Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
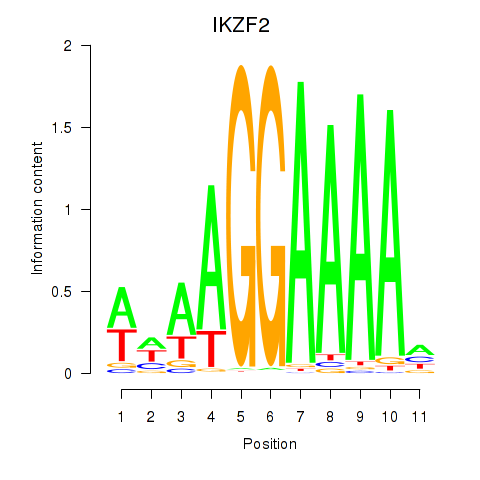
Results for IKZF2
Z-value: 1.58
Transcription factors associated with IKZF2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
IKZF2
|
ENSG00000030419.12 | IKAROS family zinc finger 2 |
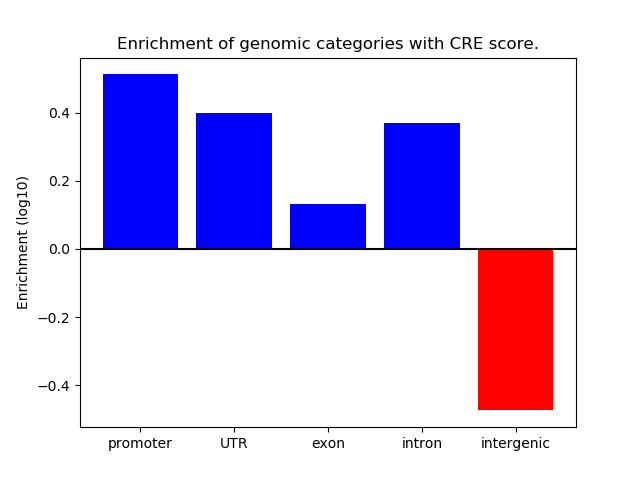
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr2_213986421_213986572 | IKZF2 | 26857 | 0.246949 | -0.71 | 3.3e-02 | Click! |
| chr2_214026642_214026793 | IKZF2 | 9566 | 0.302902 | 0.67 | 4.7e-02 | Click! |
| chr2_214010811_214010962 | IKZF2 | 2467 | 0.425227 | 0.61 | 8.3e-02 | Click! |
| chr2_213982929_213983080 | IKZF2 | 30349 | 0.234874 | -0.58 | 9.9e-02 | Click! |
| chr2_213982732_213982883 | IKZF2 | 30546 | 0.234186 | -0.53 | 1.4e-01 | Click! |
Activity of the IKZF2 motif across conditions
Conditions sorted by the z-value of the IKZF2 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
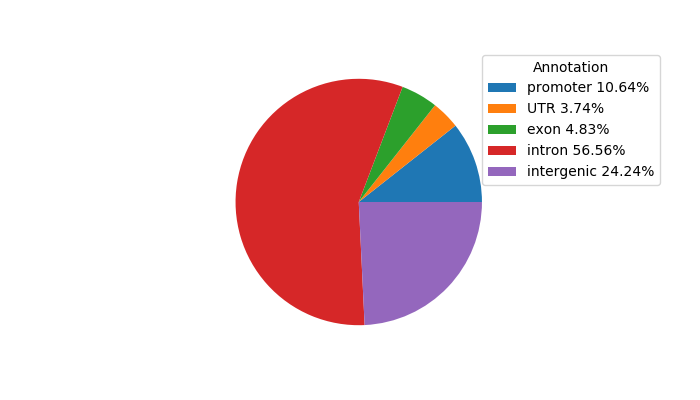
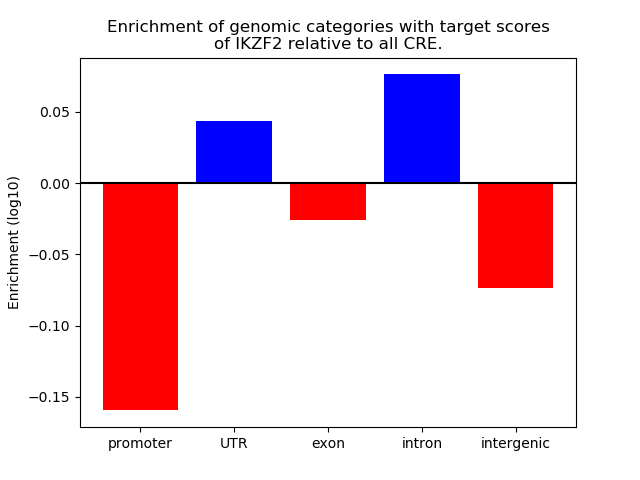
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr6_116877788_116878085 | 0.44 |
FAM26D |
family with sequence similarity 26, member D |
3023 |
0.18 |
| chr14_23006044_23006569 | 0.39 |
TRAJ15 |
T cell receptor alpha joining 15 |
7726 |
0.11 |
| chr18_20558298_20558528 | 0.38 |
RBBP8 |
retinoblastoma binding protein 8 |
9608 |
0.15 |
| chr11_3396621_3396905 | 0.36 |
ENSG00000263365 |
. |
1912 |
0.25 |
| chr13_30160711_30160862 | 0.36 |
SLC7A1 |
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
143 |
0.98 |
| chr1_65372485_65372704 | 0.36 |
JAK1 |
Janus kinase 1 |
59593 |
0.13 |
| chr2_101437963_101438114 | 0.35 |
NPAS2 |
neuronal PAS domain protein 2 |
551 |
0.67 |
| chr17_48812709_48812860 | 0.32 |
LUC7L3 |
LUC7-like 3 (S. cerevisiae) |
1212 |
0.39 |
| chr8_29109260_29109557 | 0.30 |
KIF13B |
kinesin family member 13B |
11182 |
0.17 |
| chr16_14030624_14030775 | 0.30 |
CTD-2135D7.2 |
|
1207 |
0.56 |
| chr4_170038040_170038191 | 0.28 |
RP11-327O17.2 |
|
84830 |
0.08 |
| chr9_124005105_124005256 | 0.28 |
RAB14 |
RAB14, member RAS oncogene family |
19888 |
0.13 |
| chr6_42326971_42327197 | 0.28 |
ENSG00000221252 |
. |
53050 |
0.13 |
| chr4_103998432_103998637 | 0.27 |
SLC9B2 |
solute carrier family 9, subfamily B (NHA2, cation proton antiporter 2), member 2 |
53 |
0.98 |
| chr2_30458978_30459361 | 0.27 |
LBH |
limb bud and heart development |
4123 |
0.28 |
| chr7_116270780_116270931 | 0.26 |
MET |
met proto-oncogene |
41589 |
0.14 |
| chr2_152115718_152115869 | 0.26 |
RBM43 |
RNA binding motif protein 43 |
2499 |
0.33 |
| chr22_20919404_20919765 | 0.26 |
MED15 |
mediator complex subunit 15 |
14007 |
0.14 |
| chr3_184055469_184055620 | 0.25 |
FAM131A |
family with sequence similarity 131, member A |
304 |
0.8 |
| chr18_13378456_13378780 | 0.25 |
LDLRAD4 |
low density lipoprotein receptor class A domain containing 4 |
3935 |
0.19 |
| chr9_97631859_97632254 | 0.25 |
RP11-49O14.2 |
|
30655 |
0.15 |
| chr8_38782658_38783281 | 0.25 |
PLEKHA2 |
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 2 |
24107 |
0.14 |
| chr7_55141300_55141512 | 0.25 |
EGFR |
epidermal growth factor receptor |
36010 |
0.22 |
| chr1_92220198_92220349 | 0.24 |
ENSG00000239794 |
. |
75358 |
0.11 |
| chr3_38875961_38876357 | 0.24 |
RP11-134J21.1 |
|
10766 |
0.19 |
| chr15_38779165_38779950 | 0.24 |
RP11-102L12.2 |
|
15497 |
0.17 |
| chr8_22320352_22320708 | 0.24 |
PPP3CC |
protein phosphatase 3, catalytic subunit, gamma isozyme |
12575 |
0.15 |
| chr18_55863805_55863956 | 0.24 |
RP11-718I15.1 |
|
952 |
0.42 |
| chr3_66062574_66062725 | 0.24 |
MAGI1 |
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
38424 |
0.17 |
| chr6_112078721_112079214 | 0.24 |
FYN |
FYN oncogene related to SRC, FGR, YES |
1350 |
0.56 |
| chr4_129744671_129744822 | 0.24 |
JADE1 |
jade family PHD finger 1 |
7842 |
0.3 |
| chr17_41495072_41495223 | 0.24 |
ENSG00000251763 |
. |
5352 |
0.12 |
| chr10_74004394_74004633 | 0.23 |
ENSG00000200170 |
. |
24003 |
0.12 |
| chr6_114287058_114287209 | 0.23 |
HDAC2 |
histone deacetylase 2 |
2561 |
0.27 |
| chr6_112127086_112127660 | 0.23 |
FYN |
FYN oncogene related to SRC, FGR, YES |
7747 |
0.29 |
| chr22_38281441_38281592 | 0.23 |
ENSG00000207227 |
. |
5838 |
0.1 |
| chr2_102443416_102443820 | 0.23 |
MAP4K4 |
mitogen-activated protein kinase kinase kinase kinase 4 |
12672 |
0.26 |
| chr17_47896409_47896901 | 0.23 |
RP11-304F15.3 |
|
26617 |
0.12 |
| chrX_30893495_30893646 | 0.23 |
TAB3 |
TGF-beta activated kinase 1/MAP3K7 binding protein 3 |
8251 |
0.21 |
| chr1_93399148_93399403 | 0.23 |
FAM69A |
family with sequence similarity 69, member A |
27782 |
0.11 |
| chr11_35268427_35268939 | 0.22 |
SLC1A2 |
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
18609 |
0.14 |
| chr16_87469163_87469969 | 0.22 |
ZCCHC14 |
zinc finger, CCHC domain containing 14 |
2826 |
0.22 |
| chr6_133944571_133944722 | 0.22 |
RP3-323P13.2 |
|
111780 |
0.07 |
| chr1_156099255_156099406 | 0.22 |
LMNA |
lamin A/C |
2736 |
0.16 |
| chr17_39533966_39534194 | 0.22 |
KRT34 |
keratin 34 |
4575 |
0.1 |
| chr18_7812137_7812288 | 0.22 |
RP11-678G15.1 |
|
1904 |
0.35 |
| chr9_114812031_114812182 | 0.22 |
RP11-4O1.2 |
|
12096 |
0.2 |
| chr11_128340661_128340942 | 0.21 |
ETS1 |
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
34488 |
0.2 |
| chr10_33251015_33251258 | 0.21 |
ITGB1 |
integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12) |
1075 |
0.65 |
| chr3_40505730_40505995 | 0.21 |
RPL14 |
ribosomal protein L14 |
7017 |
0.15 |
| chr11_96039540_96039791 | 0.21 |
ENSG00000266192 |
. |
34937 |
0.17 |
| chrY_21903585_21903736 | 0.21 |
KDM5D |
lysine (K)-specific demethylase 5D |
2937 |
0.4 |
| chr3_130936015_130936300 | 0.21 |
ENSG00000265235 |
. |
28915 |
0.19 |
| chr9_34203797_34204061 | 0.21 |
ENSG00000252164 |
. |
2310 |
0.22 |
| chr7_46495799_46495950 | 0.21 |
AC011294.3 |
Uncharacterized protein |
240846 |
0.02 |
| chr6_127527843_127527994 | 0.21 |
RNF146 |
ring finger protein 146 |
59837 |
0.13 |
| chr10_119300742_119300893 | 0.21 |
EMX2OS |
EMX2 opposite strand/antisense RNA |
1013 |
0.44 |
| chr19_13373156_13373307 | 0.21 |
CACNA1A |
calcium channel, voltage-dependent, P/Q type, alpha 1A subunit |
5017 |
0.22 |
| chr16_22398221_22398372 | 0.21 |
CDR2 |
cerebellar degeneration-related protein 2, 62kDa |
11885 |
0.14 |
| chr2_30477316_30477615 | 0.21 |
LBH |
limb bud and heart development |
22419 |
0.22 |
| chr8_13303389_13303540 | 0.21 |
DLC1 |
deleted in liver cancer 1 |
53286 |
0.15 |
| chr22_18405059_18405210 | 0.21 |
MICAL3 |
microtubule associated monooxygenase, calponin and LIM domain containing 3 |
15494 |
0.19 |
| chr2_238338921_238339072 | 0.21 |
AC112721.1 |
Uncharacterized protein |
5077 |
0.22 |
| chr6_143172108_143172466 | 0.21 |
HIVEP2 |
human immunodeficiency virus type I enhancer binding protein 2 |
14103 |
0.27 |
| chr3_105504443_105504594 | 0.21 |
CBLB |
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
83369 |
0.11 |
| chr2_55818169_55818320 | 0.21 |
ENSG00000212175 |
. |
25405 |
0.14 |
| chr3_23852622_23853296 | 0.21 |
UBE2E1 |
ubiquitin-conjugating enzyme E2E 1 |
10 |
0.98 |
| chr11_82780511_82780748 | 0.21 |
RAB30 |
RAB30, member RAS oncogene family |
2241 |
0.29 |
| chr1_151963809_151963960 | 0.21 |
S100A10 |
S100 calcium binding protein A10 |
1170 |
0.39 |
| chr8_117753205_117753356 | 0.21 |
EIF3H |
eukaryotic translation initiation factor 3, subunit H |
14743 |
0.21 |
| chr6_159271708_159272064 | 0.21 |
C6orf99 |
chromosome 6 open reading frame 99 |
19085 |
0.16 |
| chr11_60828326_60828529 | 0.20 |
CD5 |
CD5 molecule |
41440 |
0.11 |
| chr4_143327756_143327922 | 0.20 |
INPP4B |
inositol polyphosphate-4-phosphatase, type II, 105kDa |
24573 |
0.29 |
| chr1_202249577_202249728 | 0.20 |
ENSG00000239046 |
. |
48860 |
0.1 |
| chr2_191723875_191724026 | 0.20 |
GLS |
glutaminase |
21603 |
0.2 |
| chr15_42227548_42227790 | 0.20 |
CTD-2382E5.4 |
|
14054 |
0.11 |
| chr3_192606561_192606956 | 0.20 |
MB21D2 |
Mab-21 domain containing 2 |
29192 |
0.24 |
| chr16_24785300_24785451 | 0.20 |
TNRC6A |
trinucleotide repeat containing 6A |
17433 |
0.22 |
| chr3_141928282_141928433 | 0.20 |
GK5 |
glycerol kinase 5 (putative) |
16071 |
0.22 |
| chr4_141968010_141968161 | 0.20 |
RNF150 |
ring finger protein 150 |
6795 |
0.32 |
| chr6_148662831_148662982 | 0.20 |
SASH1 |
SAM and SH3 domain containing 1 |
823 |
0.72 |
| chr17_7213721_7213907 | 0.20 |
EIF5A |
eukaryotic translation initiation factor 5A |
2124 |
0.1 |
| chr14_52821102_52821350 | 0.20 |
PTGER2 |
prostaglandin E receptor 2 (subtype EP2), 53kDa |
40113 |
0.19 |
| chr21_36168084_36168235 | 0.20 |
AP000330.8 |
|
50025 |
0.16 |
| chr17_34909042_34909369 | 0.20 |
GGNBP2 |
gametogenetin binding protein 2 |
7852 |
0.13 |
| chr7_130587786_130587937 | 0.20 |
ENSG00000226380 |
. |
25563 |
0.22 |
| chr9_108737188_108737339 | 0.20 |
TMEM38B |
transmembrane protein 38B |
253295 |
0.02 |
| chr4_7905515_7905666 | 0.20 |
AFAP1 |
actin filament associated protein 1 |
31550 |
0.17 |
| chr12_13348032_13348183 | 0.20 |
EMP1 |
epithelial membrane protein 1 |
1543 |
0.49 |
| chr9_111858912_111859063 | 0.20 |
TMEM245 |
transmembrane protein 245 |
9414 |
0.19 |
| chr19_13132424_13132703 | 0.20 |
NFIX |
nuclear factor I/X (CCAAT-binding transcription factor) |
2242 |
0.17 |
| chr17_2310516_2310667 | 0.20 |
MNT |
MAX network transcriptional repressor |
6179 |
0.12 |
| chr6_121969345_121969496 | 0.20 |
ENSG00000222659 |
. |
67753 |
0.13 |
| chr13_100743473_100743624 | 0.20 |
PCCA |
propionyl CoA carboxylase, alpha polypeptide |
2211 |
0.41 |
| chr11_119112060_119112211 | 0.19 |
CBL |
Cbl proto-oncogene, E3 ubiquitin protein ligase |
35383 |
0.08 |
| chr20_51832864_51833015 | 0.19 |
RP11-15M15.2 |
|
7695 |
0.21 |
| chr4_143318453_143318604 | 0.19 |
INPP4B |
inositol polyphosphate-4-phosphatase, type II, 105kDa |
33884 |
0.25 |
| chr9_74375544_74375695 | 0.19 |
TMEM2 |
transmembrane protein 2 |
7683 |
0.3 |
| chr10_74004680_74004925 | 0.19 |
ENSG00000200170 |
. |
24292 |
0.12 |
| chr14_64335207_64335619 | 0.19 |
SYNE2 |
spectrin repeat containing, nuclear envelope 2 |
15681 |
0.21 |
| chr15_70991883_70992034 | 0.19 |
UACA |
uveal autoantigen with coiled-coil domains and ankyrin repeats |
2662 |
0.37 |
| chr6_42307045_42307377 | 0.19 |
ENSG00000221252 |
. |
72923 |
0.09 |
| chr10_103446662_103446813 | 0.19 |
FBXW4 |
F-box and WD repeat domain containing 4 |
8279 |
0.19 |
| chr1_86158252_86158403 | 0.19 |
ZNHIT6 |
zinc finger, HIT-type containing 6 |
15774 |
0.25 |
| chr12_69922390_69922541 | 0.19 |
FRS2 |
fibroblast growth factor receptor substrate 2 |
2138 |
0.34 |
| chr14_61932790_61932961 | 0.19 |
PRKCH |
protein kinase C, eta |
23599 |
0.2 |
| chr15_42640019_42640170 | 0.19 |
CAPN3 |
calpain 3, (p94) |
207 |
0.93 |
| chr13_24253099_24253504 | 0.19 |
AL139080.1 |
Uncharacterized protein |
68942 |
0.11 |
| chr1_218649534_218649685 | 0.19 |
C1orf143 |
chromosome 1 open reading frame 143 |
33829 |
0.2 |
| chr12_76115257_76115408 | 0.19 |
ENSG00000264062 |
. |
2989 |
0.33 |
| chr1_150662741_150662892 | 0.19 |
GOLPH3L |
golgi phosphoprotein 3-like |
6684 |
0.13 |
| chr10_33538186_33538377 | 0.19 |
NRP1 |
neuropilin 1 |
14351 |
0.21 |
| chr10_43895608_43895896 | 0.19 |
HNRNPF |
heterogeneous nuclear ribonucleoprotein F |
2958 |
0.21 |
| chr2_37686183_37686412 | 0.19 |
ENSG00000253078 |
. |
23546 |
0.21 |
| chr16_71671220_71671371 | 0.19 |
MARVELD3 |
MARVEL domain containing 3 |
8322 |
0.1 |
| chr5_143546729_143546880 | 0.19 |
YIPF5 |
Yip1 domain family, member 5 |
3390 |
0.25 |
| chr3_66027202_66027443 | 0.19 |
MAGI1 |
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
3097 |
0.3 |
| chr14_99730019_99730213 | 0.19 |
AL109767.1 |
|
831 |
0.66 |
| chr8_69838287_69838438 | 0.19 |
ENSG00000238808 |
. |
184447 |
0.03 |
| chr1_151962516_151962768 | 0.19 |
S100A10 |
S100 calcium binding protein A10 |
2412 |
0.22 |
| chr12_57628145_57628477 | 0.19 |
SHMT2 |
serine hydroxymethyltransferase 2 (mitochondrial) |
2067 |
0.16 |
| chr3_177801896_177802095 | 0.19 |
ENSG00000199858 |
. |
76428 |
0.12 |
| chr11_121973042_121973193 | 0.19 |
RP11-166D19.1 |
|
231 |
0.91 |
| chr16_68485211_68485362 | 0.19 |
SMPD3 |
sphingomyelin phosphodiesterase 3, neutral membrane (neutral sphingomyelinase II) |
2695 |
0.23 |
| chr8_32180787_32180938 | 0.19 |
ENSG00000200246 |
. |
66850 |
0.12 |
| chr10_63512082_63512376 | 0.19 |
RP11-63A2.2 |
|
67083 |
0.12 |
| chr6_112127691_112127942 | 0.19 |
FYN |
FYN oncogene related to SRC, FGR, YES |
8190 |
0.29 |
| chr10_15208005_15208246 | 0.19 |
NMT2 |
N-myristoyltransferase 2 |
2320 |
0.33 |
| chr2_172128341_172128492 | 0.19 |
TLK1 |
tousled-like kinase 1 |
40592 |
0.18 |
| chr2_225086781_225086932 | 0.19 |
ENSG00000211987 |
. |
17417 |
0.28 |
| chr1_205142673_205142824 | 0.19 |
DSTYK |
dual serine/threonine and tyrosine protein kinase |
37946 |
0.12 |
| chr5_73612868_73613019 | 0.19 |
ENSG00000244326 |
. |
868 |
0.71 |
| chr11_11867982_11868133 | 0.19 |
USP47 |
ubiquitin specific peptidase 47 |
4444 |
0.33 |
| chr9_119463950_119464101 | 0.19 |
TRIM32 |
tripartite motif containing 32 |
14403 |
0.22 |
| chr6_52416562_52416716 | 0.19 |
TRAM2 |
translocation associated membrane protein 2 |
25074 |
0.22 |
| chr1_221614582_221614778 | 0.18 |
DUSP10 |
dual specificity phosphatase 10 |
296122 |
0.01 |
| chr18_21633829_21634342 | 0.18 |
TTC39C |
tetratricopeptide repeat domain 39C |
37487 |
0.11 |
| chr13_40767790_40767941 | 0.18 |
ENSG00000207458 |
. |
33099 |
0.24 |
| chr13_101045927_101046215 | 0.18 |
PCCA |
propionyl CoA carboxylase, alpha polypeptide |
25318 |
0.23 |
| chr16_4264572_4264723 | 0.18 |
SRL |
sarcalumenin |
25097 |
0.17 |
| chr6_24883063_24883351 | 0.18 |
FAM65B |
family with sequence similarity 65, member B |
5624 |
0.23 |
| chr6_86164371_86164522 | 0.18 |
NT5E |
5'-nucleotidase, ecto (CD73) |
4619 |
0.31 |
| chr2_145853230_145853381 | 0.18 |
ENSG00000253036 |
. |
239333 |
0.02 |
| chr17_13251602_13251753 | 0.18 |
ENSG00000266115 |
. |
151968 |
0.04 |
| chr4_99444287_99444438 | 0.18 |
RP11-724M22.1 |
|
26847 |
0.24 |
| chr8_103741522_103741773 | 0.18 |
ENSG00000266799 |
. |
4300 |
0.27 |
| chr11_128372954_128373665 | 0.18 |
ETS1 |
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
1980 |
0.4 |
| chr11_128381097_128381643 | 0.18 |
ETS1 |
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
6081 |
0.25 |
| chr6_36946763_36946914 | 0.18 |
MTCH1 |
mitochondrial carrier 1 |
1389 |
0.4 |
| chr8_25117778_25117929 | 0.18 |
DOCK5 |
dedicator of cytokinesis 5 |
38605 |
0.19 |
| chr7_130630277_130630707 | 0.18 |
LINC-PINT |
long intergenic non-protein coding RNA, p53 induced transcript |
38376 |
0.18 |
| chr18_57365633_57365784 | 0.18 |
CCBE1 |
collagen and calcium binding EGF domains 1 |
1096 |
0.44 |
| chr18_55975063_55975214 | 0.18 |
NEDD4L |
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
15414 |
0.22 |
| chr6_160399730_160400221 | 0.18 |
IGF2R |
insulin-like growth factor 2 receptor |
9844 |
0.21 |
| chr12_3328021_3328172 | 0.18 |
TSPAN9 |
tetraspanin 9 |
17755 |
0.24 |
| chr11_48192181_48192553 | 0.18 |
OR4B1 |
olfactory receptor, family 4, subfamily B, member 1 |
45977 |
0.13 |
| chr1_202548605_202548756 | 0.18 |
RP11-569A11.1 |
|
24716 |
0.18 |
| chr11_92002095_92002246 | 0.18 |
FAT3 |
FAT atypical cadherin 3 |
83092 |
0.11 |
| chr12_90084691_90084842 | 0.18 |
ATP2B1 |
ATPase, Ca++ transporting, plasma membrane 1 |
17842 |
0.19 |
| chr14_64332649_64332871 | 0.18 |
SYNE2 |
spectrin repeat containing, nuclear envelope 2 |
13028 |
0.21 |
| chr8_91682668_91682819 | 0.18 |
RP11-68L18.1 |
|
21767 |
0.2 |
| chr22_20900238_20900389 | 0.18 |
MED15 |
mediator complex subunit 15 |
4961 |
0.15 |
| chr1_214646887_214647162 | 0.18 |
PTPN14 |
protein tyrosine phosphatase, non-receptor type 14 |
8878 |
0.3 |
| chr5_54892777_54892928 | 0.18 |
PPAP2A |
phosphatidic acid phosphatase type 2A |
61974 |
0.11 |
| chr1_24790818_24790969 | 0.18 |
NIPAL3 |
NIPA-like domain containing 3 |
8237 |
0.18 |
| chr2_225746364_225746903 | 0.18 |
DOCK10 |
dedicator of cytokinesis 10 |
65149 |
0.14 |
| chr11_68768633_68768911 | 0.18 |
RP11-554A11.4 |
|
744 |
0.51 |
| chr14_100434768_100434919 | 0.18 |
EVL |
Enah/Vasp-like |
2943 |
0.26 |
| chr17_57439100_57439642 | 0.18 |
ENSG00000263857 |
. |
4073 |
0.2 |
| chr11_94539031_94539182 | 0.18 |
AMOTL1 |
angiomotin like 1 |
37569 |
0.17 |
| chr9_33414190_33414341 | 0.18 |
RP11-311H10.4 |
|
11410 |
0.14 |
| chr5_54605298_54605894 | 0.18 |
SKIV2L2 |
superkiller viralicidic activity 2-like 2 (S. cerevisiae) |
1767 |
0.29 |
| chr7_27813849_27814000 | 0.18 |
TAX1BP1 |
Tax1 (human T-cell leukemia virus type I) binding protein 1 |
18833 |
0.21 |
| chr3_81265699_81265850 | 0.18 |
ENSG00000222389 |
. |
292853 |
0.01 |
| chr1_23664610_23664879 | 0.18 |
HNRNPR |
heterogeneous nuclear ribonucleoprotein R |
6011 |
0.16 |
| chr4_114679396_114679562 | 0.18 |
CAMK2D |
calcium/calmodulin-dependent protein kinase II delta |
2745 |
0.42 |
| chr5_131355905_131356056 | 0.18 |
ACSL6 |
acyl-CoA synthetase long-chain family member 6 |
8110 |
0.14 |
| chr1_12466917_12467188 | 0.18 |
VPS13D |
vacuolar protein sorting 13 homolog D (S. cerevisiae) |
2872 |
0.34 |
| chr22_42013582_42013733 | 0.18 |
DESI1 |
desumoylating isopeptidase 1 |
3443 |
0.14 |
| chr3_69834642_69834793 | 0.18 |
ENSG00000241665 |
. |
15789 |
0.23 |
| chr15_67367308_67367832 | 0.18 |
SMAD3 |
SMAD family member 3 |
9387 |
0.28 |
| chr14_96982092_96982363 | 0.18 |
PAPOLA |
poly(A) polymerase alpha |
11552 |
0.18 |
| chr2_158605215_158605366 | 0.18 |
ACVR1 |
activin A receptor, type I |
58218 |
0.13 |
| chr17_35906348_35906499 | 0.18 |
SYNRG |
synergin, gamma |
3935 |
0.23 |
| chr12_105608187_105608338 | 0.18 |
APPL2 |
adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 2 |
15376 |
0.21 |
| chr17_6527918_6528069 | 0.18 |
KIAA0753 |
KIAA0753 |
3834 |
0.13 |
| chr5_180286437_180286588 | 0.18 |
ZFP62 |
ZFP62 zinc finger protein |
1151 |
0.46 |
| chr8_42143739_42143890 | 0.18 |
IKBKB |
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase beta |
14224 |
0.15 |
| chr17_20902840_20902991 | 0.17 |
RP11-746M1.1 |
|
145 |
0.95 |
| chr4_48695143_48695294 | 0.17 |
FRYL |
FRY-like |
12030 |
0.26 |
| chr18_56529967_56530118 | 0.17 |
ZNF532 |
zinc finger protein 532 |
138 |
0.96 |
| chr2_102326009_102326243 | 0.17 |
MAP4K4 |
mitogen-activated protein kinase kinase kinase kinase 4 |
11134 |
0.3 |
| chr3_56925362_56925595 | 0.17 |
ARHGEF3 |
Rho guanine nucleotide exchange factor (GEF) 3 |
25021 |
0.21 |
| chr8_100715929_100716325 | 0.17 |
ENSG00000243254 |
. |
89821 |
0.08 |
| chr17_16133762_16133975 | 0.17 |
NCOR1 |
nuclear receptor corepressor 1 |
12369 |
0.15 |
| chr18_72662833_72662984 | 0.17 |
ZADH2 |
zinc binding alcohol dehydrogenase domain containing 2 |
254560 |
0.02 |
| chr22_24763474_24763625 | 0.17 |
ADORA2A |
adenosine A2a receptor |
56016 |
0.1 |
| chr21_44144056_44144280 | 0.17 |
AP001627.1 |
|
17700 |
0.2 |
| chr10_3929538_3929752 | 0.17 |
KLF6 |
Kruppel-like factor 6 |
102172 |
0.08 |
| chr14_98648496_98648874 | 0.17 |
ENSG00000222066 |
. |
149402 |
0.04 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0044415 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.1 | 0.3 | GO:1900120 | regulation of receptor binding(GO:1900120) negative regulation of receptor binding(GO:1900121) |
| 0.1 | 0.1 | GO:0043368 | positive T cell selection(GO:0043368) |
| 0.1 | 0.2 | GO:0010766 | negative regulation of sodium ion transport(GO:0010766) |
| 0.1 | 0.3 | GO:0002870 | lymphocyte anergy(GO:0002249) regulation of T cell anergy(GO:0002667) positive regulation of T cell anergy(GO:0002669) T cell anergy(GO:0002870) regulation of lymphocyte anergy(GO:0002911) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.1 | 0.2 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.1 | 0.3 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.1 | 0.1 | GO:0002361 | CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) |
| 0.1 | 0.2 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.1 | 0.2 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.1 | 0.2 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.1 | 0.3 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 | 0.2 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.1 | 0.3 | GO:0032909 | transforming growth factor beta2 production(GO:0032906) regulation of transforming growth factor beta2 production(GO:0032909) |
| 0.1 | 0.2 | GO:0045066 | regulatory T cell differentiation(GO:0045066) |
| 0.1 | 0.2 | GO:0048680 | positive regulation of axon regeneration(GO:0048680) positive regulation of neuron projection regeneration(GO:0070572) |
| 0.1 | 0.2 | GO:0042091 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.1 | 0.2 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.1 | 0.2 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.0 | 0.1 | GO:0000491 | small nucleolar ribonucleoprotein complex assembly(GO:0000491) box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.4 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.0 | 0.1 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.0 | 0.3 | GO:2000400 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.0 | 0.1 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.1 | GO:0001743 | optic placode formation(GO:0001743) |
| 0.0 | 0.2 | GO:0006498 | N-terminal protein lipidation(GO:0006498) |
| 0.0 | 0.6 | GO:1902749 | regulation of G2/M transition of mitotic cell cycle(GO:0010389) regulation of cell cycle G2/M phase transition(GO:1902749) |
| 0.0 | 0.2 | GO:0009169 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.0 | 0.1 | GO:0032353 | negative regulation of hormone metabolic process(GO:0032351) negative regulation of hormone biosynthetic process(GO:0032353) |
| 0.0 | 0.1 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.1 | GO:0008049 | male courtship behavior(GO:0008049) |
| 0.0 | 0.2 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.0 | 0.1 | GO:0070099 | Roundabout signaling pathway(GO:0035385) regulation of chemokine-mediated signaling pathway(GO:0070099) negative regulation of chemokine-mediated signaling pathway(GO:0070100) |
| 0.0 | 0.1 | GO:0090190 | positive regulation of mesonephros development(GO:0061213) positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.0 | 0.1 | GO:0030011 | maintenance of cell polarity(GO:0030011) |
| 0.0 | 0.1 | GO:0032825 | regulation of natural killer cell differentiation(GO:0032823) positive regulation of natural killer cell differentiation(GO:0032825) |
| 0.0 | 0.1 | GO:0071803 | regulation of podosome assembly(GO:0071801) positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.1 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
| 0.0 | 0.1 | GO:0045607 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.0 | 0.1 | GO:0046668 | retinal cell programmed cell death(GO:0046666) regulation of retinal cell programmed cell death(GO:0046668) |
| 0.0 | 0.1 | GO:0060083 | smooth muscle contraction involved in micturition(GO:0060083) |
| 0.0 | 0.3 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.1 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.1 | GO:0090206 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.0 | 0.1 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.1 | GO:0001553 | luteinization(GO:0001553) |
| 0.0 | 0.1 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.2 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.2 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.1 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 0.1 | GO:0048319 | axial mesoderm morphogenesis(GO:0048319) |
| 0.0 | 0.0 | GO:0090205 | positive regulation of cholesterol metabolic process(GO:0090205) |
| 0.0 | 0.2 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.0 | 0.1 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.1 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.0 | 0.2 | GO:0018027 | peptidyl-lysine dimethylation(GO:0018027) |
| 0.0 | 0.2 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 | 0.1 | GO:0070670 | response to interleukin-4(GO:0070670) |
| 0.0 | 0.1 | GO:0033962 | cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.0 | 0.1 | GO:0006562 | proline catabolic process(GO:0006562) |
| 0.0 | 0.0 | GO:0001779 | natural killer cell differentiation(GO:0001779) |
| 0.0 | 0.1 | GO:0036037 | CD8-positive, alpha-beta T cell activation(GO:0036037) CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.1 | GO:0032801 | receptor catabolic process(GO:0032801) |
| 0.0 | 0.1 | GO:0031282 | regulation of guanylate cyclase activity(GO:0031282) |
| 0.0 | 0.1 | GO:0002329 | immature B cell differentiation(GO:0002327) pre-B cell differentiation(GO:0002329) |
| 0.0 | 0.2 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 | 0.1 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.1 | GO:0021801 | cerebral cortex radial glia guided migration(GO:0021801) telencephalon glial cell migration(GO:0022030) |
| 0.0 | 0.2 | GO:1901797 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) negative regulation of signal transduction by p53 class mediator(GO:1901797) |
| 0.0 | 0.3 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.0 | 0.1 | GO:0050687 | negative regulation of defense response to virus(GO:0050687) |
| 0.0 | 0.1 | GO:0000470 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 0.1 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.1 | GO:0060754 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 0.1 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.0 | 0.1 | GO:0072141 | mesangial cell differentiation(GO:0072007) kidney interstitial fibroblast differentiation(GO:0072071) renal interstitial fibroblast development(GO:0072141) mesangial cell development(GO:0072143) pericyte cell differentiation(GO:1904238) |
| 0.0 | 0.1 | GO:0051409 | response to nitrosative stress(GO:0051409) |
| 0.0 | 0.0 | GO:0032714 | negative regulation of interleukin-5 production(GO:0032714) |
| 0.0 | 0.1 | GO:0021542 | dentate gyrus development(GO:0021542) |
| 0.0 | 0.1 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.0 | 0.1 | GO:0090312 | regulation of protein deacetylation(GO:0090311) positive regulation of protein deacetylation(GO:0090312) |
| 0.0 | 0.0 | GO:0072217 | negative regulation of metanephros development(GO:0072217) |
| 0.0 | 0.0 | GO:0046476 | glycosylceramide biosynthetic process(GO:0046476) |
| 0.0 | 0.2 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.0 | 0.2 | GO:0008634 | obsolete negative regulation of survival gene product expression(GO:0008634) |
| 0.0 | 0.1 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.0 | 0.0 | GO:0002713 | negative regulation of B cell mediated immunity(GO:0002713) negative regulation of immunoglobulin mediated immune response(GO:0002890) |
| 0.0 | 0.1 | GO:0042487 | regulation of odontogenesis of dentin-containing tooth(GO:0042487) |
| 0.0 | 0.1 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.0 | 0.0 | GO:0033087 | regulation of immature T cell proliferation in thymus(GO:0033084) negative regulation of immature T cell proliferation(GO:0033087) negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.0 | 0.1 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.1 | GO:0019060 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) regulation by virus of viral protein levels in host cell(GO:0046719) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.0 | 0.2 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.0 | 0.1 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.0 | 0.6 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.0 | 0.1 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 0.0 | GO:0043497 | regulation of protein heterodimerization activity(GO:0043497) |
| 0.0 | 0.0 | GO:0060789 | hair follicle placode formation(GO:0060789) |
| 0.0 | 0.1 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.1 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.0 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.0 | 0.1 | GO:0006188 | IMP biosynthetic process(GO:0006188) IMP metabolic process(GO:0046040) |
| 0.0 | 0.2 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.0 | GO:0071265 | amino acid salvage(GO:0043102) L-methionine biosynthetic process(GO:0071265) L-methionine salvage(GO:0071267) |
| 0.0 | 0.2 | GO:0001782 | B cell homeostasis(GO:0001782) |
| 0.0 | 0.0 | GO:1903054 | regulation of extracellular matrix disassembly(GO:0010715) negative regulation of extracellular matrix disassembly(GO:0010716) regulation of extracellular matrix organization(GO:1903053) negative regulation of extracellular matrix organization(GO:1903054) |
| 0.0 | 0.1 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.0 | 0.0 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.0 | 0.1 | GO:0046885 | regulation of hormone biosynthetic process(GO:0046885) |
| 0.0 | 0.1 | GO:0045540 | regulation of cholesterol biosynthetic process(GO:0045540) |
| 0.0 | 0.1 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.0 | 0.1 | GO:0002360 | T cell lineage commitment(GO:0002360) |
| 0.0 | 0.4 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.1 | GO:0016264 | gap junction assembly(GO:0016264) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.0 | GO:0021937 | cerebellar Purkinje cell-granule cell precursor cell signaling involved in regulation of granule cell precursor cell proliferation(GO:0021937) |
| 0.0 | 0.0 | GO:0031657 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G1/S transition of mitotic cell cycle(GO:0031657) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G1/S transition of mitotic cell cycle(GO:0031659) |
| 0.0 | 0.1 | GO:0060390 | regulation of SMAD protein import into nucleus(GO:0060390) |
| 0.0 | 0.2 | GO:0021527 | spinal cord association neuron differentiation(GO:0021527) |
| 0.0 | 0.1 | GO:0016553 | base conversion or substitution editing(GO:0016553) |
| 0.0 | 0.2 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.0 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.1 | GO:0003032 | detection of oxygen(GO:0003032) |
| 0.0 | 0.1 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.1 | GO:0060317 | cardiac epithelial to mesenchymal transition(GO:0060317) |
| 0.0 | 0.1 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.1 | GO:0034201 | response to oleic acid(GO:0034201) |
| 0.0 | 0.0 | GO:0072189 | ureter development(GO:0072189) kidney smooth muscle tissue development(GO:0072194) |
| 0.0 | 0.0 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.2 | GO:0045161 | neuronal ion channel clustering(GO:0045161) |
| 0.0 | 0.1 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.0 | 0.1 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.0 | 0.1 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.0 | 0.0 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 | 0.0 | GO:0060426 | lung vasculature development(GO:0060426) |
| 0.0 | 0.1 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.1 | GO:0010837 | regulation of keratinocyte proliferation(GO:0010837) |
| 0.0 | 0.0 | GO:0061054 | positive regulation of cell adhesion mediated by integrin(GO:0033630) dermatome development(GO:0061054) |
| 0.0 | 0.1 | GO:0006688 | glycosphingolipid biosynthetic process(GO:0006688) |
| 0.0 | 0.1 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.0 | 0.0 | GO:0033689 | negative regulation of osteoblast proliferation(GO:0033689) |
| 0.0 | 0.0 | GO:0072033 | renal vesicle formation(GO:0072033) |
| 0.0 | 0.0 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.0 | 0.1 | GO:0009180 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) ribonucleoside diphosphate biosynthetic process(GO:0009188) |
| 0.0 | 0.2 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.5 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.1 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.2 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.0 | 0.1 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.0 | 0.1 | GO:0042339 | keratan sulfate metabolic process(GO:0042339) |
| 0.0 | 0.0 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.1 | GO:0035511 | oxidative DNA demethylation(GO:0035511) |
| 0.0 | 0.1 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.0 | GO:0070602 | centromeric sister chromatid cohesion(GO:0070601) regulation of centromeric sister chromatid cohesion(GO:0070602) |
| 0.0 | 0.2 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.1 | GO:0098810 | neurotransmitter reuptake(GO:0098810) |
| 0.0 | 0.1 | GO:0048840 | otolith development(GO:0048840) |
| 0.0 | 0.1 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.1 | GO:0008054 | negative regulation of cyclin-dependent protein serine/threonine kinase by cyclin degradation(GO:0008054) |
| 0.0 | 0.0 | GO:0070102 | interleukin-6-mediated signaling pathway(GO:0070102) |
| 0.0 | 0.0 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.0 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.0 | 0.0 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.0 | GO:0035269 | protein mannosylation(GO:0035268) protein O-linked mannosylation(GO:0035269) mannosylation(GO:0097502) |
| 0.0 | 0.0 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.0 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.0 | 0.1 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.0 | 0.1 | GO:0032459 | regulation of protein oligomerization(GO:0032459) |
| 0.0 | 0.1 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.0 | 0.2 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.0 | 0.0 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.2 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 0.1 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.0 | GO:0002689 | negative regulation of leukocyte chemotaxis(GO:0002689) |
| 0.0 | 0.0 | GO:0046877 | regulation of saliva secretion(GO:0046877) |
| 0.0 | 0.1 | GO:0015840 | urea transport(GO:0015840) |
| 0.0 | 0.0 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.0 | GO:1901419 | regulation of response to alcohol(GO:1901419) |
| 0.0 | 0.0 | GO:0045046 | protein import into peroxisome membrane(GO:0045046) |
| 0.0 | 0.1 | GO:0042517 | positive regulation of tyrosine phosphorylation of Stat3 protein(GO:0042517) |
| 0.0 | 0.1 | GO:0031116 | positive regulation of microtubule polymerization(GO:0031116) |
| 0.0 | 0.0 | GO:0046639 | negative regulation of alpha-beta T cell differentiation(GO:0046639) |
| 0.0 | 0.0 | GO:0060124 | positive regulation of growth hormone secretion(GO:0060124) |
| 0.0 | 0.0 | GO:0060676 | ureteric bud formation(GO:0060676) |
| 0.0 | 0.1 | GO:0060710 | chorio-allantoic fusion(GO:0060710) |
| 0.0 | 0.0 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 0.0 | 0.1 | GO:0045070 | positive regulation of viral genome replication(GO:0045070) |
| 0.0 | 0.0 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.0 | 0.0 | GO:0070092 | regulation of glucagon secretion(GO:0070092) |
| 0.0 | 0.0 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.0 | 0.1 | GO:0043392 | negative regulation of DNA binding(GO:0043392) |
| 0.0 | 0.0 | GO:0002855 | natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002420) natural killer cell mediated immune response to tumor cell(GO:0002423) regulation of natural killer cell mediated immune response to tumor cell(GO:0002855) positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002858) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) |
| 0.0 | 0.2 | GO:0042745 | circadian sleep/wake cycle(GO:0042745) |
| 0.0 | 0.1 | GO:0035246 | peptidyl-arginine N-methylation(GO:0035246) |
| 0.0 | 0.0 | GO:0072202 | cell differentiation involved in metanephros development(GO:0072202) |
| 0.0 | 0.0 | GO:0034626 | fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 0.2 | GO:0045104 | intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 | 0.0 | GO:0048289 | isotype switching to IgE isotypes(GO:0048289) regulation of isotype switching to IgE isotypes(GO:0048293) |
| 0.0 | 0.0 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.0 | 0.0 | GO:0090244 | Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 0.0 | 0.0 | GO:0006537 | glutamate biosynthetic process(GO:0006537) |
| 0.0 | 0.1 | GO:0006777 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.0 | 0.1 | GO:0007090 | obsolete regulation of S phase of mitotic cell cycle(GO:0007090) |
| 0.0 | 0.1 | GO:0072111 | cell proliferation involved in kidney development(GO:0072111) |
| 0.0 | 0.0 | GO:0071635 | negative regulation of transforming growth factor beta production(GO:0071635) |
| 0.0 | 0.0 | GO:0043587 | tongue morphogenesis(GO:0043587) |
| 0.0 | 0.0 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.0 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.1 | GO:0003091 | renal water homeostasis(GO:0003091) |
| 0.0 | 0.1 | GO:0044065 | regulation of respiratory system process(GO:0044065) |
| 0.0 | 0.1 | GO:0072283 | mesenchymal to epithelial transition involved in metanephros morphogenesis(GO:0003337) metanephric renal vesicle morphogenesis(GO:0072283) |
| 0.0 | 0.0 | GO:0035581 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.1 | GO:0060396 | growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.0 | 0.1 | GO:0006930 | substrate-dependent cell migration, cell extension(GO:0006930) |
| 0.0 | 0.0 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.0 | 0.1 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.0 | 0.0 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.0 | 0.0 | GO:0046831 | regulation of RNA export from nucleus(GO:0046831) |
| 0.0 | 0.0 | GO:0080111 | DNA demethylation(GO:0080111) |
| 0.0 | 0.0 | GO:0032656 | regulation of interleukin-13 production(GO:0032656) |
| 0.0 | 0.0 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 | 0.0 | GO:0019883 | antigen processing and presentation of endogenous antigen(GO:0019883) |
| 0.0 | 0.0 | GO:0000966 | RNA 5'-end processing(GO:0000966) |
| 0.0 | 0.0 | GO:0051891 | positive regulation of cardioblast differentiation(GO:0051891) positive regulation of cardiocyte differentiation(GO:1905209) |
| 0.0 | 0.1 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 0.1 | GO:0043650 | dicarboxylic acid biosynthetic process(GO:0043650) |
| 0.0 | 0.0 | GO:0045978 | negative regulation of nucleoside metabolic process(GO:0045978) negative regulation of ATP metabolic process(GO:1903579) |
| 0.0 | 0.0 | GO:0090196 | chemokine secretion(GO:0090195) regulation of chemokine secretion(GO:0090196) positive regulation of chemokine secretion(GO:0090197) |
| 0.0 | 0.1 | GO:0032964 | collagen biosynthetic process(GO:0032964) |
| 0.0 | 0.0 | GO:0098801 | regulation of glomerular filtration(GO:0003093) glomerular filtration(GO:0003094) renal filtration(GO:0097205) regulation of renal system process(GO:0098801) |
| 0.0 | 0.0 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 0.0 | GO:0051458 | corticotropin secretion(GO:0051458) regulation of corticotropin secretion(GO:0051459) positive regulation of corticotropin secretion(GO:0051461) |
| 0.0 | 0.0 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.0 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.0 | 0.1 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.0 | GO:0060456 | positive regulation of digestive system process(GO:0060456) |
| 0.0 | 0.0 | GO:0021903 | rostrocaudal neural tube patterning(GO:0021903) |
| 0.0 | 0.0 | GO:0051305 | chromosome movement towards spindle pole(GO:0051305) |
| 0.0 | 0.0 | GO:0031958 | corticosteroid receptor signaling pathway(GO:0031958) |
| 0.0 | 0.0 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.0 | GO:0042996 | regulation of Golgi to plasma membrane protein transport(GO:0042996) |
| 0.0 | 0.0 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.0 | GO:2000189 | regulation of cholesterol homeostasis(GO:2000188) positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.0 | GO:0051590 | positive regulation of neurotransmitter transport(GO:0051590) |
| 0.0 | 0.0 | GO:0046607 | positive regulation of centrosome cycle(GO:0046607) |
| 0.0 | 0.0 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.0 | 0.0 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.0 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) regulation of gastric acid secretion(GO:0060453) |
| 0.0 | 0.0 | GO:0046621 | negative regulation of organ growth(GO:0046621) |
| 0.0 | 0.1 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.0 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 0.0 | GO:0042634 | regulation of hair cycle(GO:0042634) |
| 0.0 | 0.0 | GO:1903019 | negative regulation of glycoprotein metabolic process(GO:1903019) |
| 0.0 | 0.0 | GO:0016246 | RNA interference(GO:0016246) |
| 0.0 | 0.1 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.3 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.1 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.1 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) |
| 0.0 | 0.1 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 | 0.0 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.0 | GO:0009648 | photoperiodism(GO:0009648) |
| 0.0 | 0.0 | GO:0070669 | response to interleukin-2(GO:0070669) |
| 0.0 | 0.0 | GO:0032455 | nerve growth factor processing(GO:0032455) |
| 0.0 | 0.0 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) regulation of spindle organization(GO:0090224) |
| 0.0 | 0.0 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 | 0.0 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.0 | 0.1 | GO:0045008 | depyrimidination(GO:0045008) |
| 0.0 | 0.0 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.1 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.0 | GO:0042976 | activation of Janus kinase activity(GO:0042976) |
| 0.0 | 0.1 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.0 | GO:0070828 | heterochromatin assembly(GO:0031507) heterochromatin organization(GO:0070828) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.1 | 0.2 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.1 | 0.2 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 0.2 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.1 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 0.0 | 0.2 | GO:0005954 | calcium- and calmodulin-dependent protein kinase complex(GO:0005954) |
| 0.0 | 0.1 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.2 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.4 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.3 | GO:0030128 | AP-2 adaptor complex(GO:0030122) clathrin coat of endocytic vesicle(GO:0030128) |
| 0.0 | 0.1 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.3 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.1 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.1 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.1 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.1 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.0 | 0.1 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.0 | 0.2 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.0 | 0.1 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.3 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.2 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.1 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.1 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.0 | 0.0 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.1 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.2 | GO:0030677 | nucleolar ribonuclease P complex(GO:0005655) ribonuclease P complex(GO:0030677) multimeric ribonuclease P complex(GO:0030681) |
| 0.0 | 0.1 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.2 | GO:0045261 | proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 0.1 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.1 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.0 | 0.2 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.1 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.1 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.2 | GO:0032156 | septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.1 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.1 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.1 | GO:0042598 | obsolete vesicular fraction(GO:0042598) |
| 0.0 | 0.0 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.0 | 0.0 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.1 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.0 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.1 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.1 | GO:0005606 | laminin-1 complex(GO:0005606) |
| 0.0 | 0.3 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.6 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.2 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.1 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 0.2 | GO:0045120 | pronucleus(GO:0045120) |
| 0.0 | 0.0 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.1 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.1 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.4 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.0 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.1 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.1 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.3 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.0 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.1 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.1 | GO:0098645 | network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.0 | 0.0 | GO:0098651 | basement membrane collagen trimer(GO:0098651) |
| 0.0 | 0.0 | GO:0033185 | mannosyltransferase complex(GO:0031501) dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 0.1 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.0 | GO:0032449 | CBM complex(GO:0032449) |
| 0.0 | 0.1 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.0 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 0.1 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.1 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.1 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.1 | 0.4 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.1 | 0.6 | GO:0034593 | phosphatidylinositol bisphosphate phosphatase activity(GO:0034593) |
| 0.1 | 0.2 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.1 | 0.2 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.1 | 0.5 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.2 | GO:0005072 | transforming growth factor beta receptor, cytoplasmic mediator activity(GO:0005072) |
| 0.0 | 0.3 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.3 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.1 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.0 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.1 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.2 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.0 | 0.3 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.2 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.1 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.1 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.1 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.2 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.1 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.1 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.0 | 0.1 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.2 | GO:0034416 | bisphosphoglycerate phosphatase activity(GO:0034416) |
| 0.0 | 0.1 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.0 | 0.1 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.1 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.1 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.1 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.0 | 0.1 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.3 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.3 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.1 | GO:0032558 | purine deoxyribonucleotide binding(GO:0032554) adenyl deoxyribonucleotide binding(GO:0032558) |
| 0.0 | 0.1 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.2 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.0 | GO:0005415 | nucleoside:sodium symporter activity(GO:0005415) |
| 0.0 | 0.1 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.1 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.1 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.0 | 0.1 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.5 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.0 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.1 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.1 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.1 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.0 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.2 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.1 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.0 | 0.1 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.1 | GO:0050698 | proteoglycan sulfotransferase activity(GO:0050698) |
| 0.0 | 0.2 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.3 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.9 | GO:0001948 | glycoprotein binding(GO:0001948) |
| 0.0 | 0.1 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) |
| 0.0 | 0.1 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
| 0.0 | 0.1 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.3 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.2 | GO:0016668 | oxidoreductase activity, acting on a sulfur group of donors, NAD(P) as acceptor(GO:0016668) |
| 0.0 | 0.1 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.0 | 0.0 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.1 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.0 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.1 | GO:0016149 | translation release factor activity, codon specific(GO:0016149) |
| 0.0 | 0.3 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.1 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.1 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.2 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.1 | GO:0055106 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.1 | GO:0016595 | glutamate binding(GO:0016595) |
| 0.0 | 0.4 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.0 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.1 | GO:0008967 | phosphoglycolate phosphatase activity(GO:0008967) |
| 0.0 | 0.1 | GO:0015211 | purine nucleoside transmembrane transporter activity(GO:0015211) |
| 0.0 | 0.0 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.1 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.0 | 0.1 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.0 | 0.1 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.1 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.1 | GO:0035514 | DNA demethylase activity(GO:0035514) DNA-N1-methyladenine dioxygenase activity(GO:0043734) |
| 0.0 | 0.1 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.0 | 0.1 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.0 | 0.1 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.1 | GO:0015166 | polyol transmembrane transporter activity(GO:0015166) |
| 0.0 | 0.1 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.0 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.0 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.0 | 0.0 | GO:0019958 | C-X-C chemokine binding(GO:0019958) interleukin-8 binding(GO:0019959) |
| 0.0 | 0.0 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.1 | GO:0000036 | ACP phosphopantetheine attachment site binding involved in fatty acid biosynthetic process(GO:0000036) ACP phosphopantetheine attachment site binding(GO:0044620) prosthetic group binding(GO:0051192) |
| 0.0 | 0.1 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.0 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.0 | 0.0 | GO:0042910 | xenobiotic-transporting ATPase activity(GO:0008559) xenobiotic transporter activity(GO:0042910) |
| 0.0 | 0.2 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.2 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.0 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.1 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.1 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.0 | 0.1 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.0 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.0 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 0.1 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.3 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.1 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.0 | 0.2 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.1 | GO:0016274 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.0 | 0.1 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.0 | 0.1 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.1 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.1 | GO:0097642 | calcitonin family receptor activity(GO:0097642) |
| 0.0 | 0.1 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.0 | 0.1 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.0 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.0 | 0.2 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 0.1 | GO:0016847 | 1-aminocyclopropane-1-carboxylate synthase activity(GO:0016847) |
| 0.0 | 0.2 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 0.0 | 0.3 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.2 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.2 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.0 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.1 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.0 | 0.0 | GO:0004844 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.0 | 0.2 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 0.0 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.0 | 0.0 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.0 | 0.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.0 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.0 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.0 | 0.0 | GO:0030151 | molybdenum ion binding(GO:0030151) |
| 0.0 | 0.1 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.2 | GO:0016411 | acylglycerol O-acyltransferase activity(GO:0016411) |
| 0.0 | 0.0 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.2 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.1 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 0.1 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.0 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.0 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.0 | GO:1990939 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) ATP-dependent microtubule motor activity(GO:1990939) |
| 0.0 | 0.1 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.0 | GO:0015925 | galactosidase activity(GO:0015925) |
| 0.0 | 0.0 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.0 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.0 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.1 | GO:0048531 | beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.0 | 0.1 | GO:0090079 | translation activator activity(GO:0008494) translation regulator activity, nucleic acid binding(GO:0090079) |
| 0.0 | 0.0 | GO:0008486 | diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) |
| 0.0 | 0.0 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.1 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.1 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.0 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.4 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.0 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.6 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.6 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.0 | 0.5 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.1 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.0 | 0.2 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.1 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 0.5 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.0 | 0.6 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.2 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 0.0 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 0.9 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 0.2 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 0.2 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.3 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 0.8 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.0 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 0.1 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.0 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 0.6 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.1 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.0 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.1 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 0.2 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.1 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.4 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 0.5 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.6 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.3 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 0.3 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.0 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.0 | 0.1 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.4 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.1 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.2 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.1 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.5 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.7 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.3 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.2 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.1 | REACTOME P53 INDEPENDENT G1 S DNA DAMAGE CHECKPOINT | Genes involved in p53-Independent G1/S DNA damage checkpoint |
| 0.0 | 0.1 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.3 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.3 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.5 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.2 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.5 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.3 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.3 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.2 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.0 | 0.0 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.1 | REACTOME TRANSMISSION ACROSS CHEMICAL SYNAPSES | Genes involved in Transmission across Chemical Synapses |
| 0.0 | 0.5 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.2 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.2 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.2 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.2 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.5 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.3 | REACTOME COSTIMULATION BY THE CD28 FAMILY | Genes involved in Costimulation by the CD28 family |
| 0.0 | 0.2 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.1 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.0 | REACTOME CIRCADIAN CLOCK | Genes involved in Circadian Clock |
| 0.0 | 0.2 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.0 | 0.2 | REACTOME TRANSCRIPTIONAL ACTIVITY OF SMAD2 SMAD3 SMAD4 HETEROTRIMER | Genes involved in Transcriptional activity of SMAD2/SMAD3:SMAD4 heterotrimer |
| 0.0 | 0.1 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.2 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 0.1 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.2 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 0.0 | 0.1 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.2 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.2 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 0.3 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.0 | REACTOME PLATELET HOMEOSTASIS | Genes involved in Platelet homeostasis |
| 0.0 | 0.1 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.1 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.4 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.1 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.1 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.1 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.2 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.2 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.1 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.1 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.0 | REACTOME APC C CDC20 MEDIATED DEGRADATION OF MITOTIC PROTEINS | Genes involved in APC/C:Cdc20 mediated degradation of mitotic proteins |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.1 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.1 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.0 | 0.1 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |