Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
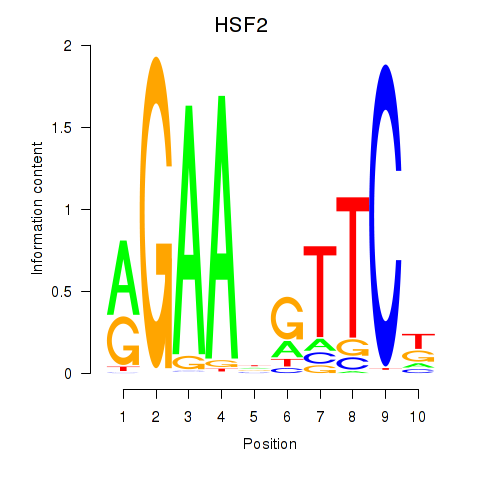
Results for HSF2
Z-value: 0.61
Transcription factors associated with HSF2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HSF2
|
ENSG00000025156.8 | heat shock transcription factor 2 |
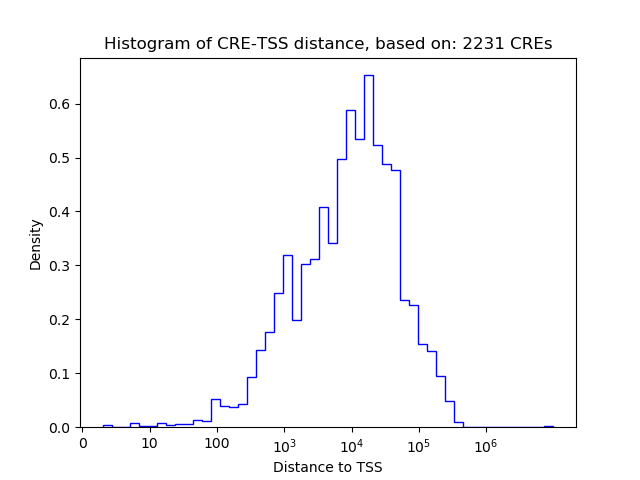
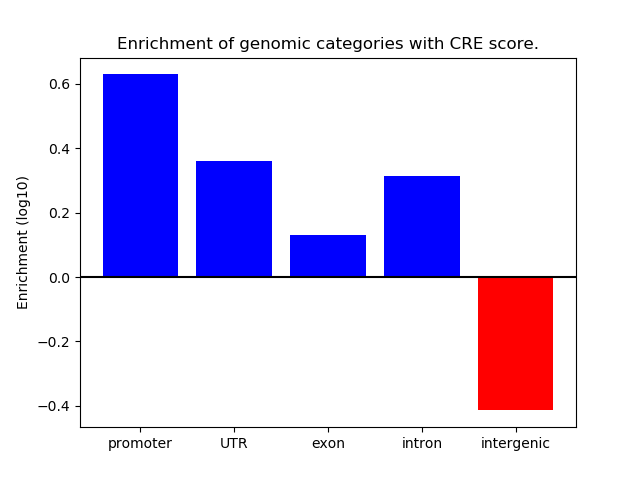
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr6_122725724_122725875 | HSF2 | 5103 | 0.289379 | 0.84 | 4.5e-03 | Click! |
| chr6_122733810_122733961 | HSF2 | 9416 | 0.254651 | 0.81 | 8.6e-03 | Click! |
| chr6_122721794_122722012 | HSF2 | 1207 | 0.589589 | 0.78 | 1.4e-02 | Click! |
| chr6_122722453_122722604 | HSF2 | 1832 | 0.452637 | 0.77 | 1.6e-02 | Click! |
| chr6_122724067_122724218 | HSF2 | 3446 | 0.326410 | 0.74 | 2.2e-02 | Click! |
Activity of the HSF2 motif across conditions
Conditions sorted by the z-value of the HSF2 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
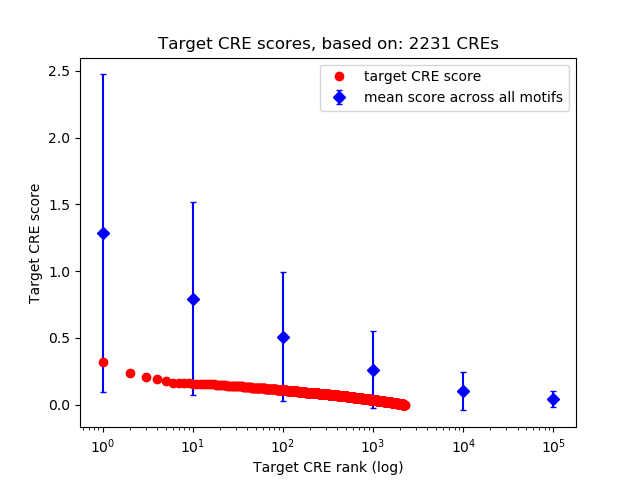
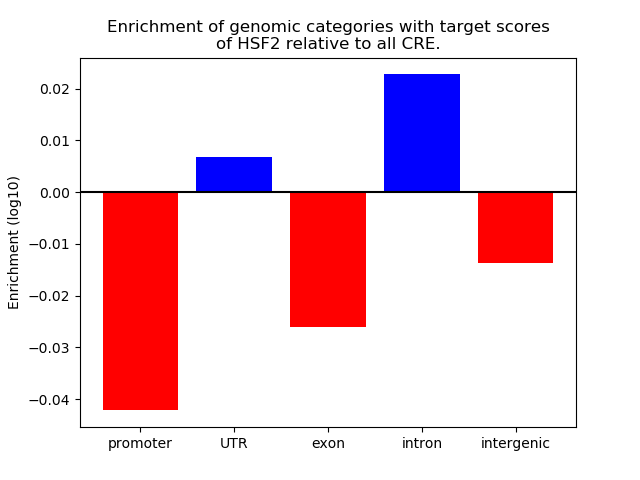
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr1_29289116_29289267 | 0.32 |
ENSG00000206704 |
. |
23031 |
0.18 |
| chr14_99670994_99671151 | 0.24 |
AL162151.4 |
|
46319 |
0.15 |
| chr2_95741879_95742163 | 0.21 |
AC103563.9 |
|
23100 |
0.15 |
| chr5_133426970_133427238 | 0.19 |
TCF7 |
transcription factor 7 (T-cell specific, HMG-box) |
23298 |
0.2 |
| chr20_57723034_57723340 | 0.18 |
ZNF831 |
zinc finger protein 831 |
42888 |
0.16 |
| chr2_98334024_98334251 | 0.17 |
ZAP70 |
zeta-chain (TCR) associated protein kinase 70kDa |
4114 |
0.2 |
| chr22_40729451_40729738 | 0.16 |
ADSL |
adenylosuccinate lyase |
12913 |
0.18 |
| chr9_95786987_95787346 | 0.16 |
FGD3 |
FYVE, RhoGEF and PH domain containing 3 |
9824 |
0.19 |
| chr19_10874164_10874345 | 0.16 |
ENSG00000265879 |
. |
16676 |
0.11 |
| chr12_55382686_55382958 | 0.16 |
TESPA1 |
thymocyte expressed, positive selection associated 1 |
4292 |
0.26 |
| chr11_60878524_60878675 | 0.16 |
CD5 |
CD5 molecule |
8618 |
0.18 |
| chr11_45856166_45856328 | 0.16 |
CRY2 |
cryptochrome 2 (photolyase-like) |
12422 |
0.15 |
| chr2_36474188_36474481 | 0.16 |
CRIM1 |
cysteine rich transmembrane BMP regulator 1 (chordin-like) |
108735 |
0.08 |
| chr2_111610099_111610292 | 0.15 |
ACOXL |
acyl-CoA oxidase-like |
47299 |
0.19 |
| chr7_149561836_149562048 | 0.15 |
RP4-751H13.7 |
|
2361 |
0.25 |
| chr22_20819732_20820026 | 0.15 |
ENSG00000255156 |
. |
9719 |
0.1 |
| chr15_44961507_44961922 | 0.15 |
PATL2 |
protein associated with topoisomerase II homolog 2 (yeast) |
1031 |
0.46 |
| chr11_121320555_121321108 | 0.15 |
SORL1 |
sortilin-related receptor, L(DLR class) A repeats containing |
2081 |
0.35 |
| chr10_14593831_14594082 | 0.15 |
FAM107B |
family with sequence similarity 107, member B |
2272 |
0.38 |
| chr15_52022956_52023209 | 0.15 |
LYSMD2 |
LysM, putative peptidoglycan-binding, domain containing 2 |
7236 |
0.14 |
| chr17_8801323_8801785 | 0.15 |
PIK3R5 |
phosphoinositide-3-kinase, regulatory subunit 5 |
13270 |
0.21 |
| chr6_128255047_128255301 | 0.15 |
THEMIS |
thymocyte selection associated |
15398 |
0.26 |
| chr1_169662886_169663277 | 0.14 |
SELL |
selectin L |
17758 |
0.18 |
| chr8_131260396_131260639 | 0.14 |
ASAP1 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1 |
1229 |
0.61 |
| chr17_38758822_38758973 | 0.14 |
SMARCE1 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily e, member 1 |
29645 |
0.12 |
| chr7_8172413_8172617 | 0.14 |
AC006042.6 |
|
18860 |
0.2 |
| chr17_49505999_49506240 | 0.14 |
UTP18 |
UTP18 small subunit (SSU) processome component homolog (yeast) |
143490 |
0.04 |
| chr10_22946008_22946242 | 0.14 |
PIP4K2A |
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha |
56912 |
0.15 |
| chr13_100039907_100040178 | 0.14 |
ENSG00000207719 |
. |
31657 |
0.16 |
| chr17_14097365_14097669 | 0.14 |
AC005224.2 |
|
16288 |
0.19 |
| chr5_39181205_39181356 | 0.14 |
FYB |
FYN binding protein |
21849 |
0.24 |
| chr19_16484143_16484894 | 0.14 |
EPS15L1 |
epidermal growth factor receptor pathway substrate 15-like 1 |
11754 |
0.14 |
| chr1_167598142_167598391 | 0.14 |
RCSD1 |
RCSD domain containing 1 |
1064 |
0.46 |
| chr20_58639264_58639415 | 0.14 |
C20orf197 |
chromosome 20 open reading frame 197 |
8359 |
0.26 |
| chr17_56422655_56423315 | 0.14 |
SUPT4H1 |
suppressor of Ty 4 homolog 1 (S. cerevisiae) |
6037 |
0.12 |
| chr13_100024049_100024200 | 0.14 |
ENSG00000207719 |
. |
15739 |
0.2 |
| chr15_38979216_38979456 | 0.13 |
C15orf53 |
chromosome 15 open reading frame 53 |
9463 |
0.29 |
| chr1_185250977_185251218 | 0.13 |
ENSG00000252612 |
. |
15430 |
0.16 |
| chr12_27124096_27124264 | 0.13 |
RP11-421F16.3 |
|
339 |
0.84 |
| chr4_68447482_68447633 | 0.13 |
STAP1 |
signal transducing adaptor family member 1 |
23111 |
0.19 |
| chr8_37134401_37134693 | 0.13 |
RP11-150O12.6 |
|
239992 |
0.02 |
| chr7_50320921_50321337 | 0.13 |
IKZF1 |
IKAROS family zinc finger 1 (Ikaros) |
23195 |
0.25 |
| chr5_140981117_140981268 | 0.13 |
DIAPH1 |
diaphanous-related formin 1 |
14140 |
0.11 |
| chr4_103510077_103510373 | 0.13 |
NFKB1 |
nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 |
11204 |
0.22 |
| chr1_206734639_206734815 | 0.13 |
RASSF5 |
Ras association (RalGDS/AF-6) domain family member 5 |
4234 |
0.2 |
| chr5_66118537_66118688 | 0.13 |
MAST4 |
microtubule associated serine/threonine kinase family member 4 |
5990 |
0.33 |
| chr8_134531655_134531880 | 0.13 |
ST3GAL1 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
20141 |
0.27 |
| chr11_117800803_117801005 | 0.13 |
TMPRSS13 |
transmembrane protease, serine 13 |
730 |
0.65 |
| chr8_61708195_61708346 | 0.13 |
RP11-33I11.2 |
|
13895 |
0.26 |
| chr21_19155941_19156118 | 0.13 |
AL109761.5 |
|
9776 |
0.23 |
| chrX_119553913_119554186 | 0.13 |
LAMP2 |
lysosomal-associated membrane protein 2 |
48798 |
0.13 |
| chr4_3204868_3205180 | 0.13 |
MSANTD1 |
Myb/SANT-like DNA-binding domain containing 1 |
41072 |
0.15 |
| chr19_3755879_3756030 | 0.13 |
AC005954.3 |
|
703 |
0.46 |
| chr19_54872936_54873267 | 0.13 |
LAIR1 |
leukocyte-associated immunoglobulin-like receptor 1 |
545 |
0.63 |
| chr20_57722445_57722676 | 0.12 |
ZNF831 |
zinc finger protein 831 |
43515 |
0.15 |
| chr2_68639578_68639939 | 0.12 |
AC015969.3 |
|
47042 |
0.11 |
| chr2_113940950_113941232 | 0.12 |
AC016683.5 |
|
8154 |
0.14 |
| chr16_68105212_68105363 | 0.12 |
NFATC3 |
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 3 |
13960 |
0.09 |
| chr22_39714376_39714672 | 0.12 |
RPL3 |
ribosomal protein L3 |
95 |
0.87 |
| chr5_39208813_39209149 | 0.12 |
FYB |
FYN binding protein |
5852 |
0.3 |
| chr17_35032252_35032424 | 0.12 |
MRM1 |
mitochondrial rRNA methyltransferase 1 homolog (S. cerevisiae) |
73947 |
0.08 |
| chr14_69280428_69280742 | 0.12 |
ZFP36L1 |
ZFP36 ring finger protein-like 1 |
17395 |
0.19 |
| chr12_55143896_55144095 | 0.12 |
MUCL1 |
mucin-like 1 |
89158 |
0.07 |
| chr14_90148267_90148437 | 0.12 |
ENSG00000200312 |
. |
30503 |
0.14 |
| chr4_164479105_164479708 | 0.12 |
ENSG00000264535 |
. |
34962 |
0.17 |
| chr13_52527282_52527433 | 0.12 |
ATP7B |
ATPase, Cu++ transporting, beta polypeptide |
8694 |
0.22 |
| chrX_71321953_71322237 | 0.12 |
RGAG4 |
retrotransposon gag domain containing 4 |
29583 |
0.14 |
| chr14_98640560_98640817 | 0.12 |
ENSG00000222066 |
. |
157399 |
0.04 |
| chr17_15493946_15494478 | 0.12 |
CDRT1 |
CMT1A duplicated region transcript 1 |
2510 |
0.2 |
| chr2_237460577_237460750 | 0.12 |
ACKR3 |
atypical chemokine receptor 3 |
15767 |
0.24 |
| chr14_107180053_107180204 | 0.12 |
IGHV2-70 |
immunoglobulin heavy variable 2-70 |
790 |
0.27 |
| chr10_14622794_14622947 | 0.12 |
FAM107B |
family with sequence similarity 107, member B |
8532 |
0.25 |
| chr19_4303888_4304210 | 0.12 |
FSD1 |
fibronectin type III and SPRY domain containing 1 |
548 |
0.54 |
| chr1_24863399_24863781 | 0.12 |
ENSG00000266551 |
. |
7386 |
0.17 |
| chr14_22392361_22392865 | 0.12 |
ENSG00000222776 |
. |
143828 |
0.04 |
| chr17_76340922_76341205 | 0.12 |
SOCS3 |
suppressor of cytokine signaling 3 |
15092 |
0.14 |
| chr2_197048278_197048429 | 0.12 |
STK17B |
serine/threonine kinase 17b |
7126 |
0.2 |
| chr3_195277399_195277835 | 0.12 |
AC091633.3 |
|
4198 |
0.18 |
| chr2_109225906_109226290 | 0.12 |
LIMS1 |
LIM and senescent cell antigen-like domains 1 |
2481 |
0.35 |
| chr8_27235111_27235488 | 0.12 |
PTK2B |
protein tyrosine kinase 2 beta |
2869 |
0.31 |
| chr7_50419027_50419178 | 0.12 |
IKZF1 |
IKAROS family zinc finger 1 (Ikaros) |
51857 |
0.15 |
| chr19_16473682_16473998 | 0.12 |
EPS15L1 |
epidermal growth factor receptor pathway substrate 15-like 1 |
1076 |
0.45 |
| chr1_109188164_109188315 | 0.11 |
HENMT1 |
HEN1 methyltransferase homolog 1 (Arabidopsis) |
15438 |
0.19 |
| chr16_23879975_23880302 | 0.11 |
PRKCB |
protein kinase C, beta |
31594 |
0.2 |
| chr20_49546677_49546828 | 0.11 |
RP5-914P20.5 |
|
769 |
0.42 |
| chr2_109646552_109646703 | 0.11 |
EDAR |
ectodysplasin A receptor |
40799 |
0.19 |
| chr2_144070446_144070597 | 0.11 |
RP11-190J23.1 |
|
140780 |
0.05 |
| chr17_18946909_18947248 | 0.11 |
GRAP |
GRB2-related adaptor protein |
1280 |
0.31 |
| chr19_47125473_47125755 | 0.11 |
PTGIR |
prostaglandin I2 (prostacyclin) receptor (IP) |
2680 |
0.13 |
| chr19_42276662_42276849 | 0.11 |
AC011513.4 |
|
14046 |
0.12 |
| chr4_82483575_82483736 | 0.11 |
RASGEF1B |
RasGEF domain family, member 1B |
90586 |
0.1 |
| chr1_160602079_160602495 | 0.11 |
SLAMF1 |
signaling lymphocytic activation molecule family member 1 |
14524 |
0.15 |
| chr1_226901230_226901381 | 0.11 |
ITPKB |
inositol-trisphosphate 3-kinase B |
23854 |
0.18 |
| chr17_47837882_47838168 | 0.11 |
FAM117A |
family with sequence similarity 117, member A |
3468 |
0.2 |
| chr15_66113575_66113726 | 0.11 |
ENSG00000252715 |
. |
22172 |
0.15 |
| chr3_177269440_177269746 | 0.11 |
ENSG00000252028 |
. |
48253 |
0.18 |
| chr13_52404399_52404550 | 0.11 |
RP11-327P2.5 |
|
26041 |
0.16 |
| chr1_229386021_229386325 | 0.11 |
TMEM78 |
transmembrane protein 78 |
790 |
0.59 |
| chrX_13100415_13100651 | 0.11 |
FAM9C |
family with sequence similarity 9, member C |
37732 |
0.2 |
| chr5_40592087_40592238 | 0.11 |
ENSG00000199552 |
. |
62903 |
0.13 |
| chr20_52302438_52302667 | 0.11 |
ENSG00000238468 |
. |
17255 |
0.24 |
| chr22_39385536_39385755 | 0.11 |
APOBEC3B |
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3B |
7231 |
0.12 |
| chr3_32413325_32413665 | 0.11 |
CMTM7 |
CKLF-like MARVEL transmembrane domain containing 7 |
19668 |
0.23 |
| chr3_18431741_18431970 | 0.11 |
RP11-158G18.1 |
|
19272 |
0.22 |
| chr1_27936707_27936953 | 0.11 |
AHDC1 |
AT hook, DNA binding motif, containing 1 |
6687 |
0.15 |
| chr6_40407629_40407780 | 0.11 |
ENSG00000252767 |
. |
32112 |
0.18 |
| chr8_2212040_2212191 | 0.11 |
MYOM2 |
myomesin 2 |
218931 |
0.02 |
| chr20_32592265_32592477 | 0.11 |
RALY |
RALY heterogeneous nuclear ribonucleoprotein |
10612 |
0.21 |
| chr2_162811958_162812253 | 0.11 |
ENSG00000253046 |
. |
36813 |
0.2 |
| chr8_125575470_125576116 | 0.11 |
MTSS1 |
metastasis suppressor 1 |
2175 |
0.26 |
| chr20_3071323_3071474 | 0.11 |
ENSG00000263905 |
. |
3463 |
0.14 |
| chr10_13160669_13160820 | 0.11 |
OPTN |
optineurin |
2476 |
0.29 |
| chr6_159139178_159139369 | 0.11 |
ENSG00000265558 |
. |
46512 |
0.12 |
| chr2_192012454_192012731 | 0.11 |
STAT4 |
signal transducer and activator of transcription 4 |
3105 |
0.29 |
| chr6_131520209_131520483 | 0.11 |
AKAP7 |
A kinase (PRKA) anchor protein 7 |
785 |
0.78 |
| chr2_65046448_65046766 | 0.11 |
ENSG00000239891 |
. |
2095 |
0.31 |
| chr2_233565834_233565985 | 0.10 |
GIGYF2 |
GRB10 interacting GYF protein 2 |
3847 |
0.18 |
| chr16_24161760_24161948 | 0.10 |
PRKCB |
protein kinase C, beta |
1152 |
0.63 |
| chr1_211503644_211504001 | 0.10 |
TRAF5 |
TNF receptor-associated factor 5 |
3643 |
0.3 |
| chr1_25319504_25319655 | 0.10 |
RUNX3 |
runt-related transcription factor 3 |
28078 |
0.16 |
| chr7_45069539_45069790 | 0.10 |
CCM2 |
cerebral cavernous malformation 2 |
2393 |
0.23 |
| chr2_7050734_7050964 | 0.10 |
RNF144A |
ring finger protein 144A |
6674 |
0.18 |
| chr21_36340846_36341061 | 0.10 |
RUNX1 |
runt-related transcription factor 1 |
78866 |
0.12 |
| chr1_206728493_206728745 | 0.10 |
RASSF5 |
Ras association (RalGDS/AF-6) domain family member 5 |
1874 |
0.31 |
| chr1_197737919_197738070 | 0.10 |
DENND1B |
DENN/MADD domain containing 1B |
6328 |
0.23 |
| chr4_153507165_153507316 | 0.10 |
ENSG00000268471 |
. |
49660 |
0.13 |
| chr3_134013773_134013924 | 0.10 |
RYK |
receptor-like tyrosine kinase |
44159 |
0.16 |
| chr8_82006436_82006760 | 0.10 |
PAG1 |
phosphoprotein associated with glycosphingolipid microdomains 1 |
17705 |
0.27 |
| chr21_19164466_19164649 | 0.10 |
AL109761.5 |
|
1248 |
0.53 |
| chr20_39784612_39784792 | 0.10 |
RP1-1J6.2 |
|
18059 |
0.18 |
| chr14_53016892_53017043 | 0.10 |
TXNDC16 |
thioredoxin domain containing 16 |
2256 |
0.3 |
| chr8_28917519_28917756 | 0.10 |
CTD-2647L4.4 |
|
4185 |
0.17 |
| chr12_32044487_32044661 | 0.10 |
ENSG00000252584 |
. |
24067 |
0.19 |
| chr7_130737156_130737472 | 0.10 |
LINC-PINT |
long intergenic non-protein coding RNA, p53 induced transcript |
5 |
0.98 |
| chr1_26861291_26861452 | 0.10 |
RPS6KA1 |
ribosomal protein S6 kinase, 90kDa, polypeptide 1 |
3552 |
0.18 |
| chr16_49517590_49517775 | 0.10 |
C16orf78 |
chromosome 16 open reading frame 78 |
109948 |
0.07 |
| chr16_12845026_12845177 | 0.10 |
CTD-2583P5.1 |
|
5371 |
0.19 |
| chr2_54226465_54226799 | 0.10 |
ACYP2 |
acylphosphatase 2, muscle type |
28387 |
0.17 |
| chr22_37518928_37519269 | 0.10 |
TMPRSS6 |
transmembrane protease, serine 6 |
13495 |
0.11 |
| chr8_38219385_38219619 | 0.10 |
WHSC1L1 |
Wolf-Hirschhorn syndrome candidate 1-like 1 |
18837 |
0.11 |
| chr3_71829654_71829923 | 0.10 |
PROK2 |
prokineticin 2 |
4424 |
0.22 |
| chr5_169693142_169693293 | 0.10 |
LCP2 |
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa) |
1083 |
0.57 |
| chr6_156719174_156719407 | 0.10 |
ENSG00000212295 |
. |
19406 |
0.3 |
| chr17_47791372_47791523 | 0.10 |
SLC35B1 |
solute carrier family 35, member B1 |
5071 |
0.15 |
| chr5_39208073_39208542 | 0.10 |
FYB |
FYN binding protein |
5178 |
0.31 |
| chr15_70795218_70795369 | 0.10 |
UACA |
uveal autoantigen with coiled-coil domains and ankyrin repeats |
199327 |
0.03 |
| chr12_9760324_9760475 | 0.10 |
KLRB1 |
killer cell lectin-like receptor subfamily B, member 1 |
83 |
0.96 |
| chr1_83973426_83973603 | 0.10 |
ENSG00000223231 |
. |
286046 |
0.01 |
| chr20_25034683_25034984 | 0.10 |
ACSS1 |
acyl-CoA synthetase short-chain family member 1 |
3985 |
0.24 |
| chr2_204974005_204974290 | 0.10 |
ICOS |
inducible T-cell co-stimulator |
172644 |
0.03 |
| chr14_68974151_68974302 | 0.10 |
RAD51B |
RAD51 paralog B |
95999 |
0.08 |
| chr18_60972547_60972701 | 0.10 |
RP11-28F1.2 |
|
8691 |
0.18 |
| chr14_66392899_66393124 | 0.10 |
CTD-2014B16.3 |
Uncharacterized protein |
78230 |
0.11 |
| chr5_40209101_40209266 | 0.10 |
ENSG00000199361 |
. |
60477 |
0.15 |
| chr9_129232305_129232865 | 0.10 |
ENSG00000252985 |
. |
43413 |
0.14 |
| chr11_118210971_118211165 | 0.10 |
CD3D |
CD3d molecule, delta (CD3-TCR complex) |
584 |
0.62 |
| chr3_16984787_16984970 | 0.10 |
ENSG00000264818 |
. |
10190 |
0.23 |
| chr12_104870761_104871014 | 0.10 |
CHST11 |
carbohydrate (chondroitin 4) sulfotransferase 11 |
20108 |
0.25 |
| chr8_68304910_68305240 | 0.10 |
ARFGEF1 |
ADP-ribosylation factor guanine nucleotide-exchange factor 1 (brefeldin A-inhibited) |
49163 |
0.18 |
| chr12_50912843_50912995 | 0.10 |
DIP2B |
DIP2 disco-interacting protein 2 homolog B (Drosophila) |
14029 |
0.22 |
| chr15_91139735_91139886 | 0.10 |
CTD-3065B20.2 |
|
131 |
0.95 |
| chrX_15768424_15768590 | 0.10 |
CA5B |
carbonic anhydrase VB, mitochondrial |
414 |
0.84 |
| chr5_141484933_141485084 | 0.10 |
NDFIP1 |
Nedd4 family interacting protein 1 |
3062 |
0.32 |
| chr1_117544919_117545104 | 0.10 |
CD101 |
CD101 molecule |
577 |
0.73 |
| chr6_158779170_158779472 | 0.10 |
RP11-732M18.2 |
|
40272 |
0.15 |
| chr17_46529942_46530093 | 0.10 |
SKAP1 |
src kinase associated phosphoprotein 1 |
22436 |
0.11 |
| chr7_101385381_101385615 | 0.10 |
CUX1 |
cut-like homeobox 1 |
73461 |
0.1 |
| chr2_12642786_12642950 | 0.10 |
ENSG00000207183 |
. |
91241 |
0.1 |
| chr2_136983452_136983659 | 0.10 |
CXCR4 |
chemokine (C-X-C motif) receptor 4 |
107820 |
0.07 |
| chr10_11193217_11193852 | 0.10 |
CELF2 |
CUGBP, Elav-like family member 2 |
13459 |
0.2 |
| chr3_9821453_9821979 | 0.09 |
CAMK1 |
calcium/calmodulin-dependent protein kinase I |
10040 |
0.09 |
| chr13_99705776_99705927 | 0.09 |
ENSG00000207298 |
. |
28363 |
0.16 |
| chr6_13679192_13679414 | 0.09 |
ENSG00000252966 |
. |
1009 |
0.51 |
| chr2_198112450_198112630 | 0.09 |
ANKRD44 |
ankyrin repeat domain 44 |
49778 |
0.11 |
| chr6_106099814_106099965 | 0.09 |
PREP |
prolyl endopeptidase |
248930 |
0.02 |
| chr1_8485207_8485538 | 0.09 |
RERE |
arginine-glutamic acid dipeptide (RE) repeats |
100 |
0.78 |
| chr10_8129790_8129941 | 0.09 |
GATA3 |
GATA binding protein 3 |
33096 |
0.25 |
| chr14_55519381_55519591 | 0.09 |
MAPK1IP1L |
mitogen-activated protein kinase 1 interacting protein 1-like |
1137 |
0.5 |
| chr10_1748390_1748541 | 0.09 |
ADARB2 |
adenosine deaminase, RNA-specific, B2 (non-functional) |
31205 |
0.25 |
| chr8_101442194_101442345 | 0.09 |
KB-1615E4.2 |
|
45634 |
0.12 |
| chr10_64978992_64979347 | 0.09 |
JMJD1C |
jumonji domain containing 1C |
11684 |
0.25 |
| chr15_45008051_45008236 | 0.09 |
B2M |
beta-2-microglobulin |
4428 |
0.17 |
| chr2_182177306_182177486 | 0.09 |
ENSG00000266705 |
. |
7017 |
0.32 |
| chr12_15116052_15116331 | 0.09 |
ARHGDIB |
Rho GDP dissociation inhibitor (GDI) beta |
1529 |
0.36 |
| chr2_136881850_136882311 | 0.09 |
CXCR4 |
chemokine (C-X-C motif) receptor 4 |
6345 |
0.3 |
| chr2_12636393_12636544 | 0.09 |
ENSG00000207183 |
. |
84841 |
0.11 |
| chr14_54905038_54905189 | 0.09 |
CNIH1 |
cornichon family AMPA receptor auxiliary protein 1 |
2945 |
0.28 |
| chr14_23035018_23035185 | 0.09 |
AE000662.93 |
|
9145 |
0.11 |
| chr12_67878065_67878216 | 0.09 |
DYRK2 |
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 2 |
163978 |
0.04 |
| chr9_92141364_92141857 | 0.09 |
SEMA4D |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D |
28565 |
0.19 |
| chr10_126347896_126348047 | 0.09 |
FAM53B-AS1 |
FAM53B antisense RNA 1 |
44223 |
0.15 |
| chr7_21393437_21393588 | 0.09 |
ENSG00000195024 |
. |
27469 |
0.2 |
| chr1_193448919_193449104 | 0.09 |
ENSG00000252241 |
. |
252063 |
0.02 |
| chr10_63792565_63792716 | 0.09 |
ARID5B |
AT rich interactive domain 5B (MRF1-like) |
16330 |
0.26 |
| chrX_64947728_64947879 | 0.09 |
MSN |
moesin |
60266 |
0.16 |
| chr11_118315811_118315962 | 0.09 |
KMT2A |
lysine (K)-specific methyltransferase 2A |
8409 |
0.11 |
| chr12_11920815_11920993 | 0.09 |
ETV6 |
ets variant 6 |
15469 |
0.28 |
| chr5_52021951_52022102 | 0.09 |
ITGA1 |
integrin, alpha 1 |
61704 |
0.12 |
| chr15_38965720_38965889 | 0.09 |
C15orf53 |
chromosome 15 open reading frame 53 |
22995 |
0.24 |
| chr5_133865434_133865585 | 0.09 |
JADE2 |
jade family PHD finger 2 |
3182 |
0.21 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.0 | 0.1 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.1 | GO:0048569 | post-embryonic organ development(GO:0048569) |
| 0.0 | 0.1 | GO:0002713 | negative regulation of B cell mediated immunity(GO:0002713) negative regulation of immunoglobulin mediated immune response(GO:0002890) |
| 0.0 | 0.2 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.0 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.0 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.1 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 0.1 | GO:0043368 | positive T cell selection(GO:0043368) |
| 0.0 | 0.1 | GO:0070670 | response to interleukin-4(GO:0070670) |
| 0.0 | 0.0 | GO:0001766 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 0.0 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) |
| 0.0 | 0.0 | GO:0072600 | protein targeting to Golgi(GO:0000042) establishment of protein localization to Golgi(GO:0072600) |
| 0.0 | 0.0 | GO:0070602 | centromeric sister chromatid cohesion(GO:0070601) regulation of centromeric sister chromatid cohesion(GO:0070602) |
| 0.0 | 0.0 | GO:0045657 | positive regulation of monocyte differentiation(GO:0045657) |
| 0.0 | 0.0 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.0 | 0.0 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 0.0 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.0 | 0.1 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.0 | GO:0002887 | negative regulation of myeloid leukocyte mediated immunity(GO:0002887) negative regulation of leukocyte degranulation(GO:0043301) |
| 0.0 | 0.0 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.0 | 0.0 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.0 | 0.1 | GO:0002478 | antigen processing and presentation of exogenous peptide antigen(GO:0002478) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.0 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.1 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.0 | GO:0000306 | extrinsic component of vacuolar membrane(GO:0000306) extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.0 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.0 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.0 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.0 | 0.1 | GO:0042598 | obsolete vesicular fraction(GO:0042598) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.1 | GO:0055106 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.0 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.0 | 0.0 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.0 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.0 | GO:0004477 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) |
| 0.0 | 0.0 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.0 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.0 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.0 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.4 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.0 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.0 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |