Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
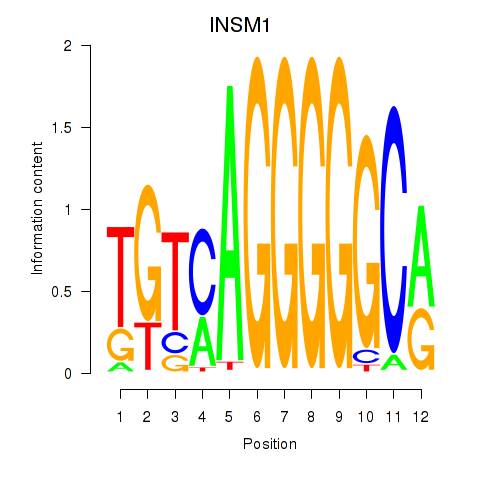
Results for INSM1
Z-value: 0.67
Transcription factors associated with INSM1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
INSM1
|
ENSG00000173404.3 | INSM transcriptional repressor 1 |
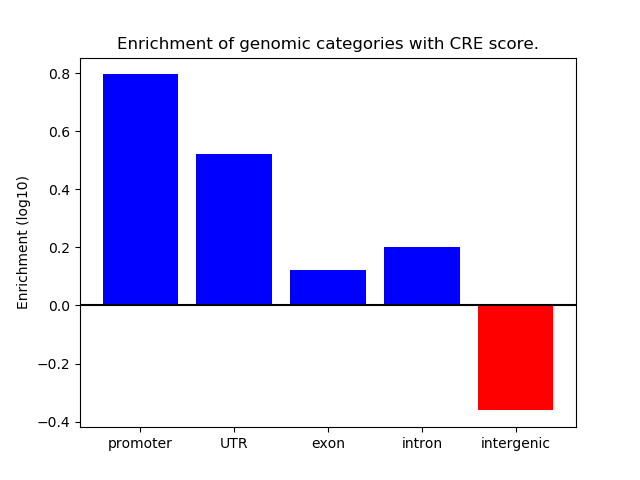
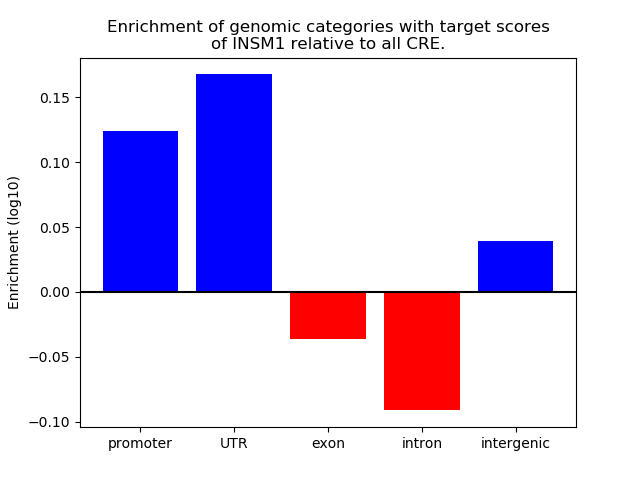
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr20_20346110_20346261 | INSM1 | 2580 | 0.343898 | -0.73 | 2.6e-02 | Click! |
| chr20_20343692_20343843 | INSM1 | 4998 | 0.266357 | -0.62 | 7.7e-02 | Click! |
| chr20_20341078_20341370 | INSM1 | 7541 | 0.244572 | -0.48 | 1.9e-01 | Click! |
| chr20_20356949_20357100 | INSM1 | 8259 | 0.255134 | 0.45 | 2.3e-01 | Click! |
| chr20_20374169_20374320 | INSM1 | 25479 | 0.218069 | 0.41 | 2.7e-01 | Click! |
Activity of the INSM1 motif across conditions
Conditions sorted by the z-value of the INSM1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
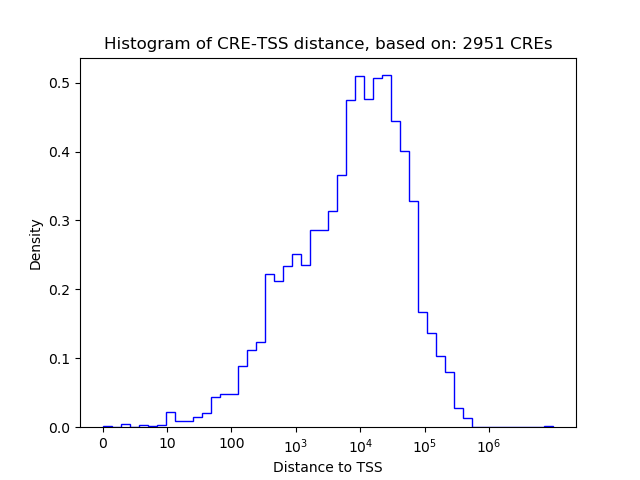
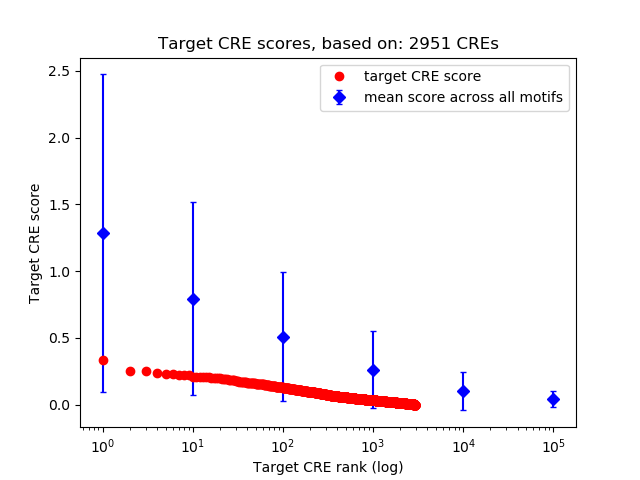
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr1_94454656_94454830 | 0.34 |
ABCA4 |
ATP-binding cassette, sub-family A (ABC1), member 4 |
22438 |
0.21 |
| chr7_1494490_1494836 | 0.26 |
MICALL2 |
MICAL-like 2 |
4299 |
0.18 |
| chr19_1839973_1840303 | 0.25 |
REXO1 |
REX1, RNA exonuclease 1 homolog (S. cerevisiae) |
8314 |
0.08 |
| chr14_103577690_103577872 | 0.24 |
EXOC3L4 |
exocyst complex component 3-like 4 |
3928 |
0.19 |
| chr8_142170805_142171062 | 0.23 |
DENND3 |
DENN/MADD domain containing 3 |
17287 |
0.18 |
| chr9_129110024_129110380 | 0.23 |
MVB12B |
multivesicular body subunit 12B |
12676 |
0.24 |
| chr10_5631913_5632190 | 0.23 |
ENSG00000240577 |
. |
41375 |
0.11 |
| chr17_27443100_27443626 | 0.23 |
MYO18A |
myosin XVIIIA |
24073 |
0.12 |
| chr6_126268678_126268829 | 0.22 |
HINT3 |
histidine triad nucleotide binding protein 3 |
9174 |
0.18 |
| chr3_194192573_194192803 | 0.21 |
ATP13A3 |
ATPase type 13A3 |
3720 |
0.19 |
| chr8_22531153_22531332 | 0.21 |
BIN3 |
bridging integrator 3 |
4581 |
0.14 |
| chr11_65340562_65341138 | 0.21 |
FAM89B |
family with sequence similarity 89, member B |
851 |
0.32 |
| chr20_19256096_19256257 | 0.21 |
RP5-1027G4.3 |
|
9064 |
0.23 |
| chr11_728177_728476 | 0.21 |
AP006621.9 |
|
1279 |
0.23 |
| chr2_10417901_10418493 | 0.20 |
ENSG00000264030 |
. |
2560 |
0.25 |
| chr10_80841245_80841683 | 0.20 |
ZMIZ1 |
zinc finger, MIZ-type containing 1 |
12672 |
0.24 |
| chr20_20299369_20299542 | 0.20 |
ENSG00000264879 |
. |
1771 |
0.4 |
| chr12_53663071_53663222 | 0.20 |
ESPL1 |
extra spindle pole bodies homolog 1 (S. cerevisiae) |
1027 |
0.36 |
| chr19_18887031_18887461 | 0.20 |
COMP |
cartilage oligomeric matrix protein |
14868 |
0.14 |
| chr12_64846967_64847658 | 0.20 |
TBK1 |
TANK-binding kinase 1 |
1057 |
0.53 |
| chr2_210337378_210337560 | 0.20 |
MAP2 |
microtubule-associated protein 2 |
48687 |
0.19 |
| chr8_145158856_145159007 | 0.19 |
SHARPIN |
SHANK-associated RH domain interactor |
162 |
0.75 |
| chr12_26304340_26304503 | 0.19 |
SSPN |
sarcospan |
16521 |
0.18 |
| chr1_226005603_226005811 | 0.19 |
EPHX1 |
epoxide hydrolase 1, microsomal (xenobiotic) |
7348 |
0.15 |
| chr1_234755000_234755251 | 0.19 |
IRF2BP2 |
interferon regulatory factor 2 binding protein 2 |
9854 |
0.19 |
| chrX_69674794_69674965 | 0.19 |
DLG3 |
discs, large homolog 3 (Drosophila) |
67 |
0.93 |
| chr8_62634134_62634577 | 0.18 |
ENSG00000264408 |
. |
7008 |
0.24 |
| chr6_144017282_144017541 | 0.18 |
PHACTR2 |
phosphatase and actin regulator 2 |
18206 |
0.25 |
| chr12_9519142_9519293 | 0.18 |
ENSG00000212432 |
. |
78437 |
0.08 |
| chr2_103103798_103104021 | 0.18 |
SLC9A4 |
solute carrier family 9, subfamily A (NHE4, cation proton antiporter 4), member 4 |
14147 |
0.18 |
| chr10_21811725_21811876 | 0.18 |
SKIDA1 |
SKI/DACH domain containing 1 |
2811 |
0.19 |
| chr19_19078219_19078600 | 0.17 |
ENSG00000241172 |
. |
9342 |
0.11 |
| chr7_48281682_48281833 | 0.17 |
ABCA13 |
ATP-binding cassette, sub-family A (ABC1), member 13 |
70700 |
0.13 |
| chr14_92956348_92956499 | 0.17 |
RIN3 |
Ras and Rab interactor 3 |
23695 |
0.23 |
| chr5_76361781_76361938 | 0.17 |
CTC-564N23.2 |
|
7847 |
0.14 |
| chr17_17109981_17110277 | 0.17 |
PLD6 |
phospholipase D family, member 6 |
500 |
0.69 |
| chr1_20926423_20926574 | 0.17 |
CDA |
cytidine deaminase |
11057 |
0.17 |
| chr6_44140870_44141021 | 0.17 |
CAPN11 |
calpain 11 |
14397 |
0.14 |
| chr14_24041269_24041503 | 0.17 |
JPH4 |
junctophilin 4 |
824 |
0.44 |
| chr15_75639915_75640464 | 0.17 |
NEIL1 |
nei endonuclease VIII-like 1 (E. coli) |
12 |
0.96 |
| chr2_20383859_20384010 | 0.16 |
ENSG00000200763 |
. |
8368 |
0.17 |
| chr1_157988129_157988280 | 0.16 |
KIRREL-IT1 |
KIRREL intronic transcript 1 (non-protein coding) |
7136 |
0.23 |
| chr4_144259118_144259477 | 0.16 |
GAB1 |
GRB2-associated binding protein 1 |
966 |
0.57 |
| chr11_12309183_12309334 | 0.16 |
MICALCL |
MICAL C-terminal like |
811 |
0.62 |
| chr17_41045106_41045352 | 0.16 |
ENSG00000252039 |
. |
1777 |
0.18 |
| chr2_60563310_60563461 | 0.16 |
ENSG00000200807 |
. |
48355 |
0.16 |
| chr5_176965938_176966089 | 0.16 |
FAM193B |
family with sequence similarity 193, member B |
339 |
0.8 |
| chr3_58036411_58036562 | 0.16 |
FLNB |
filamin B, beta |
27570 |
0.22 |
| chr1_33804015_33804326 | 0.16 |
ENSG00000222112 |
. |
1705 |
0.24 |
| chr12_52557959_52558196 | 0.16 |
KRT80 |
keratin 80 |
27707 |
0.1 |
| chr7_27193246_27193519 | 0.16 |
HOXA7 |
homeobox A7 |
2916 |
0.08 |
| chr15_70667753_70667904 | 0.16 |
ENSG00000200216 |
. |
182253 |
0.03 |
| chr4_48333635_48333967 | 0.16 |
SLAIN2 |
SLAIN motif family, member 2 |
9538 |
0.25 |
| chr19_2188884_2189035 | 0.15 |
DOT1L |
DOT1-like histone H3K79 methyltransferase |
24630 |
0.08 |
| chr20_3824293_3824586 | 0.15 |
MAVS |
mitochondrial antiviral signaling protein |
3048 |
0.18 |
| chr15_67127155_67127306 | 0.15 |
SMAD6 |
SMAD family member 6 |
123195 |
0.05 |
| chr2_172619845_172620139 | 0.15 |
AC068039.4 |
|
10000 |
0.22 |
| chr10_62214190_62214341 | 0.15 |
ANK3 |
ankyrin 3, node of Ranvier (ankyrin G) |
64777 |
0.15 |
| chr2_85895490_85895689 | 0.15 |
SFTPB |
surfactant protein B |
263 |
0.87 |
| chr8_10567948_10568099 | 0.15 |
CTD-2135J3.3 |
|
18801 |
0.12 |
| chr2_27211731_27211882 | 0.15 |
MAPRE3 |
microtubule-associated protein, RP/EB family, member 3 |
18280 |
0.1 |
| chr7_151387438_151387589 | 0.15 |
PRKAG2 |
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
45836 |
0.15 |
| chr3_128566375_128566773 | 0.15 |
RP11-723O4.2 |
|
17873 |
0.16 |
| chr3_14434671_14434822 | 0.15 |
ENSG00000199609 |
. |
1402 |
0.47 |
| chr9_131908056_131908207 | 0.15 |
PPP2R4 |
protein phosphatase 2A activator, regulatory subunit 4 |
3827 |
0.14 |
| chr2_40330130_40330281 | 0.15 |
SLC8A1-AS1 |
SLC8A1 antisense RNA 1 |
1844 |
0.51 |
| chr1_179560813_179561046 | 0.15 |
TDRD5 |
tudor domain containing 5 |
96 |
0.94 |
| chr3_124882921_124883080 | 0.15 |
ENSG00000264986 |
. |
12604 |
0.19 |
| chr5_148422708_148422859 | 0.14 |
SH3TC2 |
SH3 domain and tetratricopeptide repeats 2 |
1674 |
0.33 |
| chr12_93623599_93623820 | 0.14 |
RP11-511B23.2 |
|
14254 |
0.18 |
| chr13_114505170_114505431 | 0.14 |
GAS6-AS1 |
GAS6 antisense RNA 1 |
13303 |
0.2 |
| chr5_127915340_127915491 | 0.14 |
FBN2 |
fibrillin 2 |
41499 |
0.17 |
| chr5_14267266_14267567 | 0.14 |
TRIO |
trio Rho guanine nucleotide exchange factor |
23670 |
0.28 |
| chr9_131960663_131960907 | 0.14 |
RP11-247A12.2 |
|
2809 |
0.19 |
| chr20_31459694_31459849 | 0.14 |
EFCAB8 |
EF-hand calcium binding domain 8 |
13042 |
0.17 |
| chr1_236096877_236097091 | 0.14 |
ENSG00000206803 |
. |
18269 |
0.19 |
| chr13_33728189_33728340 | 0.14 |
STARD13-IT1 |
STARD13 intronic transcript 1 (non-protein coding) |
10282 |
0.23 |
| chr20_33945750_33945901 | 0.14 |
UQCC1 |
ubiquinol-cytochrome c reductase complex assembly factor 1 |
36149 |
0.1 |
| chr19_17202344_17202555 | 0.14 |
CTD-2528A14.5 |
|
7668 |
0.12 |
| chr17_29891815_29891977 | 0.14 |
ENSG00000221038 |
. |
983 |
0.39 |
| chr15_101789389_101789613 | 0.14 |
CHSY1 |
chondroitin sulfate synthase 1 |
2636 |
0.25 |
| chr4_41259506_41259822 | 0.14 |
UCHL1 |
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase) |
702 |
0.5 |
| chr5_88573153_88573304 | 0.14 |
MEF2C-AS1 |
MEF2C antisense RNA 1 |
141396 |
0.05 |
| chr10_12109223_12109383 | 0.14 |
DHTKD1 |
dehydrogenase E1 and transketolase domain containing 1 |
1668 |
0.33 |
| chr8_68328443_68328594 | 0.14 |
ARFGEF1 |
ADP-ribosylation factor guanine nucleotide-exchange factor 1 (brefeldin A-inhibited) |
72606 |
0.12 |
| chr6_11590304_11590455 | 0.14 |
TMEM170B |
transmembrane protein 170B |
51868 |
0.15 |
| chr12_49653889_49654155 | 0.13 |
RP11-977B10.2 |
|
4517 |
0.13 |
| chr22_39361142_39361293 | 0.13 |
APOBEC3A |
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3A |
7690 |
0.13 |
| chr16_75278250_75278858 | 0.13 |
BCAR1 |
breast cancer anti-estrogen resistance 1 |
1954 |
0.25 |
| chr8_145059566_145060066 | 0.13 |
PARP10 |
poly (ADP-ribose) polymerase family, member 10 |
782 |
0.43 |
| chr20_62169494_62169653 | 0.13 |
PTK6 |
protein tyrosine kinase 6 |
850 |
0.41 |
| chr18_43381728_43382214 | 0.13 |
SIGLEC15 |
sialic acid binding Ig-like lectin 15 |
23506 |
0.18 |
| chrX_135296533_135296684 | 0.13 |
FHL1 |
four and a half LIM domains 1 |
9728 |
0.22 |
| chr6_33806361_33806522 | 0.13 |
MLN |
motilin |
34653 |
0.15 |
| chr17_9885338_9885489 | 0.13 |
GAS7 |
growth arrest-specific 7 |
22634 |
0.21 |
| chr10_78156869_78157020 | 0.13 |
RP11-369F10.2 |
|
41589 |
0.18 |
| chr8_6786360_6786511 | 0.13 |
GS1-24F4.3 |
|
305 |
0.86 |
| chr1_244389388_244389539 | 0.13 |
ENSG00000244066 |
. |
122229 |
0.05 |
| chr2_238162134_238162403 | 0.13 |
AC112715.2 |
Uncharacterized protein |
3466 |
0.34 |
| chr2_183780593_183780744 | 0.13 |
ENSG00000207178 |
. |
42099 |
0.14 |
| chr13_30479772_30480203 | 0.13 |
LINC00572 |
long intergenic non-protein coding RNA 572 |
20801 |
0.25 |
| chr14_71108052_71108203 | 0.13 |
TTC9 |
tetratricopeptide repeat domain 9 |
377 |
0.88 |
| chr6_26481383_26481534 | 0.13 |
BTN2A1 |
butyrophilin, subfamily 2, member A1 |
16003 |
0.11 |
| chr19_42098371_42098522 | 0.13 |
CEACAMP3 |
carcinoembryonic antigen-related cell adhesion molecule pseudogene 3 |
7644 |
0.15 |
| chr21_39201745_39201896 | 0.13 |
KCNJ6 |
potassium inwardly-rectifying channel, subfamily J, member 6 |
83737 |
0.09 |
| chr1_36329942_36330109 | 0.13 |
AGO1 |
argonaute RISC catalytic component 1 |
5384 |
0.22 |
| chr1_26131534_26131685 | 0.13 |
SEPN1 |
selenoprotein N, 1 |
4925 |
0.12 |
| chr22_28114114_28114265 | 0.12 |
RP11-375H17.1 |
|
1721 |
0.5 |
| chr5_138310904_138311077 | 0.12 |
ENSG00000201532 |
. |
37028 |
0.13 |
| chr9_137334839_137335219 | 0.12 |
RXRA |
retinoid X receptor, alpha |
36601 |
0.19 |
| chr19_13041127_13041278 | 0.12 |
CTC-425F1.2 |
|
134 |
0.86 |
| chr8_8940618_8940769 | 0.12 |
ENSG00000239078 |
. |
10667 |
0.16 |
| chr16_55530572_55531230 | 0.12 |
MMP2 |
matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase) |
8327 |
0.22 |
| chr20_42161533_42161684 | 0.12 |
L3MBTL1 |
l(3)mbt-like 1 (Drosophila) |
4261 |
0.18 |
| chr19_42464027_42464203 | 0.12 |
RABAC1 |
Rab acceptor 1 (prenylated) |
573 |
0.68 |
| chr10_7899320_7899471 | 0.12 |
TAF3 |
TAF3 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 140kDa |
38928 |
0.17 |
| chr8_37683766_37684149 | 0.12 |
BRF2 |
BRF2, RNA polymerase III transcription initiation factor 50 kDa subunit |
23399 |
0.13 |
| chr1_11218024_11218175 | 0.12 |
ENSG00000253086 |
. |
5692 |
0.12 |
| chr22_30429782_30429933 | 0.12 |
HORMAD2 |
HORMA domain containing 2 |
46306 |
0.11 |
| chr6_88632888_88633039 | 0.12 |
ENSG00000211983 |
. |
75613 |
0.09 |
| chr16_88865957_88866184 | 0.12 |
CDT1 |
chromatin licensing and DNA replication factor 1 |
3551 |
0.12 |
| chr14_103488952_103489103 | 0.12 |
CDC42BPB |
CDC42 binding protein kinase beta (DMPK-like) |
34772 |
0.14 |
| chr16_8972682_8972833 | 0.12 |
RP11-77H9.6 |
|
9649 |
0.13 |
| chr7_36343566_36343768 | 0.12 |
EEPD1 |
endonuclease/exonuclease/phosphatase family domain containing 1 |
6914 |
0.18 |
| chr12_52579651_52579802 | 0.12 |
KRT80 |
keratin 80 |
6058 |
0.14 |
| chr2_9135145_9135439 | 0.12 |
MBOAT2 |
membrane bound O-acyltransferase domain containing 2 |
8502 |
0.24 |
| chr8_108506772_108506989 | 0.12 |
ANGPT1 |
angiopoietin 1 |
343 |
0.94 |
| chr3_45070851_45071181 | 0.12 |
CLEC3B |
C-type lectin domain family 3, member B |
606 |
0.71 |
| chr19_49945353_49945990 | 0.12 |
SLC17A7 |
solute carrier family 17 (vesicular glutamate transporter), member 7 |
54 |
0.92 |
| chr3_24409668_24409819 | 0.12 |
THRB-IT1 |
THRB intronic transcript 1 (non-protein coding) |
91182 |
0.09 |
| chr17_7649362_7649654 | 0.11 |
DNAH2 |
dynein, axonemal, heavy chain 2 |
26469 |
0.09 |
| chr12_6981803_6982102 | 0.11 |
SPSB2 |
splA/ryanodine receptor domain and SOCS box containing 2 |
497 |
0.44 |
| chr16_21274927_21275078 | 0.11 |
CRYM |
crystallin, mu |
1523 |
0.38 |
| chr9_139524753_139524904 | 0.11 |
ENSG00000252440 |
. |
27861 |
0.08 |
| chr2_28021717_28021927 | 0.11 |
AC110084.1 |
|
12636 |
0.16 |
| chr19_1257974_1258635 | 0.11 |
CIRBP |
cold inducible RNA binding protein |
1080 |
0.29 |
| chr6_73624153_73624304 | 0.11 |
ENSG00000266180 |
. |
53248 |
0.15 |
| chr3_151121717_151121896 | 0.11 |
P2RY12 |
purinergic receptor P2Y, G-protein coupled, 12 |
19206 |
0.19 |
| chr12_52810052_52810346 | 0.11 |
RP11-1020M18.10 |
|
8835 |
0.09 |
| chr19_52130648_52130842 | 0.11 |
SIGLEC5 |
sialic acid binding Ig-like lectin 5 |
2843 |
0.16 |
| chr1_150760534_150760685 | 0.11 |
CTSK |
cathepsin K |
19301 |
0.12 |
| chr22_46625810_46625961 | 0.11 |
CDPF1 |
cysteine-rich, DPF motif domain containing 1 |
18297 |
0.13 |
| chr8_37556598_37557288 | 0.11 |
ZNF703 |
zinc finger protein 703 |
3674 |
0.16 |
| chr1_59058556_59058707 | 0.11 |
TACSTD2 |
tumor-associated calcium signal transducer 2 |
15465 |
0.2 |
| chr12_95751456_95751607 | 0.11 |
RP11-167N24.6 |
Uncharacterized protein |
23141 |
0.15 |
| chrX_17432841_17432992 | 0.11 |
ENSG00000265465 |
. |
11088 |
0.22 |
| chr3_196354806_196354957 | 0.11 |
LINC01063 |
long intergenic non-protein coding RNA 1063 |
4577 |
0.15 |
| chr5_95070855_95071006 | 0.11 |
CTD-2154I11.2 |
|
2931 |
0.22 |
| chr1_38464859_38465107 | 0.11 |
FHL3 |
four and a half LIM domains 3 |
6194 |
0.13 |
| chr12_26430442_26430909 | 0.11 |
RP11-283G6.5 |
|
5822 |
0.22 |
| chr1_85691399_85691600 | 0.11 |
SYDE2 |
synapse defective 1, Rho GTPase, homolog 2 (C. elegans) |
24770 |
0.13 |
| chr9_127574234_127574385 | 0.11 |
OLFML2A |
olfactomedin-like 2A |
11156 |
0.14 |
| chr2_10159539_10159690 | 0.11 |
AC010969.1 |
|
13456 |
0.13 |
| chr1_36808619_36809165 | 0.11 |
STK40 |
serine/threonine kinase 40 |
18049 |
0.12 |
| chr7_75902267_75902418 | 0.11 |
SRRM3 |
serine/arginine repetitive matrix 3 |
9590 |
0.16 |
| chr14_93595134_93595285 | 0.11 |
ITPK1 |
inositol-tetrakisphosphate 1-kinase |
12544 |
0.17 |
| chr2_111228653_111228894 | 0.11 |
LIMS3 |
LIM and senescent cell antigen-like-containing domain protein 3; Uncharacterized protein; cDNA FLJ59124, highly similar to Particularly interesting newCys-His protein; cDNA, FLJ79109, highly similar to Particularly interesting newCys-His protein |
1620 |
0.3 |
| chr1_63789848_63790019 | 0.11 |
RP4-792G4.2 |
|
179 |
0.91 |
| chr10_104434298_104434771 | 0.11 |
TRIM8 |
tripartite motif containing 8 |
29890 |
0.12 |
| chr3_14424279_14424430 | 0.11 |
ENSG00000199609 |
. |
11794 |
0.22 |
| chr19_18484370_18484862 | 0.11 |
GDF15 |
growth differentiation factor 15 |
925 |
0.37 |
| chr17_38263559_38264452 | 0.11 |
NR1D1 |
nuclear receptor subfamily 1, group D, member 1 |
7027 |
0.12 |
| chr10_134389363_134389791 | 0.11 |
INPP5A |
inositol polyphosphate-5-phosphatase, 40kDa |
31853 |
0.19 |
| chr2_238783487_238783638 | 0.11 |
ENSG00000263723 |
. |
5015 |
0.22 |
| chr2_109272981_109273149 | 0.10 |
LIMS1 |
LIM and senescent cell antigen-like domains 1 |
1556 |
0.43 |
| chr11_46274872_46275023 | 0.10 |
CTD-2589M5.4 |
|
21151 |
0.17 |
| chr9_124045225_124045376 | 0.10 |
RP11-477J21.6 |
|
292 |
0.86 |
| chr2_106026820_106026993 | 0.10 |
FHL2 |
four and a half LIM domains 2 |
10782 |
0.21 |
| chr2_7150943_7151094 | 0.10 |
RNF144A |
ring finger protein 144A |
13947 |
0.24 |
| chr14_102800072_102800223 | 0.10 |
ZNF839 |
zinc finger protein 839 |
1878 |
0.25 |
| chr2_65378861_65379061 | 0.10 |
ENSG00000272025 |
. |
6835 |
0.21 |
| chr4_38671735_38672263 | 0.10 |
RP11-617D20.1 |
|
5495 |
0.18 |
| chr15_23115235_23115631 | 0.10 |
RP11-566K19.6 |
|
1172 |
0.41 |
| chr17_71301300_71301515 | 0.10 |
CDC42EP4 |
CDC42 effector protein (Rho GTPase binding) 4 |
6140 |
0.2 |
| chr5_34001183_34001334 | 0.10 |
AMACR |
alpha-methylacyl-CoA racemase |
6875 |
0.2 |
| chr13_110698625_110698925 | 0.10 |
ENSG00000265885 |
. |
51723 |
0.17 |
| chr2_113597075_113597265 | 0.10 |
IL1B |
interleukin 1, beta |
2690 |
0.24 |
| chr20_30157045_30157206 | 0.10 |
HM13-AS1 |
HM13 antisense RNA 1 |
3941 |
0.14 |
| chr22_46402087_46402238 | 0.10 |
WNT7B |
wingless-type MMTV integration site family, member 7B |
29153 |
0.1 |
| chr2_70743576_70743870 | 0.10 |
TGFA |
transforming growth factor, alpha |
36899 |
0.14 |
| chr1_172416754_172417009 | 0.10 |
C1orf105 |
chromosome 1 open reading frame 105 |
2602 |
0.26 |
| chr17_61742210_61742415 | 0.10 |
LIMD2 |
LIM domain containing 2 |
34210 |
0.11 |
| chr5_158370729_158370880 | 0.10 |
CTD-2363C16.1 |
|
39210 |
0.18 |
| chr9_72861839_72861990 | 0.10 |
SMC5-AS1 |
SMC5 antisense RNA 1 (head to head) |
11868 |
0.18 |
| chr2_223940929_223941080 | 0.10 |
KCNE4 |
potassium voltage-gated channel, Isk-related family, member 4 |
24142 |
0.25 |
| chr12_124726878_124727199 | 0.10 |
FAM101A |
family with sequence similarity 101, member A |
46672 |
0.16 |
| chr14_66170868_66171019 | 0.10 |
FUT8 |
fucosyltransferase 8 (alpha (1,6) fucosyltransferase) |
34810 |
0.24 |
| chr16_88992294_88992445 | 0.10 |
RP11-830F9.7 |
|
11795 |
0.12 |
| chr1_213132499_213132942 | 0.10 |
VASH2 |
vasohibin 2 |
7835 |
0.23 |
| chr8_142085059_142085210 | 0.10 |
DENND3 |
DENN/MADD domain containing 3 |
42243 |
0.14 |
| chr19_11372465_11372856 | 0.10 |
DOCK6 |
dedicator of cytokinesis 6 |
468 |
0.69 |
| chr20_48780646_48780797 | 0.10 |
TMEM189 |
transmembrane protein 189 |
10386 |
0.17 |
| chr2_174889480_174890119 | 0.10 |
SP3 |
Sp3 transcription factor |
59369 |
0.15 |
| chr4_13959861_13960059 | 0.10 |
ENSG00000252092 |
. |
299709 |
0.01 |
| chr1_169452783_169453112 | 0.10 |
SLC19A2 |
solute carrier family 19 (thiamine transporter), member 2 |
2222 |
0.33 |
| chr10_90791613_90792074 | 0.10 |
RP11-399O19.9 |
|
16250 |
0.14 |
| chr12_49689441_49689702 | 0.10 |
PRPH |
peripherin |
426 |
0.72 |
| chr2_7134253_7134437 | 0.10 |
RNF144A |
ring finger protein 144A |
2726 |
0.34 |
| chr20_52342252_52342403 | 0.10 |
ENSG00000238468 |
. |
57030 |
0.14 |
| chrX_109652929_109653080 | 0.10 |
RGAG1 |
retrotransposon gag domain containing 1 |
9281 |
0.23 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0007028 | cytoplasm organization(GO:0007028) |
| 0.0 | 0.2 | GO:0071731 | response to nitric oxide(GO:0071731) |
| 0.0 | 0.1 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.1 | GO:0005997 | xylulose metabolic process(GO:0005997) |
| 0.0 | 0.1 | GO:0045842 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.0 | 0.1 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.0 | 0.1 | GO:0002281 | macrophage activation involved in immune response(GO:0002281) |
| 0.0 | 0.1 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.0 | 0.1 | GO:0032069 | regulation of nuclease activity(GO:0032069) |
| 0.0 | 0.1 | GO:0032455 | nerve growth factor processing(GO:0032455) |
| 0.0 | 0.1 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.0 | GO:0070366 | regulation of hepatocyte differentiation(GO:0070366) |
| 0.0 | 0.0 | GO:0035461 | vitamin transmembrane transport(GO:0035461) |
| 0.0 | 0.0 | GO:0033603 | positive regulation of dopamine secretion(GO:0033603) |
| 0.0 | 0.1 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.0 | 0.1 | GO:0018101 | protein citrullination(GO:0018101) |
| 0.0 | 0.1 | GO:0045359 | interferon-beta biosynthetic process(GO:0045350) regulation of interferon-beta biosynthetic process(GO:0045357) positive regulation of interferon-beta biosynthetic process(GO:0045359) |
| 0.0 | 0.1 | GO:0043173 | nucleotide salvage(GO:0043173) |
| 0.0 | 0.1 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.0 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.0 | 0.0 | GO:0009169 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.0 | 0.0 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.0 | 0.0 | GO:1901534 | positive regulation of megakaryocyte differentiation(GO:0045654) positive regulation of hematopoietic progenitor cell differentiation(GO:1901534) |
| 0.0 | 0.0 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.0 | GO:0021631 | optic nerve morphogenesis(GO:0021631) |
| 0.0 | 0.0 | GO:0010193 | response to ozone(GO:0010193) |
| 0.0 | 0.0 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.0 | GO:0002903 | negative regulation of B cell apoptotic process(GO:0002903) |
| 0.0 | 0.0 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.0 | 0.0 | GO:0000101 | sulfur amino acid transport(GO:0000101) L-cystine transport(GO:0015811) |
| 0.0 | 0.0 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.0 | 0.0 | GO:0051176 | positive regulation of sulfur metabolic process(GO:0051176) |
| 0.0 | 0.0 | GO:0060083 | smooth muscle contraction involved in micturition(GO:0060083) |
| 0.0 | 0.1 | GO:0042053 | regulation of dopamine metabolic process(GO:0042053) |
| 0.0 | 0.0 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.0 | GO:0021563 | glossopharyngeal nerve development(GO:0021563) glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.0 | GO:0051852 | disruption by host of symbiont cells(GO:0051852) killing by host of symbiont cells(GO:0051873) |
| 0.0 | 0.1 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0060203 | clathrin-sculpted glutamate transport vesicle(GO:0060199) clathrin-sculpted glutamate transport vesicle membrane(GO:0060203) |
| 0.0 | 0.1 | GO:0071546 | pi-body(GO:0071546) |
| 0.0 | 0.1 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 0.1 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.0 | GO:0060200 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.0 | 0.1 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.1 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.0 | GO:0031931 | TORC1 complex(GO:0031931) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.1 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.1 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.1 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.0 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.0 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.2 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.0 | 0.2 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.1 | GO:0001640 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) G-protein coupled glutamate receptor activity(GO:0098988) |
| 0.0 | 0.1 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.1 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.0 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.0 | 0.0 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.0 | 0.1 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.0 | 0.0 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.0 | 0.1 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.0 | GO:0008518 | reduced folate carrier activity(GO:0008518) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.2 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 0.1 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |