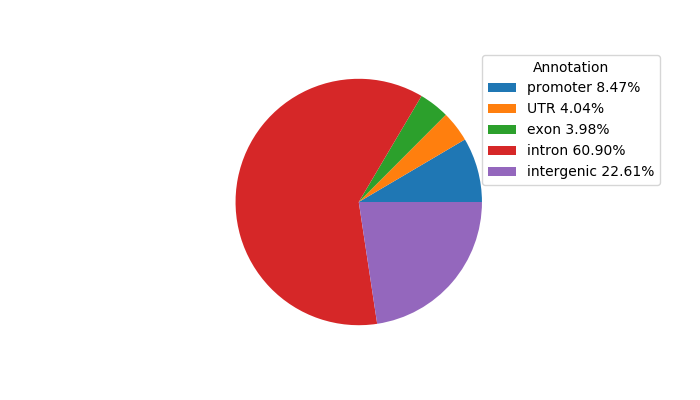
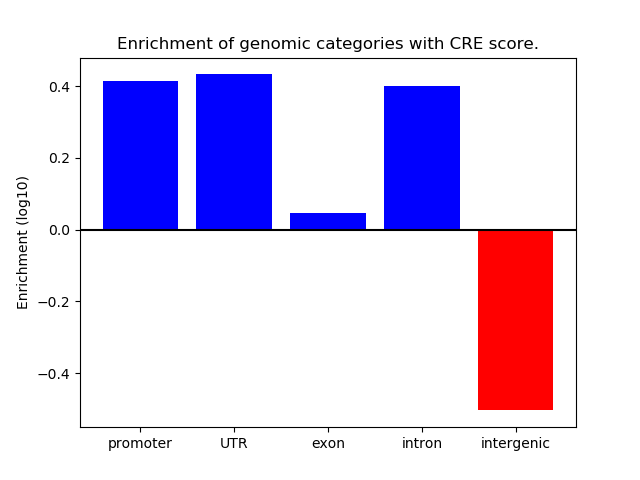
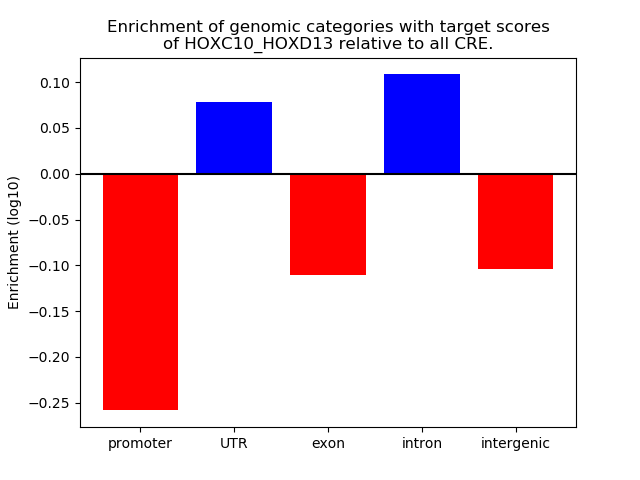
Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
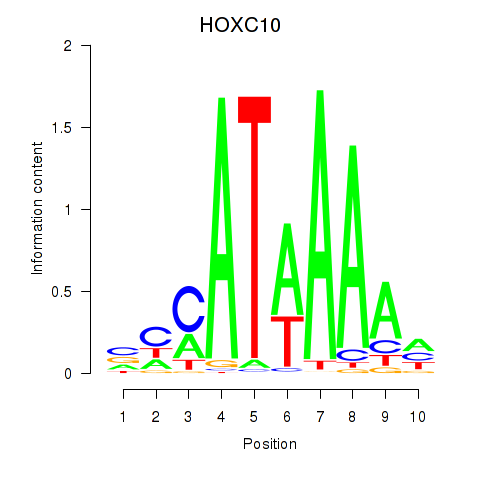
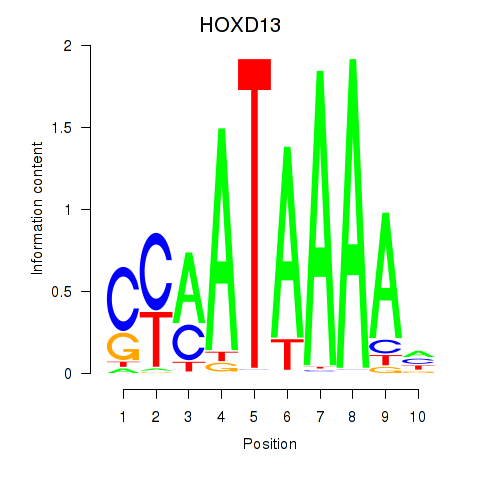
Results for HOXC10_HOXD13
Z-value: 0.71
Transcription factors associated with HOXC10_HOXD13
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXC10
|
ENSG00000180818.4 | homeobox C10 |
|
HOXD13
|
ENSG00000128714.5 | homeobox D13 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr12_54378978_54379226 | HOXC10 | 132 | 0.523416 | -0.36 | 3.4e-01 | Click! |
| chr2_176953314_176953465 | HOXD13 | 4230 | 0.101143 | 0.73 | 2.7e-02 | Click! |
| chr2_176958437_176958600 | HOXD13 | 899 | 0.355888 | 0.53 | 1.4e-01 | Click! |
| chr2_176957910_176958434 | HOXD13 | 553 | 0.559704 | -0.38 | 3.1e-01 | Click! |
| chr2_176956847_176957065 | HOXD13 | 663 | 0.484831 | 0.28 | 4.6e-01 | Click! |
| chr2_176953141_176953292 | HOXD13 | 4403 | 0.099735 | 0.25 | 5.1e-01 | Click! |
Activity of the HOXC10_HOXD13 motif across conditions
Conditions sorted by the z-value of the HOXC10_HOXD13 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
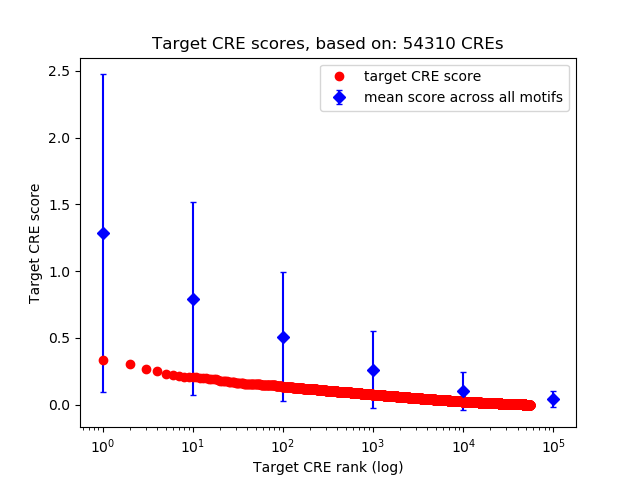
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr2_198060963_198061183 | 0.33 |
AC013264.2 |
|
1642 |
0.33 |
| chr12_837359_837675 | 0.30 |
WNK1 |
WNK lysine deficient protein kinase 1 |
24242 |
0.18 |
| chr15_60828599_60828948 | 0.26 |
CTD-2501E16.2 |
|
6601 |
0.21 |
| chr3_71461863_71462033 | 0.25 |
FOXP1 |
forkhead box P1 |
108037 |
0.07 |
| chr5_100224229_100224437 | 0.23 |
ST8SIA4 |
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 4 |
14585 |
0.28 |
| chr13_77882140_77882385 | 0.22 |
MYCBP2 |
MYC binding protein 2, E3 ubiquitin protein ligase |
18552 |
0.28 |
| chr1_184718801_184718952 | 0.22 |
EDEM3 |
ER degradation enhancer, mannosidase alpha-like 3 |
4825 |
0.26 |
| chr6_112035271_112035422 | 0.21 |
FYN |
FYN oncogene related to SRC, FGR, YES |
5919 |
0.26 |
| chr3_141270944_141271180 | 0.21 |
RASA2-IT1 |
RASA2 intronic transcript 1 (non-protein coding) |
27087 |
0.21 |
| chr9_98486374_98486879 | 0.21 |
DKFZP434H0512 |
Protein LOC100506667; Putative uncharacterized protein DKFZp434H0512 |
47979 |
0.17 |
| chr17_46375004_46375203 | 0.21 |
AC090627.1 |
|
3366 |
0.23 |
| chr1_100847278_100847500 | 0.20 |
ENSG00000216067 |
. |
3058 |
0.27 |
| chr1_86854784_86854976 | 0.20 |
ODF2L |
outer dense fiber of sperm tails 2-like |
6001 |
0.25 |
| chr11_88576113_88576264 | 0.20 |
GRM5 |
glutamate receptor, metabotropic 5 |
204350 |
0.03 |
| chr1_116931483_116931823 | 0.19 |
AL136376.1 |
Uncharacterized protein |
4935 |
0.17 |
| chr4_139937547_139937724 | 0.19 |
CCRN4L |
CCR4 carbon catabolite repression 4-like (S. cerevisiae) |
692 |
0.51 |
| chr2_198100640_198101238 | 0.19 |
ANKRD44 |
ankyrin repeat domain 44 |
38177 |
0.14 |
| chr3_53069303_53069506 | 0.19 |
SFMBT1 |
Scm-like with four mbt domains 1 |
9877 |
0.18 |
| chr7_105348761_105348986 | 0.18 |
ATXN7L1 |
ataxin 7-like 1 |
16789 |
0.26 |
| chr2_135014379_135014599 | 0.18 |
MGAT5 |
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase |
2659 |
0.35 |
| chr14_73610281_73610452 | 0.18 |
PSEN1 |
presenilin 1 |
3834 |
0.24 |
| chr13_31201174_31201431 | 0.18 |
USPL1 |
ubiquitin specific peptidase like 1 |
9382 |
0.23 |
| chr4_110612664_110612815 | 0.17 |
AC004067.5 |
|
421 |
0.84 |
| chr8_126950940_126951091 | 0.17 |
ENSG00000206695 |
. |
37820 |
0.23 |
| chr16_57021435_57021712 | 0.17 |
NLRC5 |
NLR family, CARD domain containing 5 |
1837 |
0.25 |
| chr10_63998636_63998943 | 0.17 |
RTKN2 |
rhotekin 2 |
2767 |
0.38 |
| chr14_99647139_99647315 | 0.17 |
AL162151.4 |
|
22474 |
0.23 |
| chr3_18784592_18784781 | 0.17 |
ENSG00000228956 |
. |
2550 |
0.44 |
| chr2_101918683_101918948 | 0.17 |
RNF149 |
ring finger protein 149 |
6343 |
0.18 |
| chr8_125490405_125490556 | 0.17 |
RNF139 |
ring finger protein 139 |
3501 |
0.2 |
| chr2_48543908_48544266 | 0.16 |
FOXN2 |
forkhead box N2 |
2238 |
0.35 |
| chr14_62265738_62265889 | 0.16 |
SNAPC1 |
small nuclear RNA activating complex, polypeptide 1, 43kDa |
36738 |
0.21 |
| chr16_17397269_17397420 | 0.16 |
XYLT1 |
xylosyltransferase I |
167394 |
0.04 |
| chr2_109582380_109582745 | 0.16 |
EDAR |
ectodysplasin A receptor |
23163 |
0.25 |
| chrX_135710444_135710648 | 0.16 |
ENSG00000233093 |
. |
11156 |
0.16 |
| chr20_52490835_52490986 | 0.16 |
AC005220.3 |
|
65789 |
0.13 |
| chr2_158269106_158269275 | 0.16 |
CYTIP |
cytohesin 1 interacting protein |
26736 |
0.19 |
| chr16_53498380_53498624 | 0.16 |
RBL2 |
retinoblastoma-like 2 (p130) |
14514 |
0.15 |
| chr7_105515189_105515340 | 0.16 |
ATXN7L1 |
ataxin 7-like 1 |
1659 |
0.51 |
| chr4_143621409_143621560 | 0.16 |
INPP4B |
inositol polyphosphate-4-phosphatase, type II, 105kDa |
139662 |
0.05 |
| chr15_60085307_60085458 | 0.16 |
BNIP2 |
BCL2/adenovirus E1B 19kDa interacting protein 2 |
103649 |
0.06 |
| chr2_143888983_143889411 | 0.16 |
ARHGAP15 |
Rho GTPase activating protein 15 |
2314 |
0.39 |
| chr4_47829221_47829372 | 0.16 |
CORIN |
corin, serine peptidase |
10670 |
0.21 |
| chr4_74117360_74117511 | 0.16 |
ANKRD17 |
ankyrin repeat domain 17 |
6604 |
0.19 |
| chr4_73953344_73953495 | 0.16 |
COX18 |
COX18 cytochrome C oxidase assembly factor |
17961 |
0.23 |
| chr6_35566907_35567293 | 0.16 |
ENSG00000212579 |
. |
52495 |
0.1 |
| chr6_52151518_52151818 | 0.16 |
MCM3 |
minichromosome maintenance complex component 3 |
2033 |
0.36 |
| chr1_151133033_151133283 | 0.16 |
SCNM1 |
sodium channel modifier 1 |
4018 |
0.1 |
| chr2_197014969_197015443 | 0.16 |
RP11-347P5.1 |
|
781 |
0.66 |
| chr2_204812325_204812548 | 0.15 |
ICOS |
inducible T-cell co-stimulator |
10933 |
0.28 |
| chr7_142500387_142500538 | 0.15 |
PRSS3P2 |
protease, serine, 3 pseudogene 2 |
19331 |
0.16 |
| chr17_40750860_40751042 | 0.15 |
FAM134C |
family with sequence similarity 134, member C |
10431 |
0.08 |
| chr4_78726314_78726465 | 0.15 |
CNOT6L |
CCR4-NOT transcription complex, subunit 6-like |
13828 |
0.26 |
| chr10_134069250_134069918 | 0.15 |
STK32C |
serine/threonine kinase 32C |
51134 |
0.12 |
| chr12_121323357_121323508 | 0.15 |
SPPL3 |
signal peptide peptidase like 3 |
17741 |
0.17 |
| chr8_82011673_82011842 | 0.15 |
PAG1 |
phosphoprotein associated with glycosphingolipid microdomains 1 |
12546 |
0.28 |
| chr11_96023136_96023287 | 0.15 |
ENSG00000266192 |
. |
51391 |
0.14 |
| chr12_47710625_47710974 | 0.15 |
ENSG00000199566 |
. |
29281 |
0.17 |
| chr1_66831872_66832123 | 0.15 |
