Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
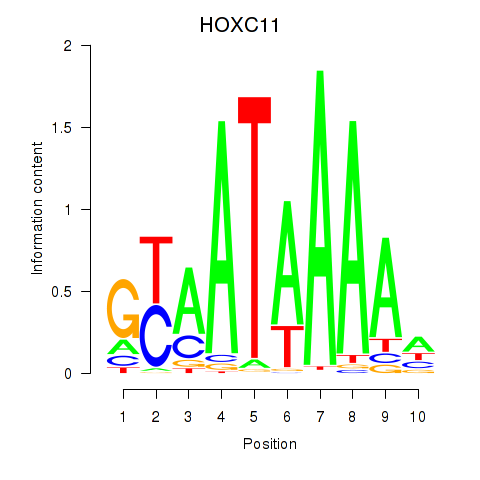
Results for HOXC11
Z-value: 0.40
Transcription factors associated with HOXC11
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXC11
|
ENSG00000123388.4 | homeobox C11 |
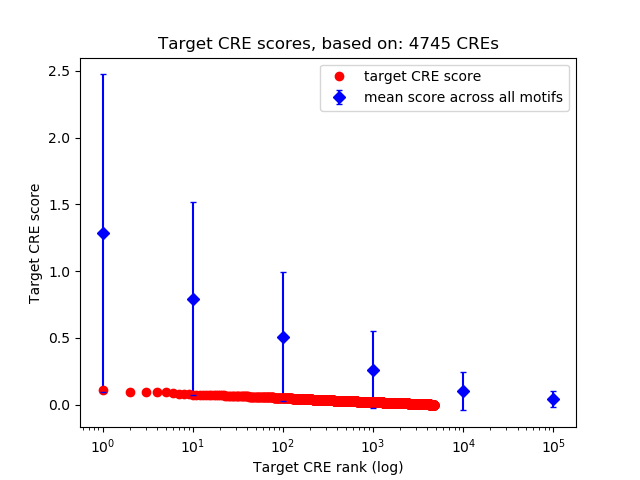
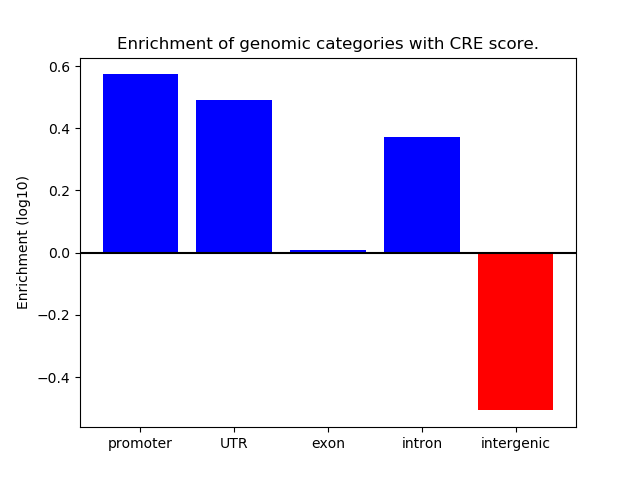
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr12_54367610_54367772 | HOXC11 | 779 | 0.289126 | 0.49 | 1.9e-01 | Click! |
| chr12_54366324_54366475 | HOXC11 | 511 | 0.503307 | 0.25 | 5.2e-01 | Click! |
Activity of the HOXC11 motif across conditions
Conditions sorted by the z-value of the HOXC11 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
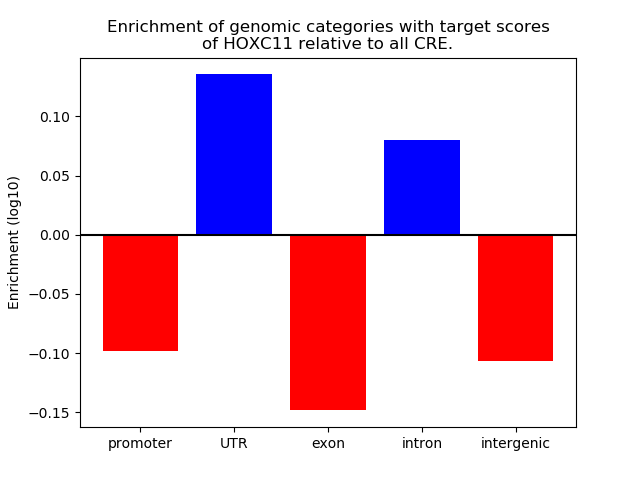
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr18_7958757_7958908 | 0.11 |
PTPRM |
protein tyrosine phosphatase, receptor type, M |
11880 |
0.23 |
| chr5_16493939_16494090 | 0.10 |
FAM134B |
family with sequence similarity 134, member B |
15093 |
0.21 |
| chr1_82265506_82265657 | 0.09 |
LPHN2 |
latrophilin 2 |
501 |
0.89 |
| chr1_170631689_170631840 | 0.09 |
PRRX1 |
paired related homeobox 1 |
511 |
0.87 |
| chr5_80263913_80264064 | 0.09 |
CTC-459I6.1 |
|
7262 |
0.24 |
| chr1_158913144_158913295 | 0.09 |
PYHIN1 |
pyrin and HIN domain family, member 1 |
11861 |
0.21 |
| chr3_73595877_73596028 | 0.08 |
PDZRN3 |
PDZ domain containing ring finger 3 |
14817 |
0.24 |
| chr10_112506565_112506716 | 0.08 |
ENSG00000252036 |
. |
9574 |
0.18 |
| chr6_45632042_45632279 | 0.08 |
ENSG00000252738 |
. |
18319 |
0.29 |
| chr6_35362532_35362683 | 0.07 |
PPARD |
peroxisome proliferator-activated receptor delta |
52205 |
0.1 |
| chr12_52673287_52673438 | 0.07 |
RP11-845M18.7 |
|
422 |
0.69 |
| chr2_69894789_69894940 | 0.07 |
ENSG00000238708 |
. |
373 |
0.88 |
| chr5_64493902_64494516 | 0.07 |
ENSG00000207439 |
. |
75013 |
0.12 |
| chr13_99961636_99961787 | 0.07 |
GPR183 |
G protein-coupled receptor 183 |
2052 |
0.34 |
| chr20_42474741_42474892 | 0.07 |
ENSG00000206801 |
. |
49633 |
0.14 |
| chr4_39531124_39531275 | 0.07 |
UGDH |
UDP-glucose 6-dehydrogenase |
1268 |
0.35 |
| chr3_73072628_73072858 | 0.07 |
ENSG00000238959 |
. |
4216 |
0.2 |
| chr2_88920682_88920833 | 0.07 |
EIF2AK3 |
eukaryotic translation initiation factor 2-alpha kinase 3 |
3690 |
0.2 |
| chr1_83031153_83031304 | 0.07 |
LPHN2 |
latrophilin 2 |
585655 |
0.0 |
| chr6_64516560_64517136 | 0.07 |
RP11-59D5__B.2 |
|
119 |
0.98 |
| chr2_192182945_192183393 | 0.07 |
MYO1B |
myosin IB |
41558 |
0.17 |
| chr2_175766480_175766631 | 0.07 |
CHN1 |
chimerin 1 |
54076 |
0.13 |
| chr6_163849459_163849641 | 0.07 |
QKI |
QKI, KH domain containing, RNA binding |
5310 |
0.35 |
| chr2_197056759_197056910 | 0.07 |
STK17B |
serine/threonine kinase 17b |
15607 |
0.18 |
| chr9_5465910_5466061 | 0.07 |
CD274 |
CD274 molecule |
15460 |
0.19 |
| chr18_56353868_56354019 | 0.07 |
RP11-126O1.4 |
|
9097 |
0.15 |
| chr7_114871692_114871916 | 0.07 |
ENSG00000199764 |
. |
23754 |
0.28 |
| chr16_27252390_27252716 | 0.07 |
NSMCE1 |
non-SMC element 1 homolog (S. cerevisiae) |
8154 |
0.17 |
| chr20_19973253_19973404 | 0.07 |
NAA20 |
N(alpha)-acetyltransferase 20, NatB catalytic subunit |
24432 |
0.19 |
| chr1_94792467_94792618 | 0.07 |
ARHGAP29 |
Rho GTPase activating protein 29 |
89353 |
0.08 |
| chr20_5755973_5756124 | 0.07 |
C20orf196 |
chromosome 20 open reading frame 196 |
24949 |
0.22 |
| chr4_38685842_38686144 | 0.06 |
RP11-617D20.1 |
|
19489 |
0.15 |
| chr3_115719451_115719602 | 0.06 |
ENSG00000243359 |
. |
162811 |
0.04 |
| chr3_142919123_142919274 | 0.06 |
ENSG00000211483 |
. |
34708 |
0.19 |
| chr16_1666082_1666233 | 0.06 |
CRAMP1L |
Crm, cramped-like (Drosophila) |
1516 |
0.23 |
| chr7_150440672_150440823 | 0.06 |
GIMAP5 |
GTPase, IMAP family member 5 |
6311 |
0.17 |
| chr3_172033208_172033359 | 0.06 |
AC092964.1 |
Uncharacterized protein |
935 |
0.62 |
| chr2_239693558_239693739 | 0.06 |
TWIST2 |
twist family bHLH transcription factor 2 |
63025 |
0.13 |
| chr15_60085307_60085458 | 0.06 |
BNIP2 |
BCL2/adenovirus E1B 19kDa interacting protein 2 |
103649 |
0.06 |
| chr2_64750645_64750930 | 0.06 |
AFTPH |
aftiphilin |
678 |
0.74 |
| chr17_55723600_55723751 | 0.06 |
ENSG00000242889 |
. |
28206 |
0.16 |
| chr8_141125038_141125212 | 0.06 |
C8orf17 |
chromosome 8 open reading frame 17 |
181709 |
0.03 |
| chr6_128529618_128529908 | 0.06 |
PTPRK |
protein tyrosine phosphatase, receptor type, K |
113563 |
0.07 |
| chr2_73334235_73334398 | 0.06 |
RAB11FIP5 |
RAB11 family interacting protein 5 (class I) |
5830 |
0.18 |
| chr5_139678742_139678893 | 0.06 |
PFDN1 |
prefoldin subunit 1 |
3851 |
0.19 |
| chrX_39176359_39176510 | 0.06 |
ENSG00000207122 |
. |
235896 |
0.02 |
| chr17_61433493_61433644 | 0.06 |
AC037445.1 |
|
17154 |
0.15 |
| chr6_18247333_18247484 | 0.06 |
DEK |
DEK oncogene |
2563 |
0.28 |
| chr2_40507984_40508135 | 0.06 |
AC007377.1 |
|
19912 |
0.23 |
| chr20_62169801_62169978 | 0.06 |
PTK6 |
protein tyrosine kinase 6 |
1166 |
0.3 |
| chr12_66005489_66005640 | 0.06 |
HMGA2 |
high mobility group AT-hook 2 |
212347 |
0.02 |
| chr16_69250892_69251043 | 0.06 |
RP11-70O5.2 |
|
23485 |
0.11 |
| chr3_180648116_180648267 | 0.06 |
FXR1 |
fragile X mental retardation, autosomal homolog 1 |
14943 |
0.21 |
| chr19_2215762_2215913 | 0.06 |
AC004490.1 |
|
273 |
0.8 |
| chr22_21978223_21978374 | 0.06 |
YDJC |
YdjC homolog (bacterial) |
6027 |
0.1 |
| chr5_38259654_38259805 | 0.06 |
EGFLAM |
EGF-like, fibronectin type III and laminin G domains |
1218 |
0.54 |
| chr20_24387296_24387447 | 0.06 |
SYNDIG1 |
synapse differentiation inducing 1 |
62464 |
0.15 |
| chr5_90476166_90476317 | 0.06 |
ENSG00000199643 |
. |
90303 |
0.1 |
| chr4_39570957_39571108 | 0.06 |
UGDH |
UDP-glucose 6-dehydrogenase |
41101 |
0.11 |
| chr17_76531986_76532137 | 0.06 |
ENSG00000243426 |
. |
2924 |
0.23 |
| chr7_104631689_104631965 | 0.06 |
ENSG00000251911 |
. |
20093 |
0.14 |
| chr6_90854294_90854445 | 0.06 |
ENSG00000222078 |
. |
143144 |
0.04 |
| chr6_158283594_158283745 | 0.06 |
RP11-52J3.2 |
|
12573 |
0.17 |
| chr17_62648244_62648405 | 0.06 |
SMURF2 |
SMAD specific E3 ubiquitin protein ligase 2 |
9706 |
0.22 |
| chr6_140084162_140084413 | 0.06 |
CITED2 |
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 |
388530 |
0.01 |
| chr2_144504368_144504519 | 0.06 |
RP11-434H14.1 |
|
5580 |
0.25 |
| chr16_86817580_86817731 | 0.06 |
FOXL1 |
forkhead box L1 |
205540 |
0.02 |
| chr3_43386050_43386201 | 0.06 |
RP11-188P20.3 |
|
3010 |
0.24 |
| chr17_45959726_45959877 | 0.06 |
SP2 |
Sp2 transcription factor |
13715 |
0.1 |
| chr5_172425652_172425803 | 0.06 |
ATP6V0E1 |
ATPase, H+ transporting, lysosomal 9kDa, V0 subunit e1 |
14893 |
0.14 |
| chr14_61125667_61125818 | 0.06 |
SIX1 |
SIX homeobox 1 |
765 |
0.7 |
| chr9_108011375_108011526 | 0.06 |
SLC44A1 |
solute carrier family 44 (choline transporter), member 1 |
4547 |
0.34 |
| chr5_172382388_172382600 | 0.06 |
RPL26L1 |
ribosomal protein L26-like 1 |
3238 |
0.19 |
| chrX_19778013_19778164 | 0.05 |
SH3KBP1 |
SH3-domain kinase binding protein 1 |
12267 |
0.29 |
| chr6_108486750_108487194 | 0.05 |
OSTM1 |
osteopetrosis associated transmembrane protein 1 |
72 |
0.78 |
| chr3_66529878_66530029 | 0.05 |
LRIG1 |
leucine-rich repeats and immunoglobulin-like domains 1 |
21403 |
0.28 |
| chr3_152808662_152808813 | 0.05 |
RP11-529G21.2 |
|
70827 |
0.11 |
| chr12_8230719_8230959 | 0.05 |
NECAP1 |
NECAP endocytosis associated 1 |
3968 |
0.17 |
| chr18_3252593_3253294 | 0.05 |
MYL12A |
myosin, light chain 12A, regulatory, non-sarcomeric |
670 |
0.64 |
| chr16_18925014_18925165 | 0.05 |
SMG1 |
SMG1 phosphatidylinositol 3-kinase-related kinase |
2054 |
0.25 |
| chr9_20459022_20459173 | 0.05 |
ENSG00000222202 |
. |
40778 |
0.14 |
| chr5_124201652_124202095 | 0.05 |
ZNF608 |
zinc finger protein 608 |
117373 |
0.06 |
| chr12_45227056_45227207 | 0.05 |
NELL2 |
NEL-like 2 (chicken) |
42120 |
0.18 |
| chr1_170562549_170562700 | 0.05 |
RP11-576I22.2 |
|
60836 |
0.12 |
| chr17_37301012_37301163 | 0.05 |
PLXDC1 |
plexin domain containing 1 |
6815 |
0.14 |
| chr1_162541630_162541823 | 0.05 |
UAP1 |
UDP-N-acteylglucosamine pyrophosphorylase 1 |
5899 |
0.17 |
| chr17_73674018_73674169 | 0.05 |
RP11-474I11.8 |
|
1954 |
0.18 |
| chr12_66548262_66548413 | 0.05 |
TMBIM4 |
transmembrane BAX inhibitor motif containing 4 |
14954 |
0.14 |
| chr12_122592230_122592666 | 0.05 |
MLXIP |
MLX interacting protein |
19979 |
0.18 |
| chr3_171812873_171813024 | 0.05 |
FNDC3B |
fibronectin type III domain containing 3B |
31816 |
0.22 |
| chr16_53809610_53810340 | 0.05 |
FTO |
fat mass and obesity associated |
71881 |
0.11 |
| chr12_123243849_123244000 | 0.05 |
DENR |
density-regulated protein |
6553 |
0.14 |
| chr17_61857649_61857869 | 0.05 |
CCDC47 |
coiled-coil domain containing 47 |
4048 |
0.14 |
| chr16_49686147_49686298 | 0.05 |
ZNF423 |
zinc finger protein 423 |
11459 |
0.24 |
| chr7_140593399_140593550 | 0.05 |
ENSG00000271932 |
. |
9602 |
0.22 |
| chr3_30668319_30669005 | 0.05 |
TGFBR2 |
transforming growth factor, beta receptor II (70/80kDa) |
20569 |
0.26 |
| chr8_48648340_48648491 | 0.05 |
CEBPD |
CCAAT/enhancer binding protein (C/EBP), delta |
3233 |
0.24 |
| chr1_198533037_198533238 | 0.05 |
ATP6V1G3 |
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G3 |
23062 |
0.24 |
| chr5_58187131_58187282 | 0.05 |
CTD-2176I21.2 |
|
27965 |
0.22 |
| chr18_61442167_61442318 | 0.05 |
SERPINB7 |
serpin peptidase inhibitor, clade B (ovalbumin), member 7 |
402 |
0.88 |
| chr14_72026196_72026347 | 0.05 |
SIPA1L1 |
signal-induced proliferation-associated 1 like 1 |
26727 |
0.25 |
| chr5_176869820_176870094 | 0.05 |
PRR7-AS1 |
PRR7 antisense RNA 1 |
1521 |
0.22 |
| chr3_73481941_73482092 | 0.05 |
PDZRN3 |
PDZ domain containing ring finger 3 |
1039 |
0.68 |
| chr6_139492633_139492784 | 0.05 |
HECA |
headcase homolog (Drosophila) |
36459 |
0.17 |
| chr12_110992885_110993169 | 0.05 |
PPTC7 |
PTC7 protein phosphatase homolog (S. cerevisiae) |
28098 |
0.12 |
| chr13_26636007_26636158 | 0.05 |
SHISA2 |
shisa family member 2 |
10913 |
0.27 |
| chr6_106643256_106643407 | 0.05 |
ENSG00000244710 |
. |
88255 |
0.07 |
| chr11_116881940_116882091 | 0.05 |
ENSG00000264344 |
. |
4230 |
0.2 |
| chr3_28391593_28391746 | 0.05 |
ZCWPW2 |
zinc finger, CW type with PWWP domain 2 |
1029 |
0.41 |
| chr2_25506534_25506685 | 0.05 |
DNMT3A |
DNA (cytosine-5-)-methyltransferase 3 alpha |
31429 |
0.17 |
| chr11_121358351_121358610 | 0.05 |
RP11-730K11.1 |
|
34758 |
0.19 |
| chr8_42052077_42052228 | 0.05 |
PLAT |
plasminogen activator, tissue |
907 |
0.54 |
| chr13_99148708_99148963 | 0.05 |
STK24 |
serine/threonine kinase 24 |
22686 |
0.19 |
| chr7_66397167_66397485 | 0.05 |
TMEM248 |
transmembrane protein 248 |
1715 |
0.35 |
| chr7_28005907_28006058 | 0.05 |
JAZF1 |
JAZF zinc finger 1 |
25693 |
0.23 |
| chr8_132857197_132857348 | 0.05 |
EFR3A |
EFR3 homolog A (S. cerevisiae) |
59063 |
0.16 |
| chr5_70778847_70779136 | 0.05 |
BDP1 |
B double prime 1, subunit of RNA polymerase III transcription initiation factor IIIB |
27549 |
0.21 |
| chr14_55140117_55140268 | 0.05 |
SAMD4A |
sterile alpha motif domain containing 4A |
81359 |
0.1 |
| chr12_67136436_67136587 | 0.05 |
GRIP1 |
glutamate receptor interacting protein 1 |
61249 |
0.16 |
| chr10_116634856_116635150 | 0.05 |
FAM160B1 |
family with sequence similarity 160, member B1 |
14453 |
0.25 |
| chr14_31663861_31664209 | 0.05 |
HECTD1 |
HECT domain containing E3 ubiquitin protein ligase 1 |
12654 |
0.17 |
| chr7_139452923_139453350 | 0.05 |
HIPK2 |
homeodomain interacting protein kinase 2 |
24381 |
0.2 |
| chr11_33769435_33769586 | 0.05 |
FBXO3 |
F-box protein 3 |
5435 |
0.16 |
| chr5_39025357_39025508 | 0.05 |
RICTOR |
RPTOR independent companion of MTOR, complex 2 |
49059 |
0.16 |
| chr12_88728604_88728755 | 0.05 |
ENSG00000199245 |
. |
95540 |
0.09 |
| chr1_230784406_230784557 | 0.05 |
COG2 |
component of oligomeric golgi complex 2 |
5924 |
0.24 |
| chr11_33295654_33295805 | 0.05 |
HIPK3 |
homeodomain interacting protein kinase 3 |
15852 |
0.21 |
| chr4_169752514_169752665 | 0.05 |
PALLD |
palladin, cytoskeletal associated protein |
567 |
0.7 |
| chr1_186271695_186271846 | 0.05 |
PRG4 |
proteoglycan 4 |
6359 |
0.19 |
| chr8_24013639_24013790 | 0.05 |
ENSG00000207201 |
. |
79043 |
0.11 |
| chr8_48254486_48254637 | 0.05 |
SPIDR |
scaffolding protein involved in DNA repair |
81004 |
0.1 |
| chrX_31284130_31284281 | 0.05 |
DMD |
dystrophin |
769 |
0.77 |
| chr1_25771368_25771519 | 0.05 |
TMEM57 |
transmembrane protein 57 |
14032 |
0.15 |
| chr7_7691133_7691284 | 0.05 |
RPA3 |
replication protein A3, 14kDa |
10605 |
0.26 |
| chr3_56501411_56501562 | 0.05 |
ERC2 |
ELKS/RAB6-interacting/CAST family member 2 |
905 |
0.72 |
| chr5_179975750_179975903 | 0.05 |
CNOT6 |
CCR4-NOT transcription complex, subunit 6 |
19551 |
0.16 |
| chrX_94314503_94314654 | 0.05 |
ENSG00000221187 |
. |
3647 |
0.36 |
| chr9_91151964_91152115 | 0.05 |
NXNL2 |
nucleoredoxin-like 2 |
2019 |
0.47 |
| chr2_10121974_10122125 | 0.05 |
GRHL1 |
grainyhead-like 1 (Drosophila) |
13385 |
0.16 |
| chr15_39923761_39923912 | 0.05 |
FSIP1 |
fibrous sheath interacting protein 1 |
30709 |
0.17 |
| chr2_230763302_230763453 | 0.05 |
ENSG00000222344 |
. |
4315 |
0.18 |
| chr9_21560365_21560626 | 0.04 |
MIR31HG |
MIR31 host gene (non-protein coding) |
827 |
0.62 |
| chr1_92040002_92040153 | 0.04 |
CDC7 |
cell division cycle 7 |
73382 |
0.12 |
| chr13_52390546_52391185 | 0.04 |
RP11-327P2.5 |
|
12432 |
0.19 |
| chr8_107160982_107161133 | 0.04 |
ENSG00000251003 |
. |
88305 |
0.1 |
| chr2_38593562_38593713 | 0.04 |
ATL2 |
atlastin GTPase 2 |
9729 |
0.29 |
| chr1_24022876_24023104 | 0.04 |
RPL11 |
ribosomal protein L11 |
3891 |
0.19 |
| chr12_67355616_67355767 | 0.04 |
GRIP1 |
glutamate receptor interacting protein 1 |
157725 |
0.04 |
| chr16_67538677_67538891 | 0.04 |
FAM65A |
family with sequence similarity 65, member A |
13537 |
0.09 |
| chr18_73122194_73122345 | 0.04 |
RP11-321M21.3 |
HCG1996301; Uncharacterized protein |
3302 |
0.31 |
| chr6_157148026_157148177 | 0.04 |
RP11-230C9.3 |
|
46512 |
0.15 |
| chr10_77879085_77879236 | 0.04 |
ENSG00000221232 |
. |
7909 |
0.3 |
| chr13_75018542_75018693 | 0.04 |
LINC00381 |
long intergenic non-protein coding RNA 381 |
25307 |
0.21 |
| chr12_78412506_78412657 | 0.04 |
NAV3 |
neuron navigator 3 |
52525 |
0.17 |
| chr2_26465301_26465452 | 0.04 |
HADHB |
hydroxyacyl-CoA dehydrogenase/3-ketoacyl-CoA thiolase/enoyl-CoA hydratase (trifunctional protein), beta subunit |
662 |
0.66 |
| chr17_41739555_41740247 | 0.04 |
MEOX1 |
mesenchyme homeobox 1 |
579 |
0.74 |
| chr10_89705727_89705964 | 0.04 |
ENSG00000200891 |
. |
48607 |
0.13 |
| chr1_113007964_113008322 | 0.04 |
WNT2B |
wingless-type MMTV integration site family, member 2B |
1020 |
0.58 |
| chr17_60132149_60132300 | 0.04 |
MED13 |
mediator complex subunit 13 |
10419 |
0.17 |
| chr17_77944274_77944425 | 0.04 |
TBC1D16 |
TBC1 domain family, member 16 |
18537 |
0.16 |
| chr12_1875760_1875911 | 0.04 |
CACNA2D4 |
calcium channel, voltage-dependent, alpha 2/delta subunit 4 |
45051 |
0.13 |
| chr13_25034322_25034473 | 0.04 |
TPTE2P6 |
transmembrane phosphoinositide 3-phosphatase and tensin homolog 2 pseudogene 6 |
52102 |
0.12 |
| chr9_137418567_137418718 | 0.04 |
COL5A1 |
collagen, type V, alpha 1 |
114978 |
0.05 |
| chr5_98261626_98261777 | 0.04 |
CHD1 |
chromodomain helicase DNA binding protein 1 |
539 |
0.8 |
| chr11_110167983_110168234 | 0.04 |
RDX |
radixin |
661 |
0.83 |
| chr16_72281999_72282150 | 0.04 |
PMFBP1 |
polyamine modulated factor 1 binding protein 1 |
71297 |
0.1 |
| chr2_118769373_118769761 | 0.04 |
CCDC93 |
coiled-coil domain containing 93 |
2142 |
0.23 |
| chr17_70358516_70358667 | 0.04 |
SOX9 |
SRY (sex determining region Y)-box 9 |
241430 |
0.02 |
| chr17_78762568_78762807 | 0.04 |
RP11-28G8.1 |
|
16745 |
0.23 |
| chr14_89078082_89078377 | 0.04 |
ZC3H14 |
zinc finger CCCH-type containing 14 |
4601 |
0.21 |
| chr16_21311524_21311721 | 0.04 |
CRYM |
crystallin, mu |
2750 |
0.29 |
| chr13_27706090_27706241 | 0.04 |
USP12-AS1 |
USP12 antisense RNA 1 |
30827 |
0.15 |
| chr13_114862694_114862923 | 0.04 |
RASA3-IT1 |
RASA3 intronic transcript 1 (non-protein coding) |
11487 |
0.22 |
| chr18_67232011_67232162 | 0.04 |
RP11-465I4.2 |
|
53844 |
0.17 |
| chr21_43061018_43061225 | 0.04 |
ENSG00000252771 |
. |
36278 |
0.17 |
| chr1_225843948_225844099 | 0.04 |
ENAH |
enabled homolog (Drosophila) |
3179 |
0.27 |
| chr19_11146514_11146699 | 0.04 |
SMARCA4 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 |
5117 |
0.16 |
| chr1_31123601_31123752 | 0.04 |
MATN1-AS1 |
MATN1 antisense RNA 1 |
67675 |
0.1 |
| chr11_95984153_95984304 | 0.04 |
ENSG00000266192 |
. |
90374 |
0.08 |
| chr14_90797503_90797880 | 0.04 |
NRDE2 |
NRDE-2, necessary for RNA interference, domain containing |
588 |
0.77 |
| chr8_714573_714724 | 0.04 |
ERICH1-AS1 |
ERICH1 antisense RNA 1 |
26906 |
0.18 |
| chr2_87552860_87553169 | 0.04 |
IGKV3OR2-268 |
immunoglobulin kappa variable 3/OR2-268 (non-functional) |
12620 |
0.29 |
| chr5_51968673_51968824 | 0.04 |
ITGA1 |
integrin, alpha 1 |
114982 |
0.06 |
| chr9_72878166_72878317 | 0.04 |
SMC5 |
structural maintenance of chromosomes 5 |
4304 |
0.21 |
| chr8_98565933_98566084 | 0.04 |
MTDH |
metadherin |
90399 |
0.09 |
| chr5_43554525_43554713 | 0.04 |
PAIP1 |
poly(A) binding protein interacting protein 1 |
1861 |
0.28 |
| chr6_44598460_44598611 | 0.04 |
ENSG00000266619 |
. |
195157 |
0.03 |
| chr2_191513228_191513379 | 0.04 |
NAB1 |
NGFI-A binding protein 1 (EGR1 binding protein 1) |
318 |
0.91 |
| chr13_33199858_33200009 | 0.04 |
PDS5B |
PDS5, regulator of cohesion maintenance, homolog B (S. cerevisiae) |
39369 |
0.19 |
| chr5_95534441_95534647 | 0.04 |
ENSG00000206997 |
. |
11387 |
0.28 |
| chr3_142754964_142755115 | 0.04 |
U2SURP |
U2 snRNP-associated SURP domain containing |
114 |
0.97 |
| chr8_61446627_61446778 | 0.04 |
RAB2A |
RAB2A, member RAS oncogene family |
16962 |
0.2 |
| chr1_107816447_107816598 | 0.04 |
NTNG1 |
netrin G1 |
125306 |
0.06 |
| chr15_78715144_78715338 | 0.04 |
IREB2 |
iron-responsive element binding protein 2 |
14532 |
0.17 |
| chr6_56598121_56598272 | 0.04 |
DST |
dystonin |
52481 |
0.16 |
| chr8_94927918_94928069 | 0.04 |
ENSG00000264448 |
. |
354 |
0.8 |
| chr5_79447122_79447273 | 0.04 |
CTC-458I2.2 |
|
23013 |
0.19 |
| chrX_72782730_72782881 | 0.04 |
CHIC1 |
cysteine-rich hydrophobic domain 1 |
231 |
0.96 |
| chr12_127073806_127073957 | 0.04 |
ENSG00000239776 |
. |
576791 |
0.0 |
| chr3_153850737_153851019 | 0.04 |
ARHGEF26 |
Rho guanine nucleotide exchange factor (GEF) 26 |
11649 |
0.22 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.0 | 0.0 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.0 | GO:0046398 | UDP-glucuronate metabolic process(GO:0046398) |
| 0.0 | 0.0 | GO:0060423 | foregut regionalization(GO:0060423) lung field specification(GO:0060424) lung induction(GO:0060492) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.0 | GO:0002162 | dystroglycan binding(GO:0002162) |