | GO:0090281 | negative regulation of calcium ion import(GO:0090281) negative regulation of cation transmembrane transport(GO:1904063) |
| 0.0 | 0.1 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.0 | 0.1 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.0 | 0.1 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.2 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.0 | 0.1 | GO:0032811 | negative regulation of epinephrine secretion(GO:0032811) |
| 0.0 | 0.0 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 0.1 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.0 | 0.0 | GO:0090083 | regulation of inclusion body assembly(GO:0090083) |
| 0.0 | 0.1 | GO:0021780 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) oligodendrocyte cell fate specification(GO:0021778) glial cell fate specification(GO:0021780) |
| 0.0 | 0.1 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.0 | 0.1 | GO:0014889 | muscle atrophy(GO:0014889) |
| 0.0 | 0.0 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.0 | 0.1 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.0 | 0.1 | GO:0032099 | negative regulation of appetite(GO:0032099) |
| 0.0 | 0.1 | GO:1901798 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) positive regulation of signal transduction by p53 class mediator(GO:1901798) |
| 0.0 | 0.1 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.1 | GO:0045197 | establishment or maintenance of epithelial cell apical/basal polarity(GO:0045197) |
| 0.0 | 0.0 | GO:0045915 | positive regulation of catecholamine metabolic process(GO:0045915) positive regulation of dopamine metabolic process(GO:0045964) |
| 0.0 | 0.0 | GO:0009111 | vitamin catabolic process(GO:0009111) fat-soluble vitamin catabolic process(GO:0042363) |
| 0.0 | 0.0 | GO:0032236 | obsolete positive regulation of calcium ion transport via store-operated calcium channel activity(GO:0032236) |
| 0.0 | 0.0 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.1 | GO:0009629 | response to gravity(GO:0009629) |
| 0.0 | 0.0 | GO:0045896 | regulation of transcription during mitosis(GO:0045896) positive regulation of transcription during mitosis(GO:0045897) |
| 0.0 | 0.0 | GO:0060029 | convergent extension involved in organogenesis(GO:0060029) |
| 0.0 | 0.1 | GO:0003084 | positive regulation of systemic arterial blood pressure(GO:0003084) |
| 0.0 | 0.1 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.1 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 0.0 | GO:0019730 | antimicrobial humoral response(GO:0019730) antibacterial humoral response(GO:0019731) |
| 0.0 | 0.4 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.0 | GO:0044321 | cellular response to leptin stimulus(GO:0044320) response to leptin(GO:0044321) |
| 0.0 | 0.1 | GO:0009103 | lipopolysaccharide biosynthetic process(GO:0009103) |
| 0.0 | 0.0 | GO:0033688 | regulation of osteoblast proliferation(GO:0033688) |
| 0.0 | 0.1 | GO:0010092 | organ induction(GO:0001759) specification of organ identity(GO:0010092) |
| 0.0 | 0.1 | GO:0000959 | mitochondrial RNA metabolic process(GO:0000959) transcription from mitochondrial promoter(GO:0006390) |
| 0.0 | 0.0 | GO:0072217 | negative regulation of metanephros development(GO:0072217) |
| 0.0 | 0.0 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.0 | 0.0 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.0 | 0.0 | GO:0051142 | regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) |
| 0.0 | 0.1 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.0 | 0.0 | GO:0042748 | circadian sleep/wake cycle, non-REM sleep(GO:0042748) regulation of circadian sleep/wake cycle, non-REM sleep(GO:0045188) |
| 0.0 | 0.1 | GO:0032455 | nerve growth factor processing(GO:0032455) |
| 0.0 | 0.0 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.0 | 0.0 | GO:0003321 | positive regulation of blood pressure by epinephrine-norepinephrine(GO:0003321) |
| 0.0 | 0.1 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.1 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.1 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.1 | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.0 | 0.1 | GO:0019098 | reproductive behavior(GO:0019098) |
| 0.0 | 0.4 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.0 | GO:0050651 | dermatan sulfate proteoglycan biosynthetic process(GO:0050651) |
| 0.0 | 0.0 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.1 | GO:0060688 | regulation of morphogenesis of a branching structure(GO:0060688) |
| 0.0 | 0.1 | GO:0061245 | establishment or maintenance of apical/basal cell polarity(GO:0035088) establishment or maintenance of bipolar cell polarity(GO:0061245) |
| 0.0 | 0.1 | GO:0048712 | negative regulation of astrocyte differentiation(GO:0048712) |
| 0.0 | 0.1 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.0 | 0.2 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.0 | GO:0010896 | regulation of triglyceride catabolic process(GO:0010896) |
| 0.0 | 0.0 | GO:0008300 | isoprenoid catabolic process(GO:0008300) |
| 0.0 | 0.0 | GO:0015864 | pyrimidine nucleoside transport(GO:0015864) |
| 0.0 | 0.0 | GO:1901978 | positive regulation of spindle checkpoint(GO:0090232) regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) positive regulation of cell cycle checkpoint(GO:1901978) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 0.0 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.0 | 0.1 | GO:0015858 | nucleoside transport(GO:0015858) |
| 0.0 | 0.1 | GO:1903963 | arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.0 | 0.0 | GO:0051386 | regulation of neurotrophin TRK receptor signaling pathway(GO:0051386) |
| 0.0 | 0.1 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.1 | GO:0032740 | positive regulation of interleukin-17 production(GO:0032740) |
| 0.0 | 0.1 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.0 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 0.1 | GO:0032964 | collagen biosynthetic process(GO:0032964) |
| 0.0 | 0.0 | GO:0032527 | protein exit from endoplasmic reticulum(GO:0032527) |
| 0.0 | 0.1 | GO:0044550 | secondary metabolite biosynthetic process(GO:0044550) |
| 0.0 | 0.0 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.0 | 0.0 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) |
| 0.0 | 0.0 | GO:0021800 | cerebral cortex tangential migration(GO:0021800) |
| 0.0 | 0.1 | GO:0042461 | photoreceptor cell development(GO:0042461) |
| 0.0 | 0.0 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.0 | 0.1 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 | 0.0 | GO:0060445 | branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.0 | 0.0 | GO:0042482 | positive regulation of odontogenesis(GO:0042482) positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 0.0 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.0 | 0.0 | GO:0006207 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.0 | 0.0 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.0 | 0.0 | GO:0090047 | obsolete positive regulation of transcription regulator activity(GO:0090047) |
| 0.0 | 0.1 | GO:0042517 | positive regulation of tyrosine phosphorylation of Stat3 protein(GO:0042517) |
| 0.0 | 0.0 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.0 | GO:0031427 | response to methotrexate(GO:0031427) |
| 0.0 | 0.0 | GO:0045822 | negative regulation of heart contraction(GO:0045822) |
| 0.0 | 0.5 | GO:0030198 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 | 0.2 | GO:0048009 | insulin-like growth factor receptor signaling pathway(GO:0048009) |
| 0.0 | 0.0 | GO:0070857 | regulation of bile acid biosynthetic process(GO:0070857) regulation of bile acid metabolic process(GO:1904251) |
| 0.0 | 0.0 | GO:0016093 | polyprenol metabolic process(GO:0016093) dolichol metabolic process(GO:0019348) |
| 0.0 | 0.0 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.0 | 0.0 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 0.0 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 0.0 | 0.0 | GO:0002023 | reduction of food intake in response to dietary excess(GO:0002023) |
| 0.0 | 0.0 | GO:0090197 | chemokine secretion(GO:0090195) regulation of chemokine secretion(GO:0090196) positive regulation of chemokine secretion(GO:0090197) |
| 0.0 | 0.0 | GO:0006498 | N-terminal protein lipidation(GO:0006498) |
| 0.0 | 0.2 | GO:0044349 | nucleotide-excision repair, DNA damage removal(GO:0000718) DNA excision(GO:0044349) |
| 0.0 | 0.1 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.0 | GO:0072205 | metanephric collecting duct development(GO:0072205) |
| 0.0 | 0.1 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.0 | 0.0 | GO:0032224 | positive regulation of synaptic transmission, cholinergic(GO:0032224) |
| 0.0 | 0.1 | GO:0051775 | response to redox state(GO:0051775) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.2 | 0.5 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.1 | 0.7 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 0.7 | GO:0071437 | invadopodium(GO:0071437) |
| 0.1 | 0.4 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 0.1 | 0.4 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.1 | 0.4 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 0.3 | GO:0005954 | calcium- and calmodulin-dependent protein kinase complex(GO:0005954) |
| 0.1 | 0.9 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 0.6 | GO:0098644 | complex of collagen trimers(GO:0098644) |
| 0.1 | 0.2 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.1 | 0.6 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 0.5 | GO:0043256 | laminin complex(GO:0043256) |
| 0.1 | 0.3 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 0.1 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.1 | 0.2 | GO:0032449 | CBM complex(GO:0032449) |
| 0.1 | 0.2 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 0.4 | GO:0030122 | AP-2 adaptor complex(GO:0030122) clathrin coat of endocytic vesicle(GO:0030128) |
| 0.1 | 0.2 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.1 | 0.2 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.1 | 0.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.3 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.4 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.3 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.4 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 1.2 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.1 | GO:0071942 | XPC complex(GO:0071942) |
| 0.0 | 0.0 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.7 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.3 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.3 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.3 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.3 | GO:0014704 | intercalated disc(GO:0014704) cell-cell contact zone(GO:0044291) |
| 0.0 | 0.1 | GO:0036454 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.0 | 0.1 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.1 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.0 | 0.2 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 1.5 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 2.9 | GO:0030016 | myofibril(GO:0030016) |
| 0.0 | 0.1 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 1.2 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.3 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.2 | GO:0016012 | dystroglycan complex(GO:0016011) sarcoglycan complex(GO:0016012) |
| 0.0 | 0.1 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.0 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 0.1 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.1 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.3 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.1 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.1 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 4.4 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.3 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.2 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.0 | 0.1 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.0 | 0.1 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.7 | GO:0044309 | neuron spine(GO:0044309) |
| 0.0 | 0.1 | GO:0042598 | obsolete vesicular fraction(GO:0042598) |
| 0.0 | 0.1 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.1 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.0 | 0.2 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.1 | GO:0031332 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 0.1 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 0.0 | 0.1 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.0 | 0.1 | GO:0031512 | motile primary cilium(GO:0031512) |
| 0.0 | 0.1 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.1 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.0 | 0.5 | GO:0008305 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 0.1 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.0 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 0.3 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.1 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 0.0 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.4 | GO:0034704 | calcium channel complex(GO:0034704) |
| 0.0 | 0.1 | GO:0030990 | intraciliary transport particle(GO:0030990) |
| 0.0 | 0.0 | GO:0033185 | mannosyltransferase complex(GO:0031501) dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 0.7 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 0.1 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 1.3 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.0 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.1 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.0 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.1 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.0 | 0.1 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.0 | 0.6 | GO:0099572 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.0 | 0.0 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.1 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.1 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.2 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.1 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 0.1 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.0 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.1 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.0 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.0 | GO:0098878 | ionotropic glutamate receptor complex(GO:0008328) neurotransmitter receptor complex(GO:0098878) |
| 0.0 | 0.1 | GO:0005678 | obsolete chromatin assembly complex(GO:0005678) |
| 0.0 | 0.0 | GO:0033268 | node of Ranvier(GO:0033268) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.2 | 0.7 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.2 | 0.5 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 0.6 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.1 | 1.0 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.1 | 0.7 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 0.3 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.1 | 0.3 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.1 | 0.1 | GO:0043559 | insulin binding(GO:0043559) |
| 0.1 | 0.4 | GO:0042805 | actinin binding(GO:0042805) |
| 0.1 | 0.3 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.1 | 0.3 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.1 | 0.3 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.1 | 0.3 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.1 | 0.2 | GO:0035514 | DNA demethylase activity(GO:0035514) DNA-N1-methyladenine dioxygenase activity(GO:0043734) |
| 0.1 | 0.9 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 0.2 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.1 | 0.2 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.1 | 3.0 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 0.4 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.1 | 0.3 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 0.3 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.1 | 1.0 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 0.2 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 1.5 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 0.2 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.1 | 0.2 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 0.2 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.1 | 0.2 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) bisphosphoglycerate 2-phosphatase activity(GO:0004083) phosphoglycerate mutase activity(GO:0004619) |
| 0.1 | 0.7 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 0.1 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.1 | 0.2 | GO:0004516 | nicotinate phosphoribosyltransferase activity(GO:0004516) |
| 0.1 | 0.3 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.1 | 0.2 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 0.2 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.1 | 0.2 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 0.4 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 0.2 | GO:0004470 | malic enzyme activity(GO:0004470) |
| 0.1 | 0.2 | GO:0016901 | oxidoreductase activity, acting on the CH-OH group of donors, quinone or similar compound as acceptor(GO:0016901) |
| 0.1 | 0.3 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.1 | 0.2 | GO:0005350 | pyrimidine nucleobase transmembrane transporter activity(GO:0005350) |
| 0.1 | 0.1 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.1 | 0.6 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.1 | 0.2 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 0.1 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.1 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.1 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.0 | 0.0 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.2 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.0 | 0.3 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.9 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.1 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) xenobiotic transporter activity(GO:0042910) |
| 0.0 | 0.2 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.3 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.0 | 0.0 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.1 | GO:0015106 | bicarbonate transmembrane transporter activity(GO:0015106) |
| 0.0 | 0.2 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.1 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.0 | 0.2 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.4 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.7 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.3 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.0 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.3 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.1 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.1 | GO:0016623 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.0 | 0.1 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.1 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.1 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.1 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.0 | 0.2 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.0 | 0.2 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.2 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.2 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.3 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.1 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.0 | 0.2 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.0 | 0.2 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.1 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.2 | GO:0016847 | 1-aminocyclopropane-1-carboxylate synthase activity(GO:0016847) |
| 0.0 | 0.2 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.1 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.0 | 0.1 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.2 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.1 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.2 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.5 | GO:0005024 | transforming growth factor beta-activated receptor activity(GO:0005024) |
| 0.0 | 0.1 | GO:0000404 | heteroduplex DNA loop binding(GO:0000404) |
| 0.0 | 0.1 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.2 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.0 | 0.3 | GO:0005283 | sodium:amino acid symporter activity(GO:0005283) |
| 0.0 | 0.2 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.1 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.1 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 0.0 | 0.1 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.0 | 0.1 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.1 | GO:0004339 | glucan 1,4-alpha-glucosidase activity(GO:0004339) |
| 0.0 | 0.1 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.2 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.4 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.1 | GO:0004946 | bombesin receptor activity(GO:0004946) |
| 0.0 | 0.2 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.1 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.0 | 0.1 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.1 | GO:0019798 | procollagen-proline 3-dioxygenase activity(GO:0019797) procollagen-proline dioxygenase activity(GO:0019798) peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.0 | 0.1 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.0 | GO:0034416 | bisphosphoglycerate phosphatase activity(GO:0034416) |
| 0.0 | 0.0 | GO:0031543 | peptidyl-proline dioxygenase activity(GO:0031543) |
| 0.0 | 0.1 | GO:0033765 | steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) |
| 0.0 | 0.1 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.1 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.1 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.0 | 0.1 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) |
| 0.0 | 0.1 | GO:0055106 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.2 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.0 | 0.1 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.2 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.1 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 0.4 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 0.0 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.0 | 0.2 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.0 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.1 | GO:0042808 | obsolete neuronal Cdc2-like kinase binding(GO:0042808) |
| 0.0 | 0.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.1 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.2 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.0 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.0 | 0.1 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 1.3 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.1 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.1 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.1 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.1 | GO:0016421 | CoA carboxylase activity(GO:0016421) |
| 0.0 | 0.2 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.1 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.1 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.1 | GO:0030354 | melanin-concentrating hormone activity(GO:0030354) |
| 0.0 | 0.2 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) lysophosphatidic acid acyltransferase activity(GO:0042171) lysophospholipid acyltransferase activity(GO:0071617) |
| 0.0 | 0.0 | GO:0005345 | purine nucleobase transmembrane transporter activity(GO:0005345) |
| 0.0 | 0.3 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 0.3 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.0 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.0 | 0.3 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.2 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.1 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.1 | GO:0015211 | purine nucleoside transmembrane transporter activity(GO:0015211) |
| 0.0 | 0.1 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.3 | GO:0016896 | exoribonuclease activity, producing 5'-phosphomonoesters(GO:0016896) |
| 0.0 | 0.2 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.0 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.0 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.1 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.0 | 0.1 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.1 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.1 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.0 | GO:0034187 | obsolete apolipoprotein E binding(GO:0034187) |
| 0.0 | 0.0 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.0 | 0.1 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 0.0 | 0.0 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.0 | 0.0 | GO:0004844 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.0 | 0.1 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.0 | GO:0030151 | molybdenum ion binding(GO:0030151) |
| 0.0 | 0.8 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 0.2 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.0 | 0.1 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.2 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 0.2 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.1 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.0 | 0.1 | GO:0016944 | obsolete RNA polymerase II transcription elongation factor activity(GO:0016944) |
| 0.0 | 0.0 | GO:1990939 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) ATP-dependent microtubule motor activity(GO:1990939) |
| 0.0 | 0.1 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0043498 | obsolete cell surface binding(GO:0043498) |
| 0.0 | 0.1 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.0 | 0.1 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 0.0 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.2 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.0 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.0 | 0.0 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 0.0 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.0 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.0 | 0.0 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.1 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.1 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.0 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.0 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.0 | 0.0 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.1 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.0 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.1 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.0 | 0.0 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.0 | GO:0098518 | polynucleotide phosphatase activity(GO:0098518) |
| 0.0 | 0.0 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.0 | 0.1 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.0 | 0.1 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.0 | 0.0 | GO:0008898 | S-adenosylmethionine-homocysteine S-methyltransferase activity(GO:0008898) |
| 0.0 | 0.1 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.0 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.0 | GO:0034618 | arginine binding(GO:0034618) |
| 0.0 | 0.0 | GO:0032296 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 1.4 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 1.1 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 0.9 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 0.6 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 2.3 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.7 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.1 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.0 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 0.9 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.0 | 0.5 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.3 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 1.2 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.0 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.0 | 0.4 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.1 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 0.1 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.7 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.1 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 4.3 | NABA CORE MATRISOME | Ensemble of genes encoding core extracellular matrix including ECM glycoproteins, collagens and proteoglycans |
| 0.0 | 0.5 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.1 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.5 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.0 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 0.0 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 0.1 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 0.1 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.2 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.5 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.3 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.2 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.4 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.3 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.6 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.5 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.7 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.1 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.2 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.2 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.3 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.1 | 0.7 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 2.0 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 0.9 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 0.1 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.1 | 0.3 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.1 | 0.5 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 0.5 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 2.7 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.0 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.5 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.8 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.6 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.4 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.4 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.1 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.3 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.1 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.0 | 0.0 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.4 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.2 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.2 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.0 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.6 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.3 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.2 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.0 | 0.0 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.5 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.3 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 0.3 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.0 | 0.0 | REACTOME SHC MEDIATED SIGNALLING | Genes involved in SHC-mediated signalling |
| 0.0 | 0.2 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.5 | REACTOME FRS2 MEDIATED CASCADE | Genes involved in FRS2-mediated cascade |
| 0.0 | 0.4 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.6 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.3 | REACTOME PROCESSIVE SYNTHESIS ON THE LAGGING STRAND | Genes involved in Processive synthesis on the lagging strand |
| 0.0 | 0.4 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.5 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.1 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.2 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.0 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.0 | 0.1 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 0.1 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.0 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.0 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.1 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.1 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.1 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.1 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 0.6 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.1 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.8 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 0.1 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 0.2 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.1 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.2 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.2 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.0 | REACTOME CELL DEATH SIGNALLING VIA NRAGE NRIF AND NADE | Genes involved in Cell death signalling via NRAGE, NRIF and NADE |
| 0.0 | 0.1 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.1 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 0.0 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.0 | 0.0 | REACTOME NRIF SIGNALS CELL DEATH FROM THE NUCLEUS | Genes involved in NRIF signals cell death from the nucleus |
| 0.0 | 0.1 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.1 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.0 | 0.0 | REACTOME CA DEPENDENT EVENTS | Genes involved in Ca-dependent events |