PDE4B |
phosphodiesterase 4B, cAMP-specific |
11932 |
0.3 |
| chr2_204596528_204596679 | 0.15 |
CD28 |
CD28 molecule |
25187 |
0.19 |
| chr3_151911039_151911356 | 0.15 |
MBNL1 |
muscleblind-like splicing regulator 1 |
74632 |
0.11 |
| chr11_10819600_10819751 | 0.15 |
EIF4G2 |
eukaryotic translation initiation factor 4 gamma, 2 |
2354 |
0.19 |
| chr2_32036582_32036828 | 0.15 |
ENSG00000238448 |
. |
1348 |
0.49 |
| chr6_109357989_109358140 | 0.15 |
ENSG00000199944 |
. |
22753 |
0.16 |
| chr15_38904317_38904468 | 0.15 |
RASGRP1 |
RAS guanyl releasing protein 1 (calcium and DAG-regulated) |
46616 |
0.14 |
| chr2_192505177_192505550 | 0.15 |
NABP1 |
nucleic acid binding protein 1 |
37499 |
0.21 |
| chr3_107697281_107697722 | 0.15 |
CD47 |
CD47 molecule |
79707 |
0.11 |
| chr8_48594509_48594782 | 0.15 |
ENSG00000207369 |
. |
14312 |
0.19 |
| chr4_100765453_100765604 | 0.15 |
DAPP1 |
dual adaptor of phosphotyrosine and 3-phosphoinositides |
27525 |
0.18 |
| chr5_59892964_59893115 | 0.15 |
DEPDC1B |
DEP domain containing 1B |
102922 |
0.07 |
| chr1_160682851_160683057 | 0.15 |
CD48 |
CD48 molecule |
1313 |
0.4 |
| chr1_171470969_171471120 | 0.15 |
ENSG00000239373 |
. |
9494 |
0.15 |
| chr20_51025367_51025552 | 0.15 |
ZFP64 |
ZFP64 zinc finger protein |
216934 |
0.02 |
| chrX_123476453_123476604 | 0.15 |
SH2D1A |
SH2 domain containing 1A |
3666 |
0.37 |
| chr8_29964950_29965250 | 0.15 |
LEPROTL1 |
leptin receptor overlapping transcript-like 1 |
5644 |
0.16 |
| chr14_98606379_98606534 | 0.15 |
C14orf64 |
chromosome 14 open reading frame 64 |
161995 |
0.04 |
| chr1_92942408_92942722 | 0.14 |
GFI1 |
growth factor independent 1 transcription repressor |
6946 |
0.27 |
| chr2_198150239_198150434 | 0.14 |
ANKRD44-IT1 |
ANKRD44 intronic transcript 1 (non-protein coding) |
16907 |
0.16 |
| chr14_38540644_38540853 | 0.14 |
CTD-2058B24.2 |
|
19615 |
0.24 |
| chr15_60843099_60843250 | 0.14 |
CTD-2501E16.2 |
|
21002 |
0.18 |
| chr9_97347238_97347389 | 0.14 |
FBP2 |
fructose-1,6-bisphosphatase 2 |
8762 |
0.24 |
| chrX_56594702_56594894 | 0.14 |
UBQLN2 |
ubiquilin 2 |
4772 |
0.35 |
| chr4_114614482_114614847 | 0.14 |
CAMK2D |
calcium/calmodulin-dependent protein kinase II delta |
67560 |
0.14 |
| chr3_56729846_56730094 | 0.14 |
FAM208A |
family with sequence similarity 208, member A |
12705 |
0.27 |
| chr15_81586884_81587161 | 0.14 |
IL16 |
interleukin 16 |
2232 |
0.32 |
| chr2_99278059_99278217 | 0.14 |
MGAT4A |
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A |
1798 |
0.39 |
| chr11_14721093_14721244 | 0.14 |
PDE3B |
phosphodiesterase 3B, cGMP-inhibited |
55791 |
0.14 |
| chr6_16420917_16421068 | 0.14 |
ENSG00000265642 |
. |
7762 |
0.31 |
| chr19_2850075_2850226 | 0.14 |
AC006130.3 |
|
8564 |
0.1 |
| chr1_226067346_226067612 | 0.14 |
TMEM63A |
transmembrane protein 63A |
2100 |
0.22 |
| chr1_198578902_198579207 | 0.14 |
PTPRC |
protein tyrosine phosphatase, receptor type, C |
28747 |
0.21 |
| chr3_71127524_71127675 | 0.14 |
FOXP1 |
forkhead box P1 |
12681 |
0.31 |
| chr9_2053303_2053454 | 0.14 |
RP11-264I13.2 |
|
7355 |
0.25 |
| chr6_135333699_135333925 | 0.14 |
HBS1L |
HBS1-like (S. cerevisiae) |
14482 |
0.21 |
| chr6_108958559_108958710 | 0.14 |
FOXO3 |
forkhead box O3 |
18915 |
0.27 |
| chr8_20131803_20131954 | 0.14 |
ENSG00000253733 |
. |
15807 |
0.18 |
| chr12_55129725_55129876 | 0.14 |
DCD |
dermcidin |
87523 |
0.07 |
| chr6_149793703_149793934 | 0.14 |
ZC3H12D |
zinc finger CCCH-type containing 12D |
10451 |
0.17 |
| chr10_14618640_14618843 | 0.14 |
FAM107B |
family with sequence similarity 107, member B |
4403 |
0.28 |
| chr15_94815185_94815463 | 0.14 |
MCTP2 |
multiple C2 domains, transmembrane 2 |
26106 |
0.28 |
| chr2_97620034_97620317 | 0.14 |
ENSG00000252845 |
. |
2377 |
0.24 |
| chr2_8616066_8616523 | 0.14 |
AC011747.7 |
|
199602 |
0.03 |
| chr1_89751943_89752616 | 0.14 |
GBP5 |
guanylate binding protein 5 |
13735 |
0.19 |
| chr7_114567304_114567455 | 0.14 |
MDFIC |
MyoD family inhibitor domain containing |
4248 |
0.36 |
| chr12_46607511_46607662 | 0.14 |
SLC38A1 |
solute carrier family 38, member 1 |
53898 |
0.17 |
| chr2_58136289_58136440 | 0.13 |
VRK2 |
vaccinia related kinase 2 |
1578 |
0.56 |
| chr9_4598144_4598295 | 0.13 |
SLC1A1 |
solute carrier family 1 (neuronal/epithelial high affinity glutamate transporter, system Xag), member 1 |
25884 |
0.18 |
| chr14_68734669_68735014 | 0.13 |
ENSG00000243546 |
. |
31585 |
0.22 |
| chr18_2950520_2950671 | 0.13 |
RP11-737O24.2 |
|
2256 |
0.25 |
| chr2_144990703_144990900 | 0.13 |
GTDC1 |
glycosyltransferase-like domain containing 1 |
2503 |
0.44 |
| chr11_116796998_116797221 | 0.13 |
SIK3 |
SIK family kinase 3 |
30651 |
0.13 |
| chr1_158905295_158905446 | 0.13 |
PYHIN1 |
pyrin and HIN domain family, member 1 |
4012 |
0.26 |
| chr12_12679407_12679558 | 0.13 |
DUSP16 |
dual specificity phosphatase 16 |
5423 |
0.27 |
| chr1_90186329_90186568 | 0.13 |
ENSG00000239176 |
. |
47429 |
0.13 |
| chr1_117311065_117311639 | 0.13 |
CD2 |
CD2 molecule |
14263 |
0.21 |
| chr3_46031859_46032010 | 0.13 |
FYCO1 |
FYVE and coiled-coil domain containing 1 |
5373 |
0.2 |
| chr11_58329457_58329608 | 0.13 |
LPXN |
leupaxin |
13802 |
0.15 |
| chr8_131255625_131255776 | 0.13 |
ASAP1 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1 |
3588 |
0.34 |
| chr12_76941839_76941990 | 0.13 |
OSBPL8 |
oxysterol binding protein-like 8 |
11370 |
0.3 |
| chr6_112310464_112310615 | 0.13 |
WISP3 |
WNT1 inducible signaling pathway protein 3 |
64736 |
0.11 |
| chr7_8022696_8022847 | 0.13 |
AC006042.7 |
|
13237 |
0.18 |
| chr6_130923499_130923650 | 0.13 |
ENSG00000202438 |
. |
28317 |
0.25 |
| chr1_183444780_183444944 | 0.13 |
SMG7 |
SMG7 nonsense mediated mRNA decay factor |
2795 |
0.29 |
| chr20_47786456_47786607 | 0.13 |
STAU1 |
staufen double-stranded RNA binding protein 1 |
4269 |
0.24 |
| chr22_42623963_42624114 | 0.13 |
TCF20 |
transcription factor 20 (AR1) |
12590 |
0.19 |
| chr10_32619705_32619856 | 0.13 |
RP11-135A24.4 |
|
15647 |
0.13 |
| chr2_148053777_148053928 | 0.13 |
ENSG00000238860 |
. |
27687 |
0.24 |
| chr2_202093371_202093641 | 0.13 |
CASP8 |
caspase 8, apoptosis-related cysteine peptidase |
4660 |
0.21 |
| chr18_2961462_2961796 | 0.13 |
RP11-737O24.1 |
|
5387 |
0.16 |
| chr3_107672504_107672666 | 0.13 |
CD47 |
CD47 molecule |
104623 |
0.08 |
| chr16_16053619_16053929 | 0.13 |
ABCC1 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 1 |
10340 |
0.22 |
| chr13_100033049_100033200 | 0.13 |
ENSG00000207719 |
. |
24739 |
0.18 |
| chr14_68752319_68752470 | 0.13 |
ENSG00000243546 |
. |
49138 |
0.17 |
| chr1_160676517_160676733 | 0.13 |
CD48 |
CD48 molecule |
4968 |
0.18 |
| chr7_150453228_150453688 | 0.13 |
GIMAP5 |
GTPase, IMAP family member 5 |
19022 |
0.14 |
| chr11_88579489_88579640 | 0.13 |
GRM5 |
glutamate receptor, metabotropic 5 |
200974 |
0.03 |
| chr12_55364288_55364490 | 0.13 |
TESPA1 |
thymocyte expressed, positive selection associated 1 |
2949 |
0.31 |
| chr3_111854142_111854385 | 0.13 |
RP11-757F18.5 |
|
1993 |
0.26 |
| chr2_191280247_191280414 | 0.13 |
MFSD6 |
major facilitator superfamily domain containing 6 |
7249 |
0.21 |
| chr5_79436518_79436669 | 0.13 |
CTC-458I2.2 |
|
12409 |
0.21 |
| chr2_224822884_224823038 | 0.13 |
MRPL44 |
mitochondrial ribosomal protein L44 |
840 |
0.61 |
| chr10_104685349_104685500 | 0.13 |
CNNM2 |
cyclin M2 |
7310 |
0.19 |
| chr4_4527024_4527175 | 0.13 |
STX18 |
syntaxin 18 |
16638 |
0.19 |
| chr15_85240505_85240759 | 0.13 |
RP11-245C17.2 |
|
11517 |
0.12 |
| chr2_16106193_16106413 | 0.13 |
ENSG00000243541 |
. |
15200 |
0.18 |
| chr20_5717080_5717231 | 0.13 |
C20orf196 |
chromosome 20 open reading frame 196 |
13884 |
0.26 |
| chr10_112000954_112001185 | 0.13 |
MXI1 |
MAX interactor 1, dimerization protein |
13632 |
0.21 |
| chr1_121311422_121311588 | 0.13 |
FCGR1B |
Fc fragment of IgG, high affinity Ib, receptor (CD64) |
375568 |
0.01 |
| chr4_102204361_102204644 | 0.12 |
ENSG00000221265 |
. |
47069 |
0.15 |
| chr3_12601064_12601266 | 0.12 |
MKRN2 |
makorin ring finger protein 2 |
2576 |
0.21 |
| chr4_140150628_140150779 | 0.12 |
ENSG00000252362 |
. |
1570 |
0.3 |
| chr12_8861700_8861896 | 0.12 |
RIMKLB |
ribosomal modification protein rimK-like family member B |
9356 |
0.17 |
| chr9_80730925_80731076 | 0.12 |
ENSG00000222452 |
. |
49381 |
0.17 |
| chr9_21331248_21331399 | 0.12 |
KLHL9 |
kelch-like family member 9 |
3917 |
0.16 |
| chr12_40004458_40004609 | 0.12 |
ABCD2 |
ATP-binding cassette, sub-family D (ALD), member 2 |
9020 |
0.24 |
| chr4_122836640_122836791 | 0.12 |
TRPC3 |
transient receptor potential cation channel, subfamily C, member 3 |
17478 |
0.21 |
| chr6_6609840_6610222 | 0.12 |
LY86-AS1 |
LY86 antisense RNA 1 |
12897 |
0.24 |
| chr15_31134475_31134626 | 0.12 |
ENSG00000221379 |
. |
41041 |
0.12 |
| chr10_8130727_8131157 | 0.12 |
GATA3 |
GATA binding protein 3 |
34173 |
0.24 |
| chr15_52860223_52860374 | 0.12 |
ARPP19 |
cAMP-regulated phosphoprotein, 19kDa |
731 |
0.65 |
| chr6_99938487_99938691 | 0.12 |
USP45 |
ubiquitin specific peptidase 45 |
7848 |
0.18 |
| chr2_55278350_55278727 | 0.12 |
RTN4 |
reticulon 4 |
574 |
0.78 |
| chr3_197317837_197317988 | 0.12 |
BDH1 |
3-hydroxybutyrate dehydrogenase, type 1 |
17718 |
0.2 |
| chr22_40314840_40314991 | 0.12 |
GRAP2 |
GRB2-related adaptor protein 2 |
7680 |
0.18 |
| chr10_22938945_22939188 | 0.12 |
PIP4K2A |
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha |
58424 |
0.15 |
| chr3_152018760_152019009 | 0.12 |
MBNL1 |
muscleblind-like splicing regulator 1 |
897 |
0.64 |
| chrY_1549050_1549214 | 0.12 |
NA |
NA |
> 106 |
NA |
| chrX_1599047_1599214 | 0.12 |
ASMTL |
acetylserotonin O-methyltransferase-like |
26475 |
0.16 |
| chr1_167407962_167408153 | 0.12 |
RP11-104L21.2 |
|
19841 |
0.19 |
| chr2_235204073_235204224 | 0.12 |
ARL4C |
ADP-ribosylation factor-like 4C |
201096 |
0.03 |
| chr18_51745857_51746008 | 0.12 |
ENSG00000207233 |
. |
2850 |
0.28 |
| chr2_99345745_99345966 | 0.12 |
MGAT4A |
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A |
856 |
0.67 |
| chr2_149481240_149481391 | 0.12 |
EPC2 |
enhancer of polycomb homolog 2 (Drosophila) |
33532 |
0.21 |
| chr7_117827399_117827550 | 0.12 |
NAA38 |
N(alpha)-acetyltransferase 38, NatC auxiliary subunit |
3253 |
0.34 |
| chr1_167434236_167434536 | 0.12 |
RP11-104L21.2 |
|
6488 |
0.23 |
| chr13_80024271_80024542 | 0.12 |
NDFIP2 |
Nedd4 family interacting protein 2 |
30881 |
0.15 |
| chr20_5565865_5566016 | 0.12 |
GPCPD1 |
glycerophosphocholine phosphodiesterase GDE1 homolog (S. cerevisiae) |
9786 |
0.26 |
| chr5_35671011_35671203 | 0.12 |
SPEF2 |
sperm flagellar 2 |
4597 |
0.28 |
| chr11_96064460_96064611 | 0.12 |
ENSG00000266192 |
. |
10067 |
0.2 |
| chr18_67566005_67566307 | 0.12 |
CD226 |
CD226 molecule |
48499 |
0.18 |
| chr2_97477573_97477724 | 0.12 |
CNNM3 |
cyclin M3 |
4334 |
0.15 |
| chr8_98697623_98697774 | 0.12 |
MTDH |
metadherin |
5661 |
0.27 |
| chr14_91876116_91876296 | 0.12 |
CCDC88C |
coiled-coil domain containing 88C |
7484 |
0.25 |
| chr11_95949552_95949826 | 0.12 |
ENSG00000266192 |
. |
124913 |
0.05 |
| chr5_55773462_55773845 | 0.12 |
CTC-236F12.4 |
Uncharacterized protein |
3943 |
0.25 |
| chr19_36420808_36421170 | 0.12 |
LRFN3 |
leucine rich repeat and fibronectin type III domain containing 3 |
5463 |
0.1 |
| chrX_15773519_15773681 | 0.12 |
CA5B |
carbonic anhydrase VB, mitochondrial |
5507 |
0.2 |
| chr9_100692681_100693016 | 0.12 |
C9orf156 |
chromosome 9 open reading frame 156 |
7996 |
0.15 |
| chr6_34283479_34283630 | 0.12 |
C6orf1 |
chromosome 6 open reading frame 1 |
66307 |
0.09 |
| chr2_235125609_235126070 | 0.12 |
SPP2 |
secreted phosphoprotein 2, 24kDa |
166408 |
0.04 |
| chr2_62445208_62445359 | 0.12 |
B3GNT2 |
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 2 |
2630 |
0.25 |
| chr17_46435794_46435945 | 0.12 |
RP11-433M22.1 |
|
21863 |
0.14 |
| chr7_36820218_36820369 | 0.12 |
AOAH |
acyloxyacyl hydrolase (neutrophil) |
56139 |
0.13 |
| chr16_79124068_79124219 | 0.12 |
RP11-556H2.3 |
|
196 |
0.94 |
| chr13_99931416_99931591 | 0.12 |
GPR18 |
G protein-coupled receptor 18 |
17505 |
0.18 |
| chr5_170740649_170740930 | 0.12 |
TLX3 |
T-cell leukemia homeobox 3 |
4501 |
0.2 |
| chr14_104113341_104113492 | 0.12 |
KLC1 |
kinesin light chain 1 |
17782 |
0.09 |
| chr7_130701521_130701683 | 0.12 |
LINC-PINT |
long intergenic non-protein coding RNA, p53 induced transcript |
32734 |
0.18 |
| chr3_43043421_43043572 | 0.12 |
FAM198A |
family with sequence similarity 198, member A |
22456 |
0.17 |
| chr1_182555451_182555602 | 0.12 |
RNASEL |
ribonuclease L (2',5'-oligoisoadenylate synthetase-dependent) |
582 |
0.78 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0001911 | negative regulation of leukocyte mediated cytotoxicity(GO:0001911) |
| 0.1 | 0.3 | GO:0046137 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.1 | 0.3 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.1 | 0.3 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.1 | 0.3 | GO:1901339 | activation of store-operated calcium channel activity(GO:0032237) regulation of store-operated calcium channel activity(GO:1901339) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.1 | 0.2 | GO:0045589 | regulation of regulatory T cell differentiation(GO:0045589) |
| 0.1 | 0.5 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 0.2 | GO:0033629 | negative regulation of cell adhesion mediated by integrin(GO:0033629) |
| 0.1 | 0.2 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.1 | 0.2 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.1 | 0.1 | GO:0042091 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.1 | 0.2 | GO:0002664 | T cell tolerance induction(GO:0002517) regulation of T cell tolerance induction(GO:0002664) positive regulation of T cell tolerance induction(GO:0002666) |
| 0.1 | 0.5 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 0.3 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) negative regulation of signal transduction by p53 class mediator(GO:1901797) |
| 0.1 | 0.2 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.1 | 0.2 | GO:0002335 | mature B cell differentiation(GO:0002335) |
| 0.1 | 0.2 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.1 | 0.3 | GO:0045348 | positive regulation of MHC class II biosynthetic process(GO:0045348) |
| 0.1 | 0.1 | GO:0002507 | tolerance induction(GO:0002507) |
| 0.1 | 0.2 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.1 | 0.2 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.1 | 0.2 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.2 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.0 | 0.1 | GO:0045343 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) |
| 0.0 | 0.2 | GO:0019987 | obsolete negative regulation of anti-apoptosis(GO:0019987) |
| 0.0 | 0.1 | GO:0002890 | negative regulation of B cell mediated immunity(GO:0002713) negative regulation of immunoglobulin mediated immune response(GO:0002890) |
| 0.0 | 0.1 | GO:0032506 | cytokinetic process(GO:0032506) |
| 0.0 | 0.2 | GO:0043368 | positive T cell selection(GO:0043368) |
| 0.0 | 0.2 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.1 | GO:0003323 | type B pancreatic cell development(GO:0003323) |
| 0.0 | 0.6 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.1 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.2 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.1 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.1 | GO:0045066 | regulatory T cell differentiation(GO:0045066) |
| 0.0 | 0.3 | GO:0050856 | regulation of T cell receptor signaling pathway(GO:0050856) |
| 0.0 | 0.1 | GO:0042510 | regulation of tyrosine phosphorylation of Stat1 protein(GO:0042510) |
| 0.0 | 0.1 | GO:0030011 | maintenance of cell polarity(GO:0030011) |
| 0.0 | 0.1 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.0 | GO:0051665 | membrane raft localization(GO:0051665) |
| 0.0 | 0.1 | GO:0006154 | adenosine catabolic process(GO:0006154) |
| 0.0 | 0.2 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.2 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 1.9 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.0 | 0.2 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.1 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 | 0.1 | GO:0034392 | negative regulation of smooth muscle cell apoptotic process(GO:0034392) |
| 0.0 | 0.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.1 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 | 0.1 | GO:0032875 | regulation of DNA endoreduplication(GO:0032875) negative regulation of DNA endoreduplication(GO:0032876) DNA endoreduplication(GO:0042023) |
| 0.0 | 0.2 | GO:0002063 | chondrocyte development(GO:0002063) |
| 0.0 | 0.1 | GO:0045354 | interferon-alpha biosynthetic process(GO:0045349) regulation of interferon-alpha biosynthetic process(GO:0045354) positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.0 | 0.1 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.0 | 0.1 | GO:0015917 | aminophospholipid transport(GO:0015917) |
| 0.0 | 0.2 | GO:0048679 | regulation of axon regeneration(GO:0048679) regulation of neuron projection regeneration(GO:0070570) |
| 0.0 | 0.1 | GO:0001779 | natural killer cell differentiation(GO:0001779) |
| 0.0 | 0.2 | GO:0032733 | regulation of interleukin-10 production(GO:0032653) positive regulation of interleukin-10 production(GO:0032733) |
| 0.0 | 0.2 | GO:0035246 | peptidyl-arginine N-methylation(GO:0035246) |
| 0.0 | 0.1 | GO:0015801 | aromatic amino acid transport(GO:0015801) |
| 0.0 | 0.1 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.0 | 0.1 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.1 | GO:0006681 | galactosylceramide metabolic process(GO:0006681) |
| 0.0 | 0.1 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.0 | 0.1 | GO:0042996 | regulation of Golgi to plasma membrane protein transport(GO:0042996) |
| 0.0 | 0.1 | GO:0043320 | natural killer cell activation involved in immune response(GO:0002323) natural killer cell degranulation(GO:0043320) |
| 0.0 | 0.1 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.0 | GO:0034139 | regulation of toll-like receptor 3 signaling pathway(GO:0034139) |
| 0.0 | 0.1 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.1 | GO:0070266 | necroptotic process(GO:0070266) programmed necrotic cell death(GO:0097300) |
| 0.0 | 0.1 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 0.0 | 0.1 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.0 | 0.1 | GO:0021530 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) oligodendrocyte cell fate specification(GO:0021778) glial cell fate specification(GO:0021780) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.0 | GO:0030581 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) regulation by virus of viral protein levels in host cell(GO:0046719) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.0 | 0.1 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.0 | 0.0 | GO:0071224 | cellular response to peptidoglycan(GO:0071224) |
| 0.0 | 0.1 | GO:0071922 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.0 | 0.1 | GO:0002836 | regulation of response to tumor cell(GO:0002834) positive regulation of response to tumor cell(GO:0002836) regulation of immune response to tumor cell(GO:0002837) positive regulation of immune response to tumor cell(GO:0002839) |
| 0.0 | 0.2 | GO:0070723 | response to sterol(GO:0036314) response to cholesterol(GO:0070723) |
| 0.0 | 0.2 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.0 | 0.1 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.2 | GO:0003084 | positive regulation of systemic arterial blood pressure(GO:0003084) |
| 0.0 | 0.3 | GO:0060330 | regulation of response to interferon-gamma(GO:0060330) regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.0 | 0.1 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.3 | GO:0060324 | face development(GO:0060324) |
| 0.0 | 0.1 | GO:0006065 | UDP-glucuronate biosynthetic process(GO:0006065) UDP-glucuronate metabolic process(GO:0046398) |
| 0.0 | 0.1 | GO:0008291 | acetylcholine metabolic process(GO:0008291) acetate ester metabolic process(GO:1900619) |
| 0.0 | 0.0 | GO:0046958 | nonassociative learning(GO:0046958) |
| 0.0 | 0.1 | GO:0050687 | negative regulation of defense response to virus(GO:0050687) |
| 0.0 | 0.0 | GO:0002643 | regulation of tolerance induction(GO:0002643) positive regulation of tolerance induction(GO:0002645) |
| 0.0 | 0.1 | GO:0045672 | positive regulation of osteoclast differentiation(GO:0045672) |
| 0.0 | 0.1 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.0 | 0.1 | GO:0030187 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.0 | 0.1 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.0 | 0.0 | GO:0002538 | arachidonic acid metabolite production involved in inflammatory response(GO:0002538) leukotriene production involved in inflammatory response(GO:0002540) |
| 0.0 | 0.1 | GO:0032814 | regulation of natural killer cell activation(GO:0032814) |
| 0.0 | 0.1 | GO:0009048 | dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 0.0 | GO:0002282 | microglial cell activation involved in immune response(GO:0002282) |
| 0.0 | 0.1 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.0 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.0 | 0.1 | GO:0045954 | positive regulation of natural killer cell mediated immunity(GO:0002717) positive regulation of natural killer cell mediated cytotoxicity(GO:0045954) |
| 0.0 | 0.0 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.1 | GO:0045910 | negative regulation of DNA recombination(GO:0045910) |
| 0.0 | 0.0 | GO:0042531 | positive regulation of tyrosine phosphorylation of STAT protein(GO:0042531) |
| 0.0 | 0.1 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.0 | GO:0010533 | regulation of activation of Janus kinase activity(GO:0010533) positive regulation of activation of Janus kinase activity(GO:0010536) |
| 0.0 | 0.1 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.0 | 0.2 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.0 | 0.1 | GO:0006978 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) |
| 0.0 | 0.1 | GO:0042518 | negative regulation of tyrosine phosphorylation of Stat3 protein(GO:0042518) |
| 0.0 | 0.0 | GO:0010988 | regulation of low-density lipoprotein particle clearance(GO:0010988) |
| 0.0 | 0.1 | GO:0090322 | regulation of superoxide metabolic process(GO:0090322) |
| 0.0 | 0.1 | GO:0001913 | T cell mediated cytotoxicity(GO:0001913) regulation of T cell mediated cytotoxicity(GO:0001914) |
| 0.0 | 0.0 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.0 | 0.0 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.0 | GO:0010587 | miRNA metabolic process(GO:0010586) miRNA catabolic process(GO:0010587) |
| 0.0 | 0.2 | GO:0045022 | early endosome to late endosome transport(GO:0045022) vesicle-mediated transport between endosomal compartments(GO:0098927) |
| 0.0 | 0.1 | GO:0045197 | establishment or maintenance of epithelial cell apical/basal polarity(GO:0045197) |
| 0.0 | 0.1 | GO:0043628 | ncRNA 3'-end processing(GO:0043628) |
| 0.0 | 0.2 | GO:0044409 | entry into host cell(GO:0030260) entry into host(GO:0044409) entry into cell of other organism involved in symbiotic interaction(GO:0051806) entry into other organism involved in symbiotic interaction(GO:0051828) |
| 0.0 | 0.0 | GO:0060586 | multicellular organismal iron ion homeostasis(GO:0060586) |
| 0.0 | 0.1 | GO:0032988 | ribonucleoprotein complex disassembly(GO:0032988) |
| 0.0 | 0.1 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.0 | 0.1 | GO:0033261 | obsolete regulation of S phase(GO:0033261) |
| 0.0 | 0.0 | GO:0034626 | fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.0 | GO:0033632 | regulation of cell-cell adhesion mediated by integrin(GO:0033632) |
| 0.0 | 0.0 | GO:0060916 | mesenchymal cell proliferation involved in lung development(GO:0060916) |
| 0.0 | 0.0 | GO:0046476 | glycosylceramide biosynthetic process(GO:0046476) |
| 0.0 | 0.1 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.2 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.0 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 0.0 | GO:0031054 | pre-miRNA processing(GO:0031054) |
| 0.0 | 0.0 | GO:0021860 | pyramidal neuron differentiation(GO:0021859) pyramidal neuron development(GO:0021860) |
| 0.0 | 0.0 | GO:0032494 | response to peptidoglycan(GO:0032494) |
| 0.0 | 0.0 | GO:0033160 | positive regulation of protein import into nucleus, translocation(GO:0033160) |
| 0.0 | 0.0 | GO:0042253 | granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0042253) regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045423) |
| 0.0 | 0.1 | GO:0051642 | centrosome localization(GO:0051642) |
| 0.0 | 0.1 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.0 | 0.1 | GO:0051103 | DNA ligation involved in DNA repair(GO:0051103) |
| 0.0 | 0.1 | GO:0042523 | positive regulation of tyrosine phosphorylation of Stat5 protein(GO:0042523) |
| 0.0 | 0.1 | GO:0002483 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 0.1 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.1 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.1 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.0 | GO:0070669 | response to interleukin-2(GO:0070669) |
| 0.0 | 0.0 | GO:0032075 | positive regulation of nuclease activity(GO:0032075) |
| 0.0 | 0.0 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.1 | GO:0008608 | attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.0 | 0.0 | GO:0051570 | regulation of histone H3-K9 methylation(GO:0051570) |
| 0.0 | 0.0 | GO:0052255 | modulation by symbiont of host defense response(GO:0052031) modulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052255) positive regulation by symbiont of host defense response(GO:0052509) positive regulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052510) modulation by organism of immune response of other organism involved in symbiotic interaction(GO:0052552) modulation by symbiont of host immune response(GO:0052553) |
| 0.0 | 0.0 | GO:0009180 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) ribonucleoside diphosphate biosynthetic process(GO:0009188) |
| 0.0 | 0.0 | GO:0035066 | positive regulation of histone acetylation(GO:0035066) positive regulation of protein acetylation(GO:1901985) positive regulation of peptidyl-lysine acetylation(GO:2000758) |
| 0.0 | 0.0 | GO:0002921 | negative regulation of humoral immune response(GO:0002921) |
| 0.0 | 0.0 | GO:1903313 | positive regulation of mRNA 3'-end processing(GO:0031442) positive regulation of mRNA processing(GO:0050685) positive regulation of mRNA metabolic process(GO:1903313) |
| 0.0 | 0.0 | GO:0097709 | connective tissue replacement involved in inflammatory response wound healing(GO:0002248) connective tissue replacement(GO:0097709) |
| 0.0 | 0.0 | GO:0032613 | interleukin-10 production(GO:0032613) |
| 0.0 | 0.0 | GO:0046877 | regulation of saliva secretion(GO:0046877) |
| 0.0 | 0.1 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.1 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.0 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.0 | 0.0 | GO:0046185 | aldehyde catabolic process(GO:0046185) |
| 0.0 | 0.0 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.0 | GO:0007343 | egg activation(GO:0007343) |
| 0.0 | 0.1 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) |
| 0.0 | 0.0 | GO:0045842 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.0 | 0.1 | GO:0048194 | COPI-coated vesicle budding(GO:0035964) Golgi vesicle budding(GO:0048194) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.0 | GO:0046607 | positive regulation of centrosome cycle(GO:0046607) |
| 0.0 | 0.1 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.2 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 | 0.0 | GO:0061030 | epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.0 | 0.0 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.0 | 0.0 | GO:0009822 | alkaloid catabolic process(GO:0009822) |
| 0.0 | 0.0 | GO:0001821 | histamine secretion(GO:0001821) |
| 0.0 | 0.0 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.0 | GO:0032292 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.0 | 0.1 | GO:0006983 | ER overload response(GO:0006983) |
| 0.0 | 0.1 | GO:0070265 | necrotic cell death(GO:0070265) |
| 0.0 | 0.0 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.0 | 0.0 | GO:0042516 | regulation of tyrosine phosphorylation of Stat3 protein(GO:0042516) |
| 0.0 | 0.0 | GO:0070670 | response to interleukin-4(GO:0070670) |
| 0.0 | 0.1 | GO:0050851 | antigen receptor-mediated signaling pathway(GO:0050851) |
| 0.0 | 0.0 | GO:0007042 | lysosomal lumen acidification(GO:0007042) regulation of lysosomal lumen pH(GO:0035751) |
| 0.0 | 0.0 | GO:0060206 | estrous cycle phase(GO:0060206) |
| 0.0 | 0.0 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.1 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.0 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.1 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.0 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.0 | GO:0070245 | positive regulation of T cell apoptotic process(GO:0070234) positive regulation of thymocyte apoptotic process(GO:0070245) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.1 | 0.2 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.1 | 0.2 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 0.5 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 0.2 | GO:0072487 | MSL complex(GO:0072487) |
| 0.1 | 0.6 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.1 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.2 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 0.2 | GO:0042598 | obsolete vesicular fraction(GO:0042598) |
| 0.0 | 0.1 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.1 | GO:0031264 | death-inducing signaling complex(GO:0031264) |
| 0.0 | 0.1 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.1 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.0 | 0.0 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.0 | 0.1 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.3 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.1 | GO:0000300 | obsolete peripheral to membrane of membrane fraction(GO:0000300) |
| 0.0 | 0.1 | GO:0071942 | XPC complex(GO:0071942) |
| 0.0 | 0.1 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.0 | GO:0000306 | extrinsic component of vacuolar membrane(GO:0000306) extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.2 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.1 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.1 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.0 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.0 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.1 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.1 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.1 | GO:0070688 | MLL5-L complex(GO:0070688) |
| 0.0 | 0.2 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.1 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.2 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.1 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.1 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.0 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.1 | GO:0005954 | calcium- and calmodulin-dependent protein kinase complex(GO:0005954) |
| 0.0 | 0.0 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.1 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.1 | GO:0000153 | cytoplasmic ubiquitin ligase complex(GO:0000153) |
| 0.0 | 0.2 | GO:0042627 | chylomicron(GO:0042627) |
| 0.0 | 0.1 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.1 | GO:0044453 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 0.2 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.0 | 0.1 | GO:0019908 | nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.0 | 0.2 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.0 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.0 | 0.0 | GO:0001740 | Barr body(GO:0001740) |
| 0.0 | 0.1 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.1 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.1 | GO:0045120 | pronucleus(GO:0045120) |
| 0.0 | 0.1 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.1 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.2 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.0 | GO:0000791 | euchromatin(GO:0000791) |
| 0.0 | 0.1 | GO:0032838 | cell projection cytoplasm(GO:0032838) |
| 0.0 | 0.1 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.0 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.0 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.0 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.0 | 0.0 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.1 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 0.2 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.1 | 0.2 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.1 | 0.2 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.1 | 0.2 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 0.2 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.1 | 0.2 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.4 | GO:0034593 | phosphatidylinositol bisphosphate phosphatase activity(GO:0034593) |
| 0.0 | 0.0 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.1 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.1 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.0 | 0.1 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.2 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.2 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.1 | GO:0010861 | thyroid hormone receptor activator activity(GO:0010861) |
| 0.0 | 0.4 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.1 | GO:0004525 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.0 | 0.8 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.1 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.0 | 0.1 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.1 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.0 | 0.1 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.1 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.0 | 0.2 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.1 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.2 | GO:0052813 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.1 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.1 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.1 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) |
| 0.0 | 0.2 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.1 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.7 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.2 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.1 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.1 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.0 | 0.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.1 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.1 | GO:0015925 | galactosidase activity(GO:0015925) |
| 0.0 | 0.0 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.0 | 0.1 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.2 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.2 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.2 | GO:0005283 | sodium:amino acid symporter activity(GO:0005283) |
| 0.0 | 0.1 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.0 | 0.4 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.2 | GO:0016565 | obsolete general transcriptional repressor activity(GO:0016565) |
| 0.0 | 0.5 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.2 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.1 | GO:0016362 | activin receptor activity, type II(GO:0016362) |
| 0.0 | 0.1 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.1 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.0 | 0.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.1 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.1 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.0 | 0.0 | GO:0031701 | angiotensin receptor binding(GO:0031701) |
| 0.0 | 0.2 | GO:0016274 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.0 | 0.1 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.0 | 0.1 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.3 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.1 | GO:0003909 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.1 | GO:0051998 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 0.0 | 0.1 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.6 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.1 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.0 | 0.2 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.1 | GO:0043175 | RNA polymerase core enzyme binding(GO:0043175) |
| 0.0 | 0.1 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 0.1 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.0 | 0.1 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.1 | GO:0004904 | interferon receptor activity(GO:0004904) |
| 0.0 | 0.0 | GO:0000739 | obsolete DNA strand annealing activity(GO:0000739) |
| 0.0 | 0.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.1 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.4 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.1 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.1 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.0 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) |
| 0.0 | 0.1 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.0 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.7 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.1 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.0 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.0 | 0.2 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.0 | GO:0004946 | bombesin receptor activity(GO:0004946) |
| 0.0 | 0.0 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 0.0 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.0 | 0.5 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.1 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.0 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.0 | 0.0 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.0 | GO:0042156 | obsolete zinc-mediated transcriptional activator activity(GO:0042156) |
| 0.0 | 0.1 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.1 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.1 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.0 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.2 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.0 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.0 | 0.0 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.0 | 0.1 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.2 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.0 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.0 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
| 0.0 | 0.0 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.1 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 0.0 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) |
| 0.0 | 0.1 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.0 | 0.4 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.1 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.0 | 0.0 | GO:0000404 | heteroduplex DNA loop binding(GO:0000404) |
| 0.0 | 0.0 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.3 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.0 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.1 | GO:0016520 | growth hormone-releasing hormone receptor activity(GO:0016520) |
| 0.0 | 0.0 | GO:0046625 | sphingolipid binding(GO:0046625) |
| 0.0 | 0.0 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.1 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.2 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.1 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.0 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.0 | GO:0045309 | protein phosphorylated amino acid binding(GO:0045309) |
| 0.0 | 0.0 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.2 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.0 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.1 | GO:0004952 | dopamine neurotransmitter receptor activity(GO:0004952) |
| 0.0 | 0.3 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.1 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.0 | GO:0019959 | C-X-C chemokine binding(GO:0019958) interleukin-8 binding(GO:0019959) |
| 0.0 | 0.0 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.5 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.5 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.3 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 0.0 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.7 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.5 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.6 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.0 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 0.2 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.0 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 0.1 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.6 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.0 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.7 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 0.1 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.2 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.3 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.8 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.4 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.3 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.1 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 0.3 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.2 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.2 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.2 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 0.2 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.0 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.1 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 0.2 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.3 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.0 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.0 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 0.4 | PID PLK1 PATHWAY | PLK1 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.1 | 0.8 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 0.6 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.1 | 0.8 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.4 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.9 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.1 | REACTOME AQUAPORIN MEDIATED TRANSPORT | Genes involved in Aquaporin-mediated transport |
| 0.0 | 0.0 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.2 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.5 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.3 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.2 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.3 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.3 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.0 | 0.0 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.0 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 0.1 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.1 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.1 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.1 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.1 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.1 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 0.4 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.3 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.1 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.0 | 0.3 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.1 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 0.2 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.0 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.0 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.1 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.2 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.1 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.2 | REACTOME TCR SIGNALING | Genes involved in TCR signaling |
| 0.0 | 0.2 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.5 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.1 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.6 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.2 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.3 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.1 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.1 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.0 | 0.1 | REACTOME IL 2 SIGNALING | Genes involved in Interleukin-2 signaling |
| 0.0 | 0.2 | REACTOME NUCLEOTIDE BINDING DOMAIN LEUCINE RICH REPEAT CONTAINING RECEPTOR NLR SIGNALING PATHWAYS | Genes involved in Nucleotide-binding domain, leucine rich repeat containing receptor (NLR) signaling pathways |
| 0.0 | 0.2 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.1 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.2 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.0 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 0.2 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.1 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |