Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
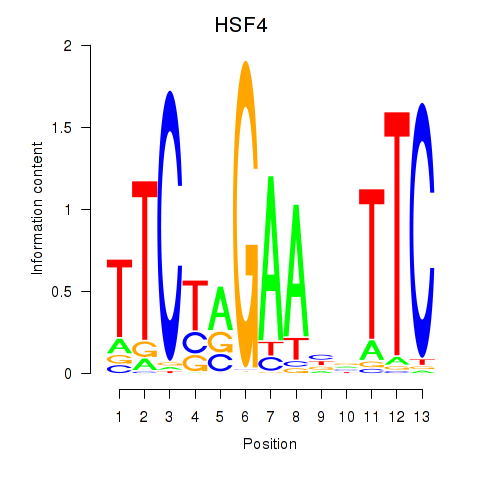
Results for HSF4
Z-value: 1.06
Transcription factors associated with HSF4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HSF4
|
ENSG00000102878.11 | heat shock transcription factor 4 |
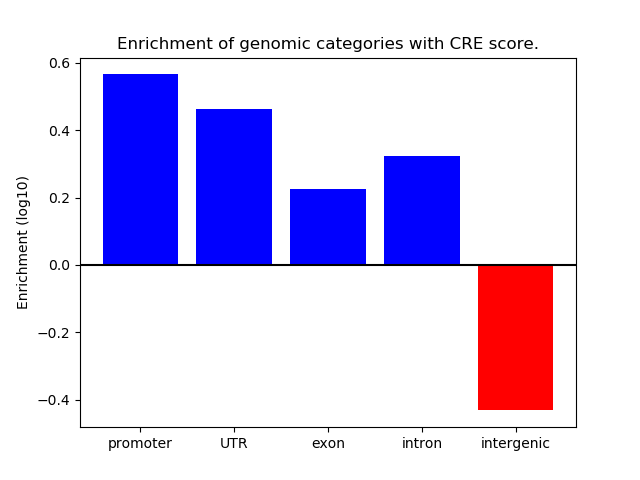
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr16_67203074_67203225 | HSF4 | 172 | 0.812528 | -0.67 | 4.7e-02 | Click! |
| chr16_67196662_67196813 | HSF4 | 551 | 0.445907 | 0.61 | 8.0e-02 | Click! |
| chr16_67198740_67198891 | HSF4 | 100 | 0.901692 | 0.58 | 1.0e-01 | Click! |
| chr16_67199421_67199612 | HSF4 | 88 | 0.908316 | 0.53 | 1.4e-01 | Click! |
| chr16_67197789_67198350 | HSF4 | 646 | 0.420290 | 0.38 | 3.1e-01 | Click! |
Activity of the HSF4 motif across conditions
Conditions sorted by the z-value of the HSF4 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
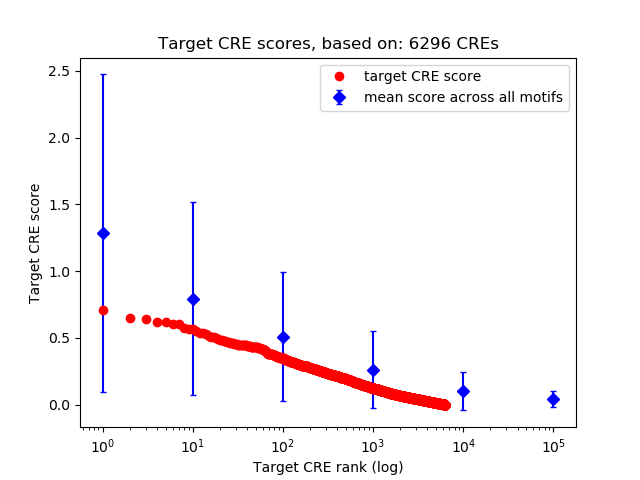
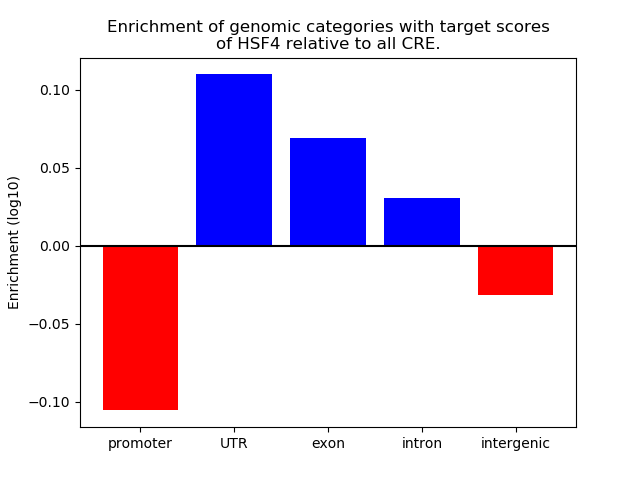
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr7_139312464_139312655 | 0.71 |
CLEC2L |
C-type lectin domain family 2, member L |
103731 |
0.06 |
| chr14_99670994_99671151 | 0.65 |
AL162151.4 |
|
46319 |
0.15 |
| chr7_1087128_1087349 | 0.65 |
GPR146 |
G protein-coupled receptor 146 |
3026 |
0.13 |
| chr21_36340846_36341061 | 0.62 |
RUNX1 |
runt-related transcription factor 1 |
78866 |
0.12 |
| chr5_156681732_156681896 | 0.62 |
CYFIP2 |
cytoplasmic FMR1 interacting protein 2 |
11277 |
0.13 |
| chr7_2739468_2739742 | 0.60 |
AMZ1 |
archaelysin family metallopeptidase 1 |
11769 |
0.21 |
| chr14_98657577_98657728 | 0.60 |
ENSG00000222066 |
. |
140435 |
0.05 |
| chr22_27043526_27043866 | 0.57 |
CRYBA4 |
crystallin, beta A4 |
25768 |
0.16 |
| chr12_47758069_47758220 | 0.57 |
ENSG00000264906 |
. |
92 |
0.98 |
| chr5_150597516_150598012 | 0.57 |
CCDC69 |
coiled-coil domain containing 69 |
5942 |
0.19 |
| chr8_37134401_37134693 | 0.55 |
RP11-150O12.6 |
|
239992 |
0.02 |
| chr16_3838912_3839326 | 0.54 |
CREBBP |
CREB binding protein |
8251 |
0.23 |
| chr1_100856884_100857083 | 0.54 |
ENSG00000216067 |
. |
12652 |
0.2 |
| chr22_40327971_40328220 | 0.53 |
GRAP2 |
GRB2-related adaptor protein 2 |
5454 |
0.19 |
| chr10_14702537_14702914 | 0.51 |
ENSG00000201766 |
. |
2768 |
0.28 |
| chr13_114914753_114915138 | 0.51 |
RASA3 |
RAS p21 protein activator 3 |
16859 |
0.2 |
| chr1_25350610_25350934 | 0.51 |
ENSG00000264371 |
. |
778 |
0.7 |
| chr11_128598285_128598783 | 0.50 |
SENCR |
smooth muscle and endothelial cell enriched migration/differentiation-associated long non-coding RNA |
32616 |
0.15 |
| chr16_67837738_67838160 | 0.49 |
RANBP10 |
RAN binding protein 10 |
2511 |
0.15 |
| chr1_118197732_118198071 | 0.49 |
ENSG00000212266 |
. |
33463 |
0.17 |
| chr1_111437508_111437815 | 0.48 |
CD53 |
CD53 molecule |
21885 |
0.15 |
| chr2_205833792_205833953 | 0.48 |
PARD3B |
par-3 family cell polarity regulator beta |
423149 |
0.01 |
| chr22_27071165_27071316 | 0.48 |
CRYBA4 |
crystallin, beta A4 |
53312 |
0.12 |
| chr8_27235111_27235488 | 0.47 |
PTK2B |
protein tyrosine kinase 2 beta |
2869 |
0.31 |
| chr13_114912317_114912766 | 0.47 |
RASA3 |
RAS p21 protein activator 3 |
14455 |
0.21 |
| chr1_24851023_24851224 | 0.46 |
ENSG00000266551 |
. |
5081 |
0.18 |
| chrX_154417614_154417799 | 0.46 |
VBP1 |
von Hippel-Lindau binding protein 1 |
7578 |
0.2 |
| chr3_15332186_15332608 | 0.46 |
SH3BP5 |
SH3-domain binding protein 5 (BTK-associated) |
14845 |
0.14 |
| chrX_64947728_64947879 | 0.46 |
MSN |
moesin |
60266 |
0.16 |
| chr19_4084516_4084732 | 0.46 |
MAP2K2 |
mitogen-activated protein kinase kinase 2 |
16421 |
0.11 |
| chr19_3757648_3757799 | 0.45 |
AC005954.3 |
|
1066 |
0.31 |
| chr16_68105212_68105363 | 0.45 |
NFATC3 |
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 3 |
13960 |
0.09 |
| chr17_63025333_63025500 | 0.45 |
RP11-583F2.5 |
|
22611 |
0.15 |
| chr7_50422851_50423205 | 0.45 |
IKZF1 |
IKAROS family zinc finger 1 (Ikaros) |
55783 |
0.14 |
| chr1_203255177_203255328 | 0.45 |
BTG2 |
BTG family, member 2 |
19412 |
0.16 |
| chr2_62430640_62430961 | 0.45 |
ENSG00000266097 |
. |
2161 |
0.29 |
| chr9_82248211_82248468 | 0.45 |
TLE4 |
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
19169 |
0.31 |
| chr8_28917519_28917756 | 0.44 |
CTD-2647L4.4 |
|
4185 |
0.17 |
| chr4_154449092_154449243 | 0.44 |
KIAA0922 |
KIAA0922 |
27881 |
0.22 |
| chr12_122228197_122228738 | 0.44 |
RHOF |
ras homolog family member F (in filopodia) |
2799 |
0.22 |
| chr15_81576710_81577129 | 0.44 |
IL16 |
interleukin 16 |
12335 |
0.21 |
| chr5_169702138_169702842 | 0.44 |
LCP2 |
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa) |
8159 |
0.23 |
| chr1_185250977_185251218 | 0.44 |
ENSG00000252612 |
. |
15430 |
0.16 |
| chr14_69280428_69280742 | 0.43 |
ZFP36L1 |
ZFP36 ring finger protein-like 1 |
17395 |
0.19 |
| chr21_47972699_47973025 | 0.43 |
ENSG00000272283 |
. |
17153 |
0.17 |
| chr17_63022687_63022838 | 0.43 |
RP11-583F2.5 |
|
25265 |
0.14 |
| chr8_22496270_22496646 | 0.43 |
BIN3 |
bridging integrator 3 |
5859 |
0.12 |
| chr19_42399773_42399960 | 0.43 |
ARHGEF1 |
Rho guanine nucleotide exchange factor (GEF) 1 |
414 |
0.76 |
| chr19_3756961_3757129 | 0.43 |
AC005954.3 |
|
388 |
0.71 |
| chr19_3813101_3813594 | 0.43 |
MATK |
megakaryocyte-associated tyrosine kinase |
11220 |
0.11 |
| chr2_3225421_3225572 | 0.43 |
TSSC1-IT1 |
TSSC1 intronic transcript 1 (non-protein coding) |
79740 |
0.1 |
| chr21_45575872_45576023 | 0.43 |
AP001055.1 |
|
17633 |
0.13 |
| chr14_53211245_53211397 | 0.43 |
STYX |
serine/threonine/tyrosine interacting protein |
14421 |
0.14 |
| chr3_48551527_48551686 | 0.42 |
SHISA5 |
shisa family member 5 |
9347 |
0.1 |
| chr1_117544919_117545104 | 0.42 |
CD101 |
CD101 molecule |
577 |
0.73 |
| chr1_154376165_154376624 | 0.42 |
IL6R |
interleukin 6 receptor |
1275 |
0.33 |
| chr8_59930806_59931005 | 0.42 |
RP11-328K2.1 |
|
25408 |
0.23 |
| chr11_6762047_6762301 | 0.42 |
GVINP1 |
GTPase, very large interferon inducible pseudogene 1 |
19063 |
0.11 |
| chr16_79320751_79320982 | 0.42 |
ENSG00000222244 |
. |
22515 |
0.27 |
| chr2_135005025_135005319 | 0.42 |
MGAT5 |
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase |
6658 |
0.26 |
| chr19_41850700_41850906 | 0.41 |
TGFB1 |
transforming growth factor, beta 1 |
2868 |
0.13 |
| chr2_182177306_182177486 | 0.41 |
ENSG00000266705 |
. |
7017 |
0.32 |
| chr1_25421803_25422058 | 0.41 |
ENSG00000264371 |
. |
71936 |
0.09 |
| chr10_7201395_7201546 | 0.40 |
SFMBT2 |
Scm-like with four mbt domains 2 |
249237 |
0.02 |
| chr22_20819732_20820026 | 0.40 |
ENSG00000255156 |
. |
9719 |
0.1 |
| chr17_55681912_55682272 | 0.39 |
RP11-118E18.4 |
|
3681 |
0.24 |
| chr10_26753045_26753285 | 0.39 |
APBB1IP |
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein |
25811 |
0.22 |
| chr6_45462657_45462916 | 0.38 |
RUNX2 |
runt-related transcription factor 2 |
72564 |
0.11 |
| chr5_39155242_39155486 | 0.38 |
FYB |
FYN binding protein |
47765 |
0.15 |
| chr1_115009857_115010035 | 0.38 |
TRIM33 |
tripartite motif containing 33 |
3898 |
0.27 |
| chr20_50019481_50019826 | 0.38 |
ENSG00000263645 |
. |
25795 |
0.22 |
| chr19_16379790_16380087 | 0.38 |
CTD-2562J15.6 |
|
24448 |
0.13 |
| chr9_132794767_132794918 | 0.38 |
FNBP1 |
formin binding protein 1 |
10599 |
0.2 |
| chr3_3213200_3213369 | 0.38 |
CRBN |
cereblon |
8074 |
0.19 |
| chr5_151175034_151175185 | 0.38 |
ATOX1 |
antioxidant 1 copper chaperone |
23016 |
0.15 |
| chr16_85337462_85337809 | 0.38 |
ENSG00000266307 |
. |
2296 |
0.37 |
| chr6_159139178_159139369 | 0.37 |
ENSG00000265558 |
. |
46512 |
0.12 |
| chrX_6978429_6978580 | 0.37 |
ENSG00000264268 |
. |
87397 |
0.09 |
| chr1_38138940_38139091 | 0.37 |
C1orf109 |
chromosome 1 open reading frame 109 |
17161 |
0.12 |
| chr22_40729451_40729738 | 0.37 |
ADSL |
adenylosuccinate lyase |
12913 |
0.18 |
| chr6_52150845_52151081 | 0.37 |
MCM3 |
minichromosome maintenance complex component 3 |
1328 |
0.48 |
| chr2_204865819_204865980 | 0.37 |
ICOS |
inducible T-cell co-stimulator |
64396 |
0.14 |
| chr3_12991701_12991859 | 0.37 |
IQSEC1 |
IQ motif and Sec7 domain 1 |
17388 |
0.22 |
| chr7_50321524_50321798 | 0.36 |
IKZF1 |
IKAROS family zinc finger 1 (Ikaros) |
22663 |
0.25 |
| chr3_105552050_105552274 | 0.36 |
CBLB |
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
35725 |
0.25 |
| chr5_158838800_158839019 | 0.36 |
IL12B |
interleukin 12B (natural killer cell stimulatory factor 2, cytotoxic lymphocyte maturation factor 2, p40) |
81014 |
0.09 |
| chr3_15319262_15319413 | 0.36 |
SH3BP5 |
SH3-domain binding protein 5 (BTK-associated) |
1785 |
0.29 |
| chr2_109225906_109226290 | 0.36 |
LIMS1 |
LIM and senescent cell antigen-like domains 1 |
2481 |
0.35 |
| chr11_121452316_121452687 | 0.36 |
SORL1 |
sortilin-related receptor, L(DLR class) A repeats containing |
4954 |
0.34 |
| chr12_6542028_6542189 | 0.36 |
CD27 |
CD27 molecule |
11925 |
0.09 |
| chr19_9972790_9973038 | 0.35 |
OLFM2 |
olfactomedin 2 |
4070 |
0.12 |
| chr17_5292178_5292426 | 0.35 |
RABEP1 |
rabaptin, RAB GTPase binding effector protein 1 |
11796 |
0.12 |
| chr16_68453137_68453288 | 0.35 |
SMPD3 |
sphingomyelin phosphodiesterase 3, neutral membrane (neutral sphingomyelinase II) |
29183 |
0.12 |
| chr20_31555391_31555580 | 0.35 |
EFCAB8 |
EF-hand calcium binding domain 8 |
7834 |
0.17 |
| chr7_21393437_21393588 | 0.35 |
ENSG00000195024 |
. |
27469 |
0.2 |
| chrX_129222828_129223069 | 0.35 |
ELF4 |
E74-like factor 4 (ets domain transcription factor) |
21388 |
0.19 |
| chr1_65528171_65528405 | 0.35 |
ENSG00000199135 |
. |
4097 |
0.22 |
| chr1_24235021_24235316 | 0.35 |
RP11-4M23.3 |
|
1567 |
0.24 |
| chr11_117821389_117821783 | 0.35 |
TMPRSS13 |
transmembrane protease, serine 13 |
21412 |
0.15 |
| chr4_40251320_40251540 | 0.35 |
RHOH |
ras homolog family member H |
49466 |
0.13 |
| chr15_81593592_81593958 | 0.35 |
IL16 |
interleukin 16 |
2018 |
0.33 |
| chr6_39195665_39195856 | 0.35 |
KCNK5 |
potassium channel, subfamily K, member 5 |
1466 |
0.51 |
| chr6_134953210_134953361 | 0.35 |
ALDH8A1 |
aldehyde dehydrogenase 8 family, member A1 |
297014 |
0.01 |
| chr1_183447249_183447596 | 0.34 |
SMG7 |
SMG7 nonsense mediated mRNA decay factor |
5355 |
0.23 |
| chr1_169672251_169672700 | 0.34 |
SELL |
selectin L |
8364 |
0.2 |
| chr17_75959968_75960181 | 0.34 |
TNRC6C |
trinucleotide repeat containing 6C |
40175 |
0.14 |
| chr5_35788787_35788938 | 0.34 |
SPEF2 |
sperm flagellar 2 |
9592 |
0.22 |
| chr5_55351651_55351834 | 0.34 |
ENSG00000238326 |
. |
55293 |
0.09 |
| chr1_26696466_26696677 | 0.34 |
ZNF683 |
zinc finger protein 683 |
665 |
0.58 |
| chr12_104880310_104880679 | 0.33 |
CHST11 |
carbohydrate (chondroitin 4) sulfotransferase 11 |
29715 |
0.22 |
| chr19_10254387_10254800 | 0.33 |
EIF3G |
eukaryotic translation initiation factor 3, subunit G |
24015 |
0.07 |
| chr4_8349309_8349460 | 0.33 |
ENSG00000202054 |
. |
45358 |
0.14 |
| chr17_75395164_75395315 | 0.33 |
SEPT9 |
septin 9 |
1401 |
0.34 |
| chr9_78642998_78643161 | 0.33 |
PCSK5 |
proprotein convertase subtilisin/kexin type 5 |
67814 |
0.13 |
| chr16_85768216_85768412 | 0.33 |
ENSG00000222190 |
. |
6992 |
0.12 |
| chr18_60911782_60912122 | 0.33 |
ENSG00000238988 |
. |
50054 |
0.12 |
| chr13_114991565_114991854 | 0.33 |
CDC16 |
cell division cycle 16 |
8653 |
0.18 |
| chr12_121848379_121848530 | 0.33 |
RNF34 |
ring finger protein 34, E3 ubiquitin protein ligase |
1176 |
0.51 |
| chr15_26056534_26056770 | 0.33 |
ENSG00000199214 |
. |
6478 |
0.21 |
| chr9_112906499_112906706 | 0.32 |
AKAP2 |
A kinase (PRKA) anchor protein 2 |
18821 |
0.24 |
| chr9_112713247_112713398 | 0.32 |
PALM2 |
paralemmin 2 |
84011 |
0.09 |
| chr6_139452567_139452918 | 0.32 |
HECA |
headcase homolog (Drosophila) |
3507 |
0.32 |
| chr10_51517238_51517411 | 0.32 |
AGAP7 |
ArfGAP with GTPase domain, ankyrin repeat and PH domain 7 |
30997 |
0.14 |
| chrX_41775628_41775779 | 0.32 |
ENSG00000251807 |
. |
2899 |
0.31 |
| chr5_130597752_130597932 | 0.32 |
CDC42SE2 |
CDC42 small effector 2 |
1860 |
0.47 |
| chr10_27087573_27087735 | 0.32 |
ENSG00000206605 |
. |
8079 |
0.19 |
| chr2_48078802_48078953 | 0.32 |
FBXO11 |
F-box protein 11 |
15776 |
0.21 |
| chr1_226916534_226916814 | 0.32 |
ITPKB |
inositol-trisphosphate 3-kinase B |
8485 |
0.23 |
| chr10_11213546_11213708 | 0.32 |
RP3-323N1.2 |
|
288 |
0.92 |
| chr17_7456942_7457093 | 0.32 |
TNFSF12 |
tumor necrosis factor (ligand) superfamily, member 12 |
4546 |
0.06 |
| chr13_77793771_77793922 | 0.32 |
MYCBP2 |
MYC binding protein 2, E3 ubiquitin protein ligase |
106968 |
0.07 |
| chr6_24955515_24955958 | 0.32 |
FAM65B |
family with sequence similarity 65, member B |
19548 |
0.21 |
| chr5_72528452_72528679 | 0.32 |
TMEM174 |
transmembrane protein 174 |
59543 |
0.12 |
| chrX_135709367_135709640 | 0.32 |
ENSG00000233093 |
. |
12199 |
0.16 |
| chr2_158750571_158750722 | 0.32 |
UPP2 |
uridine phosphorylase 2 |
17432 |
0.16 |
| chr19_5077571_5077794 | 0.32 |
KDM4B |
lysine (K)-specific demethylase 4B |
4781 |
0.27 |
| chr3_32498174_32498359 | 0.31 |
CMTM6 |
CKLF-like MARVEL transmembrane domain containing 6 |
46634 |
0.14 |
| chr20_58639264_58639415 | 0.31 |
C20orf197 |
chromosome 20 open reading frame 197 |
8359 |
0.26 |
| chr15_32651213_32651761 | 0.31 |
ENSG00000221444 |
. |
34186 |
0.1 |
| chr11_121332248_121332530 | 0.31 |
RP11-730K11.1 |
|
8667 |
0.24 |
| chr15_59836576_59836759 | 0.31 |
ENSG00000201704 |
. |
29754 |
0.14 |
| chr22_27040755_27040920 | 0.31 |
CRYBA4 |
crystallin, beta A4 |
22909 |
0.17 |
| chr1_226901230_226901381 | 0.31 |
ITPKB |
inositol-trisphosphate 3-kinase B |
23854 |
0.18 |
| chr14_64963769_64963936 | 0.31 |
ZBTB1 |
zinc finger and BTB domain containing 1 |
6591 |
0.12 |
| chr22_45068838_45068989 | 0.31 |
PRR5 |
proline rich 5 (renal) |
4024 |
0.25 |
| chr2_197015707_197015858 | 0.30 |
RP11-347P5.1 |
|
205 |
0.94 |
| chr2_231521971_231522122 | 0.30 |
CAB39 |
calcium binding protein 39 |
55514 |
0.11 |
| chr11_114086673_114087539 | 0.30 |
NNMT |
nicotinamide N-methyltransferase |
41447 |
0.17 |
| chr1_231748922_231749511 | 0.30 |
LINC00582 |
long intergenic non-protein coding RNA 582 |
1380 |
0.44 |
| chr1_206556159_206556344 | 0.30 |
SRGAP2 |
SLIT-ROBO Rho GTPase activating protein 2 |
1115 |
0.4 |
| chr2_12642786_12642950 | 0.30 |
ENSG00000207183 |
. |
91241 |
0.1 |
| chr6_88453159_88453310 | 0.30 |
ENSG00000238628 |
. |
22601 |
0.18 |
| chr17_47813077_47813254 | 0.30 |
FAM117A |
family with sequence similarity 117, member A |
11276 |
0.14 |
| chr16_84628580_84628866 | 0.30 |
RP11-61F12.1 |
|
724 |
0.65 |
| chr7_101385381_101385615 | 0.30 |
CUX1 |
cut-like homeobox 1 |
73461 |
0.1 |
| chrX_135851684_135852073 | 0.30 |
ARHGEF6 |
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6 |
1092 |
0.49 |
| chr2_235335702_235336092 | 0.30 |
ARL4C |
ADP-ribosylation factor-like 4C |
69347 |
0.14 |
| chr15_30471500_30471800 | 0.30 |
ENSG00000221785 |
. |
34570 |
0.11 |
| chr1_42123736_42123959 | 0.29 |
HIVEP3 |
human immunodeficiency virus type I enhancer binding protein 3 |
42825 |
0.18 |
| chr3_15709764_15709915 | 0.29 |
ENSG00000264354 |
. |
29039 |
0.15 |
| chr2_233179198_233179349 | 0.29 |
DIS3L2 |
DIS3 mitotic control homolog (S. cerevisiae)-like 2 |
19380 |
0.17 |
| chr13_28030262_28030413 | 0.29 |
MTIF3 |
mitochondrial translational initiation factor 3 |
5598 |
0.19 |
| chr6_28473877_28474028 | 0.29 |
GPX6 |
glutathione peroxidase 6 (olfactory) |
9612 |
0.17 |
| chr10_115441935_115442086 | 0.29 |
CASP7 |
caspase 7, apoptosis-related cysteine peptidase |
2311 |
0.29 |
| chr1_26865494_26865771 | 0.29 |
RPS6KA1 |
ribosomal protein S6 kinase, 90kDa, polypeptide 1 |
3971 |
0.17 |
| chr11_95375181_95375332 | 0.29 |
FAM76B |
family with sequence similarity 76, member B |
144713 |
0.04 |
| chr5_527247_527623 | 0.29 |
RP11-310P5.2 |
|
2615 |
0.18 |
| chr19_50835735_50835918 | 0.29 |
KCNC3 |
potassium voltage-gated channel, Shaw-related subfamily, member 3 |
946 |
0.37 |
| chr2_28572176_28572390 | 0.29 |
AC093690.1 |
|
38957 |
0.13 |
| chr10_14698882_14699236 | 0.29 |
RP11-7C6.1 |
|
4318 |
0.23 |
| chr1_779209_779379 | 0.29 |
RP11-206L10.8 |
|
33753 |
0.1 |
| chr18_72075391_72075542 | 0.29 |
FAM69C |
family with sequence similarity 69, member C |
49037 |
0.14 |
| chr10_11252452_11252842 | 0.29 |
RP3-323N1.2 |
|
39308 |
0.17 |
| chr17_56456497_56456648 | 0.29 |
RNF43 |
ring finger protein 43 |
23510 |
0.11 |
| chr2_13037487_13037811 | 0.29 |
ENSG00000264370 |
. |
160156 |
0.04 |
| chr3_107673374_107673767 | 0.29 |
CD47 |
CD47 molecule |
103638 |
0.08 |
| chr19_4641704_4641855 | 0.29 |
TNFAIP8L1 |
tumor necrosis factor, alpha-induced protein 8-like 1 |
1750 |
0.24 |
| chr10_13160669_13160820 | 0.29 |
OPTN |
optineurin |
2476 |
0.29 |
| chr2_241564217_241564368 | 0.29 |
GPR35 |
G protein-coupled receptor 35 |
370 |
0.81 |
| chr5_156644620_156644840 | 0.29 |
CTB-4E7.1 |
|
6660 |
0.14 |
| chr8_28936608_28936759 | 0.29 |
CTD-2647L4.5 |
|
11887 |
0.14 |
| chr3_3233085_3233387 | 0.29 |
CRBN |
cereblon |
11842 |
0.2 |
| chr9_92044873_92045111 | 0.29 |
SEMA4D |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D |
6386 |
0.26 |
| chr16_4725775_4726545 | 0.29 |
NUDT16L1 |
nudix (nucleoside diphosphate linked moiety X)-type motif 16-like 1 |
17535 |
0.1 |
| chr19_42276662_42276849 | 0.29 |
AC011513.4 |
|
14046 |
0.12 |
| chr7_157263857_157264008 | 0.29 |
AC006372.6 |
|
42056 |
0.16 |
| chr3_18540522_18540673 | 0.29 |
ENSG00000228956 |
. |
27291 |
0.22 |
| chr2_85101975_85102126 | 0.29 |
TRABD2A |
TraB domain containing 2A |
6156 |
0.2 |
| chr4_144441518_144441669 | 0.28 |
SMARCA5-AS1 |
SMARCA5 antisense RNA 1 |
5805 |
0.23 |
| chr20_44832267_44832487 | 0.28 |
CDH22 |
cadherin 22, type 2 |
47957 |
0.13 |
| chr14_99645282_99645517 | 0.28 |
AL162151.4 |
|
20646 |
0.24 |
| chr1_160612345_160612670 | 0.28 |
SLAMF1 |
signaling lymphocytic activation molecule family member 1 |
4304 |
0.19 |
| chr3_42388906_42389324 | 0.28 |
LYZL4 |
lysozyme-like 4 |
62935 |
0.1 |
| chr1_25436980_25437131 | 0.28 |
ENSG00000264371 |
. |
87061 |
0.07 |
| chr12_133289810_133290003 | 0.28 |
PGAM5 |
phosphoglycerate mutase family member 5 |
1433 |
0.3 |
| chr19_5952060_5952229 | 0.28 |
RANBP3 |
RAN binding protein 3 |
25946 |
0.08 |
| chr5_118648731_118648896 | 0.28 |
ENSG00000243333 |
. |
6487 |
0.21 |
| chr14_25140513_25140664 | 0.28 |
GZMB |
granzyme B (granzyme 2, cytotoxic T-lymphocyte-associated serine esterase 1) |
37115 |
0.14 |
| chr14_104858355_104858716 | 0.28 |
ENSG00000222761 |
. |
8850 |
0.27 |
| chr5_39160522_39160673 | 0.28 |
FYB |
FYN binding protein |
42532 |
0.17 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0033689 | negative regulation of osteoblast proliferation(GO:0033689) |
| 0.1 | 0.2 | GO:0060766 | negative regulation of androgen receptor signaling pathway(GO:0060766) |
| 0.1 | 0.6 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.1 | 0.2 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.1 | 0.2 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.1 | 0.2 | GO:0072600 | protein targeting to Golgi(GO:0000042) establishment of protein localization to Golgi(GO:0072600) |
| 0.1 | 0.1 | GO:0002890 | negative regulation of B cell mediated immunity(GO:0002713) negative regulation of immunoglobulin mediated immune response(GO:0002890) |
| 0.1 | 0.3 | GO:0006515 | misfolded or incompletely synthesized protein catabolic process(GO:0006515) |
| 0.1 | 0.3 | GO:0019987 | obsolete negative regulation of anti-apoptosis(GO:0019987) |
| 0.1 | 0.2 | GO:0032713 | negative regulation of interleukin-4 production(GO:0032713) |
| 0.1 | 0.2 | GO:0030952 | establishment or maintenance of cytoskeleton polarity(GO:0030952) |
| 0.1 | 0.2 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.0 | 0.1 | GO:0010882 | regulation of cardiac muscle contraction by calcium ion signaling(GO:0010882) |
| 0.0 | 0.3 | GO:0045066 | regulatory T cell differentiation(GO:0045066) |
| 0.0 | 0.1 | GO:0002870 | lymphocyte anergy(GO:0002249) regulation of T cell anergy(GO:0002667) positive regulation of T cell anergy(GO:0002669) T cell anergy(GO:0002870) regulation of lymphocyte anergy(GO:0002911) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.0 | 0.1 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 | 0.5 | GO:0045061 | thymic T cell selection(GO:0045061) |
| 0.0 | 0.2 | GO:0050685 | positive regulation of mRNA 3'-end processing(GO:0031442) positive regulation of mRNA processing(GO:0050685) positive regulation of mRNA metabolic process(GO:1903313) |
| 0.0 | 0.0 | GO:2000041 | regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 0.0 | 0.2 | GO:0070102 | interleukin-6-mediated signaling pathway(GO:0070102) |
| 0.0 | 0.2 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.2 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.1 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.2 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.0 | 0.1 | GO:0006772 | thiamine metabolic process(GO:0006772) |
| 0.0 | 0.1 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 0.1 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.2 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) |
| 0.0 | 0.1 | GO:0001911 | negative regulation of leukocyte mediated cytotoxicity(GO:0001911) |
| 0.0 | 0.1 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.1 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.0 | 0.1 | GO:2000380 | regulation of mesodermal cell fate specification(GO:0042661) regulation of mesoderm development(GO:2000380) |
| 0.0 | 0.1 | GO:0032506 | cytokinetic process(GO:0032506) |
| 0.0 | 0.1 | GO:0010766 | negative regulation of sodium ion transport(GO:0010766) |
| 0.0 | 0.1 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.1 | GO:0048087 | positive regulation of developmental pigmentation(GO:0048087) |
| 0.0 | 0.1 | GO:0045091 | regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045091) |
| 0.0 | 0.3 | GO:0019059 | obsolete initiation of viral infection(GO:0019059) |
| 0.0 | 0.1 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.0 | 0.3 | GO:0002418 | immune response to tumor cell(GO:0002418) |
| 0.0 | 0.1 | GO:0006925 | inflammatory cell apoptotic process(GO:0006925) |
| 0.0 | 0.0 | GO:0002693 | positive regulation of cellular extravasation(GO:0002693) |
| 0.0 | 0.9 | GO:0050772 | positive regulation of axonogenesis(GO:0050772) |
| 0.0 | 0.1 | GO:0032100 | positive regulation of response to food(GO:0032097) positive regulation of appetite(GO:0032100) |
| 0.0 | 0.1 | GO:0010511 | regulation of phosphatidylinositol biosynthetic process(GO:0010511) |
| 0.0 | 0.2 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 0.1 | GO:0060710 | chorio-allantoic fusion(GO:0060710) |
| 0.0 | 0.1 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 0.1 | GO:1901534 | positive regulation of megakaryocyte differentiation(GO:0045654) positive regulation of hematopoietic progenitor cell differentiation(GO:1901534) |
| 0.0 | 0.0 | GO:0019042 | viral latency(GO:0019042) |
| 0.0 | 0.3 | GO:0006911 | phagocytosis, engulfment(GO:0006911) membrane invagination(GO:0010324) |
| 0.0 | 0.1 | GO:0007343 | egg activation(GO:0007343) |
| 0.0 | 0.1 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 0.0 | GO:0071354 | cellular response to interleukin-6(GO:0071354) |
| 0.0 | 0.2 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.1 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.0 | 0.0 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 0.0 | GO:0033629 | negative regulation of cell adhesion mediated by integrin(GO:0033629) |
| 0.0 | 0.1 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.1 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.0 | 0.1 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.0 | 0.2 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.0 | 0.0 | GO:0046101 | hypoxanthine biosynthetic process(GO:0046101) |
| 0.0 | 0.1 | GO:0045056 | transcytosis(GO:0045056) |
| 0.0 | 0.1 | GO:0046831 | regulation of RNA export from nucleus(GO:0046831) |
| 0.0 | 0.1 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.1 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.0 | 0.1 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.1 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) |
| 0.0 | 0.1 | GO:0090292 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.0 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.0 | 0.2 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.0 | 0.2 | GO:0050684 | regulation of mRNA processing(GO:0050684) regulation of mRNA metabolic process(GO:1903311) |
| 0.0 | 0.1 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.1 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.1 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 0.0 | GO:0071379 | cellular response to prostaglandin stimulus(GO:0071379) |
| 0.0 | 0.1 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.0 | 0.2 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.1 | GO:0001866 | NK T cell proliferation(GO:0001866) |
| 0.0 | 0.1 | GO:0018106 | peptidyl-histidine phosphorylation(GO:0018106) |
| 0.0 | 0.1 | GO:0002360 | T cell lineage commitment(GO:0002360) |
| 0.0 | 0.0 | GO:0070672 | response to interleukin-15(GO:0070672) |
| 0.0 | 0.0 | GO:0045586 | regulation of gamma-delta T cell differentiation(GO:0045586) positive regulation of gamma-delta T cell differentiation(GO:0045588) |
| 0.0 | 0.1 | GO:0033033 | negative regulation of myeloid cell apoptotic process(GO:0033033) |
| 0.0 | 0.0 | GO:0003161 | cardiac conduction system development(GO:0003161) |
| 0.0 | 0.1 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.0 | GO:0060976 | coronary vasculature development(GO:0060976) |
| 0.0 | 0.0 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.0 | GO:0002032 | desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 0.0 | 0.1 | GO:0002538 | arachidonic acid metabolite production involved in inflammatory response(GO:0002538) leukotriene production involved in inflammatory response(GO:0002540) |
| 0.0 | 0.1 | GO:0042996 | regulation of Golgi to plasma membrane protein transport(GO:0042996) |
| 0.0 | 0.1 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.0 | 0.1 | GO:0010887 | negative regulation of cholesterol storage(GO:0010887) |
| 0.0 | 0.1 | GO:0003056 | regulation of vascular smooth muscle contraction(GO:0003056) |
| 0.0 | 0.0 | GO:0010216 | maintenance of DNA methylation(GO:0010216) |
| 0.0 | 0.0 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.0 | 0.0 | GO:0007195 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.0 | 0.1 | GO:0060272 | embryonic skeletal joint morphogenesis(GO:0060272) embryonic skeletal joint development(GO:0072498) |
| 0.0 | 0.1 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) regulation of store-operated calcium channel activity(GO:1901339) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.1 | GO:0033522 | histone H2A ubiquitination(GO:0033522) |
| 0.0 | 0.0 | GO:0002691 | regulation of cellular extravasation(GO:0002691) |
| 0.0 | 0.1 | GO:0002063 | chondrocyte development(GO:0002063) |
| 0.0 | 0.0 | GO:0001743 | optic placode formation(GO:0001743) |
| 0.0 | 0.1 | GO:0060765 | regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.0 | 0.0 | GO:0030910 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.0 | 0.1 | GO:0033262 | regulation of nuclear cell cycle DNA replication(GO:0033262) |
| 0.0 | 0.0 | GO:0010661 | positive regulation of muscle cell apoptotic process(GO:0010661) positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.0 | 0.1 | GO:0060242 | contact inhibition(GO:0060242) |
| 0.0 | 0.1 | GO:0045759 | negative regulation of action potential(GO:0045759) |
| 0.0 | 0.1 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 0.0 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 0.1 | GO:0048563 | post-embryonic organ morphogenesis(GO:0048563) |
| 0.0 | 0.0 | GO:0032817 | regulation of natural killer cell proliferation(GO:0032817) |
| 0.0 | 0.0 | GO:0051788 | response to misfolded protein(GO:0051788) |
| 0.0 | 0.1 | GO:0034626 | fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:0032695 | negative regulation of interleukin-12 production(GO:0032695) |
| 0.0 | 0.1 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.1 | GO:1902222 | L-phenylalanine metabolic process(GO:0006558) L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.0 | 0.0 | GO:1904376 | negative regulation of establishment of protein localization to plasma membrane(GO:0090005) negative regulation of protein localization to plasma membrane(GO:1903077) negative regulation of protein localization to cell periphery(GO:1904376) |
| 0.0 | 0.0 | GO:0003402 | planar cell polarity pathway involved in axis elongation(GO:0003402) |
| 0.0 | 0.0 | GO:0060789 | hair follicle placode formation(GO:0060789) |
| 0.0 | 0.0 | GO:0022038 | corpus callosum development(GO:0022038) |
| 0.0 | 0.0 | GO:0032212 | positive regulation of telomere maintenance via telomerase(GO:0032212) positive regulation of telomere maintenance via telomere lengthening(GO:1904358) |
| 0.0 | 0.0 | GO:0061054 | dermatome development(GO:0061054) |
| 0.0 | 0.0 | GO:0008615 | pyridoxine metabolic process(GO:0008614) pyridoxine biosynthetic process(GO:0008615) vitamin B6 metabolic process(GO:0042816) vitamin B6 biosynthetic process(GO:0042819) |
| 0.0 | 0.1 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.1 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) |
| 0.0 | 0.2 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.0 | 0.1 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.0 | 0.0 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.0 | GO:0048143 | astrocyte activation(GO:0048143) |
| 0.0 | 0.0 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 | 0.0 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.0 | 0.1 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.0 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.0 | GO:0034145 | positive regulation of toll-like receptor 4 signaling pathway(GO:0034145) |
| 0.0 | 0.0 | GO:0006922 | obsolete cleavage of lamin involved in execution phase of apoptosis(GO:0006922) |
| 0.0 | 0.0 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.0 | 0.0 | GO:2000344 | positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.1 | GO:0043049 | otic placode formation(GO:0043049) |
| 0.0 | 0.1 | GO:0006546 | glycine catabolic process(GO:0006546) |
| 0.0 | 0.2 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.1 | GO:0043558 | regulation of translational initiation in response to stress(GO:0043558) |
| 0.0 | 0.0 | GO:0032455 | nerve growth factor processing(GO:0032455) |
| 0.0 | 0.2 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.0 | 0.1 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.0 | GO:0045343 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) |
| 0.0 | 0.0 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.0 | 0.1 | GO:0035246 | peptidyl-arginine N-methylation(GO:0035246) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.0 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.0 | 0.0 | GO:0043320 | natural killer cell activation involved in immune response(GO:0002323) natural killer cell degranulation(GO:0043320) |
| 0.0 | 0.2 | GO:0060330 | regulation of response to interferon-gamma(GO:0060330) regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.0 | 0.1 | GO:0046618 | drug export(GO:0046618) |
| 0.0 | 0.1 | GO:0045356 | positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.0 | 0.1 | GO:0060287 | epithelial cilium movement(GO:0003351) epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.1 | GO:0006734 | NADH metabolic process(GO:0006734) |
| 0.0 | 0.1 | GO:0003100 | regulation of systemic arterial blood pressure by endothelin(GO:0003100) |
| 0.0 | 0.1 | GO:0070670 | response to interleukin-4(GO:0070670) |
| 0.0 | 0.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.2 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.0 | GO:0090192 | regulation of glomerulus development(GO:0090192) |
| 0.0 | 0.1 | GO:0036230 | granulocyte activation(GO:0036230) |
| 0.0 | 0.1 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 | 0.0 | GO:0032898 | neurotrophin production(GO:0032898) |
| 0.0 | 0.0 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.0 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.0 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.0 | GO:0033194 | response to hydroperoxide(GO:0033194) |
| 0.0 | 0.1 | GO:0043628 | ncRNA 3'-end processing(GO:0043628) |
| 0.0 | 0.0 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 0.0 | GO:1901984 | negative regulation of histone acetylation(GO:0035067) negative regulation of protein acetylation(GO:1901984) negative regulation of peptidyl-lysine acetylation(GO:2000757) |
| 0.0 | 0.0 | GO:0060972 | left/right pattern formation(GO:0060972) |
| 0.0 | 0.1 | GO:0002052 | positive regulation of neuroblast proliferation(GO:0002052) |
| 0.0 | 0.0 | GO:0032959 | inositol trisphosphate biosynthetic process(GO:0032959) |
| 0.0 | 0.1 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.0 | 0.0 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 0.0 | GO:0010533 | regulation of activation of Janus kinase activity(GO:0010533) regulation of activation of JAK2 kinase activity(GO:0010534) positive regulation of activation of JAK2 kinase activity(GO:0010535) positive regulation of activation of Janus kinase activity(GO:0010536) |
| 0.0 | 0.0 | GO:0043368 | positive T cell selection(GO:0043368) |
| 0.0 | 0.0 | GO:1902803 | regulation of synaptic vesicle transport(GO:1902803) regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.0 | 0.1 | GO:0060438 | trachea development(GO:0060438) |
| 0.0 | 0.0 | GO:0051569 | regulation of histone H3-K4 methylation(GO:0051569) |
| 0.0 | 0.0 | GO:0045634 | regulation of melanocyte differentiation(GO:0045634) regulation of pigment cell differentiation(GO:0050932) |
| 0.0 | 0.0 | GO:0071922 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.0 | 0.0 | GO:0032693 | negative regulation of interleukin-10 production(GO:0032693) |
| 0.0 | 0.1 | GO:0042159 | lipoprotein catabolic process(GO:0042159) |
| 0.0 | 0.1 | GO:0045197 | establishment or maintenance of epithelial cell apical/basal polarity(GO:0045197) |
| 0.0 | 0.1 | GO:0042435 | indole-containing compound biosynthetic process(GO:0042435) |
| 0.0 | 0.2 | GO:0043526 | obsolete neuroprotection(GO:0043526) |
| 0.0 | 0.0 | GO:0070570 | regulation of axon regeneration(GO:0048679) regulation of neuron projection regeneration(GO:0070570) |
| 0.0 | 0.2 | GO:0097581 | lamellipodium organization(GO:0097581) |
| 0.0 | 0.0 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.0 | 0.0 | GO:0017085 | response to insecticide(GO:0017085) |
| 0.0 | 0.1 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.0 | 0.1 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.0 | 0.1 | GO:0036465 | synaptic vesicle recycling(GO:0036465) synaptic vesicle endocytosis(GO:0048488) clathrin-mediated endocytosis(GO:0072583) |
| 0.0 | 0.0 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.0 | 0.0 | GO:0060677 | ureteric bud elongation(GO:0060677) |
| 0.0 | 0.0 | GO:0042253 | granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0042253) regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045423) |
| 0.0 | 0.0 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.0 | 0.2 | GO:0046834 | lipid phosphorylation(GO:0046834) |
| 0.0 | 0.0 | GO:0043374 | CD8-positive, alpha-beta T cell activation(GO:0036037) CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.0 | GO:0044332 | Wnt signaling pathway involved in dorsal/ventral axis specification(GO:0044332) regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000053) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.1 | 0.3 | GO:0043218 | compact myelin(GO:0043218) |
| 0.1 | 0.2 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.2 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.5 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.2 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.1 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.0 | 0.1 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 0.1 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.1 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.0 | 0.1 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.1 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.3 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.0 | 0.1 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.1 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.1 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.3 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.2 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.1 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.0 | 0.3 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.1 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.2 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.1 | GO:0000791 | euchromatin(GO:0000791) |
| 0.0 | 0.1 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.6 | GO:0009295 | nucleoid(GO:0009295) |
| 0.0 | 0.0 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.1 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.0 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.0 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.1 | GO:0033011 | perinuclear theca(GO:0033011) |
| 0.0 | 0.0 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.1 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.1 | GO:0045261 | proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 0.1 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.1 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.0 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.1 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.5 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.1 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.2 | GO:0035267 | NuA4 histone acetyltransferase complex(GO:0035267) |
| 0.0 | 0.1 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.1 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.2 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.1 | GO:0034518 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.0 | 0.1 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.0 | 0.0 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.0 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.0 | GO:0000300 | obsolete peripheral to membrane of membrane fraction(GO:0000300) |
| 0.0 | 0.1 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.0 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 0.1 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) nuclear transcriptional repressor complex(GO:0090568) |
| 0.0 | 0.1 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.1 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.1 | GO:0098984 | asymmetric synapse(GO:0032279) neuron to neuron synapse(GO:0098984) |
| 0.0 | 0.0 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.0 | 0.1 | GO:0031010 | ISWI-type complex(GO:0031010) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.1 | 0.2 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.1 | 0.2 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.1 | 0.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.2 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.2 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.2 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.0 | 0.2 | GO:0050542 | icosanoid binding(GO:0050542) fatty acid derivative binding(GO:1901567) |
| 0.0 | 0.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.4 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.3 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.2 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.1 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.0 | 0.1 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.1 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 0.2 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.1 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.0 | 0.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.2 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.0 | 0.1 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.2 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.3 | GO:0099604 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.0 | 0.2 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.1 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.5 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.4 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.1 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.0 | 0.2 | GO:0051766 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) inositol trisphosphate kinase activity(GO:0051766) |
| 0.0 | 0.1 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.1 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.0 | 0.1 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.1 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.0 | 0.1 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.0 | 0.1 | GO:0030955 | potassium ion binding(GO:0030955) |
| 0.0 | 0.1 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.0 | 0.2 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.0 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.2 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.0 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.4 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.0 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.1 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.1 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.1 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.1 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.0 | 0.0 | GO:0009008 | DNA-methyltransferase activity(GO:0009008) |
| 0.0 | 0.1 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.1 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.0 | 0.1 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.1 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.1 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.2 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.0 | 0.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.7 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.1 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.1 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.1 | GO:0016715 | dopamine beta-monooxygenase activity(GO:0004500) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.0 | 0.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.1 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.1 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.1 | GO:0004075 | biotin carboxylase activity(GO:0004075) |
| 0.0 | 0.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.1 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.1 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 0.2 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.1 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 0.0 | 0.0 | GO:0031708 | bombesin receptor binding(GO:0031705) endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.1 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.2 | GO:0016409 | palmitoyltransferase activity(GO:0016409) |
| 0.0 | 0.3 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.2 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.1 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.1 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.4 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.1 | GO:0016778 | diphosphotransferase activity(GO:0016778) |
| 0.0 | 0.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.1 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.0 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.7 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.1 | GO:0048531 | beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.0 | 0.1 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 0.1 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 0.1 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.0 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.0 | 0.1 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.0 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.1 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.4 | GO:0016814 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amidines(GO:0016814) |
| 0.0 | 0.0 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.0 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.1 | GO:0034593 | phosphatidylinositol bisphosphate phosphatase activity(GO:0034593) |
| 0.0 | 0.1 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.1 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.1 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.1 | GO:0016274 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.0 | 0.1 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.0 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.0 | 0.0 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.1 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 0.0 | GO:0034187 | obsolete apolipoprotein E binding(GO:0034187) |
| 0.0 | 0.0 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) |
| 0.0 | 0.1 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.1 | GO:0005451 | monovalent cation:proton antiporter activity(GO:0005451) |
| 0.0 | 0.0 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) xenobiotic transporter activity(GO:0042910) |
| 0.0 | 0.0 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.0 | 0.1 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.1 | GO:0015924 | mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.0 | 0.0 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.0 | 0.0 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.0 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.0 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.0 | GO:0046980 | tapasin binding(GO:0046980) |
| 0.0 | 0.0 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.0 | 0.0 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.1 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.2 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.1 | GO:0016653 | oxidoreductase activity, acting on NAD(P)H, heme protein as acceptor(GO:0016653) |
| 0.0 | 0.0 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 0.1 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.2 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.0 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.0 | 0.1 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.1 | GO:0015166 | polyol transmembrane transporter activity(GO:0015166) |
| 0.0 | 0.0 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.1 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.0 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.1 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.2 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.0 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.0 | 0.1 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
| 0.0 | 0.0 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.1 | GO:0031996 | thioesterase binding(GO:0031996) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 1.5 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.2 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.2 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 1.6 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.3 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.7 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.0 | 0.2 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.6 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.3 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.2 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.2 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.7 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 0.0 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.2 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 0.7 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.3 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.2 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.0 | 0.3 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.2 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 0.0 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 0.3 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.1 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.3 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.3 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 0.1 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.3 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.4 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 0.1 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.1 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.0 | 0.1 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.0 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.0 | PID S1P S1P2 PATHWAY | S1P2 pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.6 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.5 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 0.0 | REACTOME SCFSKP2 MEDIATED DEGRADATION OF P27 P21 | Genes involved in SCF(Skp2)-mediated degradation of p27/p21 |
| 0.0 | 0.3 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.7 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 1.0 | REACTOME TCR SIGNALING | Genes involved in TCR signaling |
| 0.0 | 0.5 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.1 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.0 | 0.3 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.2 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.0 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.1 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.2 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.0 | 0.0 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.0 | 0.3 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 0.3 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.2 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.1 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.2 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 0.1 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.8 | REACTOME G ALPHA1213 SIGNALLING EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.0 | 0.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.2 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.1 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.1 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 0.1 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.0 | 0.0 | REACTOME SIGNALLING BY NGF | Genes involved in Signalling by NGF |
| 0.0 | 0.3 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.2 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.0 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.0 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.0 | 0.1 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.0 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.6 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.1 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.2 | REACTOME PROLONGED ERK ACTIVATION EVENTS | Genes involved in Prolonged ERK activation events |
| 0.0 | 0.0 | REACTOME SIGNALING BY SCF KIT | Genes involved in Signaling by SCF-KIT |
| 0.0 | 0.1 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.1 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 0.1 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.2 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.1 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |