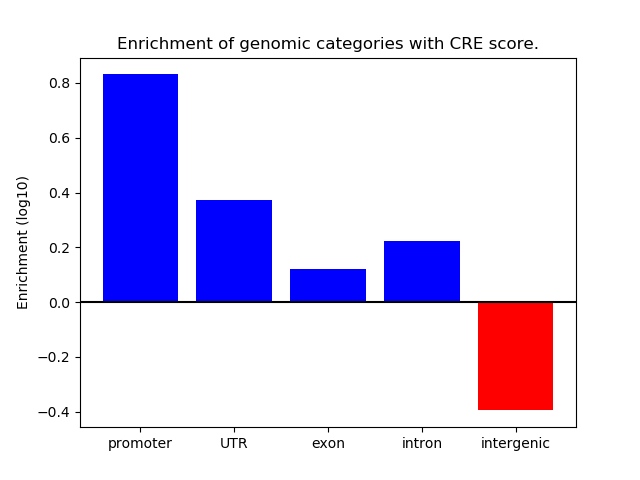
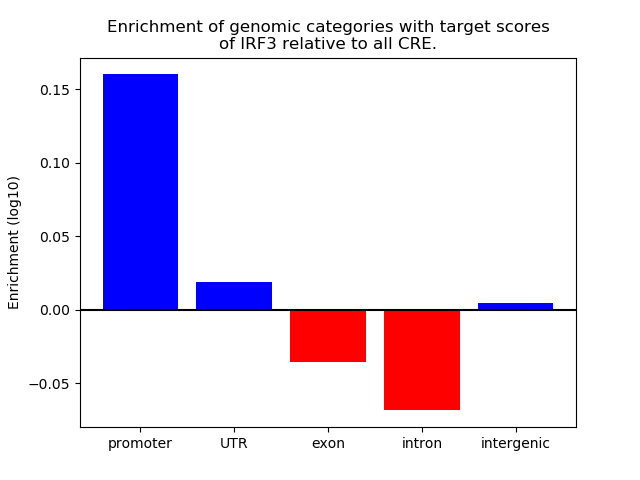
Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
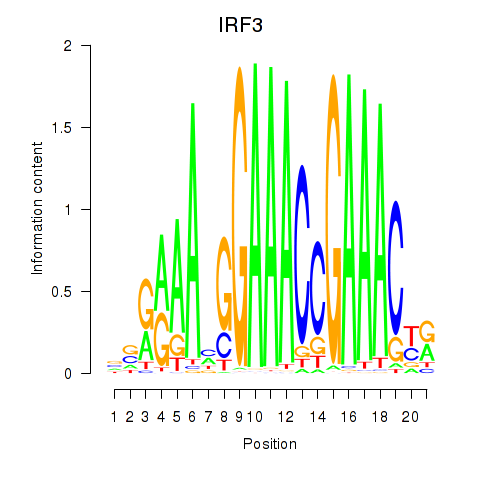
Results for IRF3
Z-value: 1.31
Transcription factors associated with IRF3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
IRF3
|
ENSG00000126456.11 | interferon regulatory factor 3 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr19_50168266_50168469 | IRF3 | 59 | 0.836579 | 0.64 | 6.1e-02 | Click! |
| chr19_50168006_50168157 | IRF3 | 14 | 0.899155 | 0.53 | 1.4e-01 | Click! |
| chr19_50167443_50167632 | IRF3 | 558 | 0.457522 | 0.50 | 1.7e-01 | Click! |
| chr19_50162147_50162298 | IRF3 | 5873 | 0.071742 | -0.30 | 4.4e-01 | Click! |
| chr19_50166639_50166988 | IRF3 | 1282 | 0.192093 | 0.27 | 4.9e-01 | Click! |
Activity of the IRF3 motif across conditions
Conditions sorted by the z-value of the IRF3 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr2_198480069_198480220 | 0.71 |
RFTN2 |
raftlin family member 2 |
2475 |
0.31 |
| chr11_33182270_33182533 | 0.48 |
CSTF3 |
cleavage stimulation factor, 3' pre-RNA, subunit 3, 77kDa |
605 |
0.51 |
| chr19_47217852_47218003 | 0.45 |
PRKD2 |
protein kinase D2 |
71 |
0.94 |
| chr6_152270159_152270310 | 0.40 |
ESR1 |
estrogen receptor 1 |
68442 |
0.13 |
| chr10_126223396_126223623 | 0.40 |
LHPP |
phospholysine phosphohistidine inorganic pyrophosphate phosphatase |
73097 |
0.09 |
| chr22_34249799_34250025 | 0.39 |
LARGE |
like-glycosyltransferase |
7700 |
0.27 |
| chr9_20623530_20623717 | 0.39 |
MLLT3 |
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
1081 |
0.55 |
| chr1_47060166_47060317 | 0.39 |
MKNK1 |
MAP kinase interacting serine/threonine kinase 1 |
289 |
0.88 |
| chr22_19417405_19417667 | 0.39 |
HIRA |
histone cell cycle regulator |
1683 |
0.2 |
| chr16_19083888_19084039 | 0.39 |
COQ7 |
coenzyme Q7 homolog, ubiquinone (yeast) |
4652 |
0.17 |
| chr10_43429375_43429752 | 0.38 |
ENSG00000263795 |
. |
63448 |
0.12 |
| chr5_176938568_176938719 | 0.36 |
DOK3 |
docking protein 3 |
368 |
0.75 |
| chr2_27108887_27109038 | 0.35 |
DPYSL5 |
dihydropyrimidinase-like 5 |
37586 |
0.13 |
| chr20_58712679_58712887 | 0.34 |
C20orf197 |
chromosome 20 open reading frame 197 |
81803 |
0.1 |
| chr12_89551166_89551590 | 0.34 |
ENSG00000238302 |
. |
124684 |
0.06 |
| chr18_9700449_9700600 | 0.34 |
RAB31 |
RAB31, member RAS oncogene family |
7638 |
0.23 |
| chr13_29041064_29041377 | 0.34 |
FLT1 |
fms-related tyrosine kinase 1 |
28012 |
0.24 |
| chr16_23540221_23540372 | 0.33 |
GGA2 |
golgi-associated, gamma adaptin ear containing, ARF binding protein 2 |
6980 |
0.14 |
| chr7_8007598_8007760 | 0.32 |
GLCCI1 |
glucocorticoid induced transcript 1 |
746 |
0.59 |
| chr10_13142624_13142986 | 0.32 |
OPTN |
optineurin |
580 |
0.64 |
| chr11_33895099_33895282 | 0.32 |
LMO2 |
LIM domain only 2 (rhombotin-like 1) |
3828 |
0.24 |
| chr8_104090645_104090882 | 0.32 |
ENSG00000265667 |
. |
14186 |
0.14 |
| chr4_5035629_5036207 | 0.32 |
CYTL1 |
cytokine-like 1 |
14719 |
0.22 |
| chr1_17636167_17636318 | 0.31 |
PADI4 |
peptidyl arginine deiminase, type IV |
1550 |
0.36 |
| chr7_25898359_25898510 | 0.31 |
ENSG00000199085 |
. |
91172 |
0.09 |
| chr6_139693993_139694144 | 0.31 |
CITED2 |
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 |
1282 |
0.57 |
| chr6_20207196_20207347 | 0.31 |
RP11-239H6.2 |
|
5047 |
0.25 |
| chr13_29106267_29106418 | 0.31 |
FLT1 |
fms-related tyrosine kinase 1 |
37077 |
0.21 |
| chr10_82358758_82358909 | 0.31 |
SH2D4B |
SH2 domain containing 4B |
58258 |
0.13 |
| chr12_113415972_113416272 | 0.30 |
OAS2 |
2'-5'-oligoadenylate synthetase 2, 69/71kDa |
78 |
0.97 |
| chr5_137801729_137801883 | 0.30 |
EGR1 |
early growth response 1 |
627 |
0.69 |
| chr1_247336437_247336716 | 0.30 |
ZNF124 |
zinc finger protein 124 |
1258 |
0.39 |
| chr12_7053100_7053251 | 0.30 |
C12orf57 |
chromosome 12 open reading frame 57 |
11 |
0.66 |
| chr17_33570615_33570828 | 0.30 |
SLFN5 |
schlafen family member 5 |
613 |
0.47 |
| chr14_90033233_90033446 | 0.29 |
FOXN3-AS2 |
FOXN3 antisense RNA 2 |
9221 |
0.17 |
| chr8_126497882_126498033 | 0.29 |
ENSG00000266452 |
. |
41150 |
0.18 |
| chr2_136780779_136780951 | 0.29 |
AC093391.2 |
|
10830 |
0.23 |
| chr17_8024292_8024443 | 0.29 |
ALOXE3 |
arachidonate lipoxygenase 3 |
2002 |
0.15 |
| chr4_140426120_140426271 | 0.28 |
RP11-83A24.2 |
|
51007 |
0.1 |
| chr20_39571883_39572034 | 0.28 |
ENSG00000222612 |
. |
80887 |
0.09 |
| chr22_17588430_17588590 | 0.28 |
CECR6 |
cat eye syndrome chromosome region, candidate 6 |
13633 |
0.14 |
| chr18_3877337_3877666 | 0.28 |
DLGAP1-AS3 |
DLGAP1 antisense RNA 3 |
679 |
0.65 |
| chr1_192829951_192830102 | 0.28 |
ENSG00000223075 |
. |
15089 |
0.24 |
| chr8_24206460_24207135 | 0.28 |
ADAM28 |
ADAM metallopeptidase domain 28 |
7632 |
0.22 |
| chr11_111101268_111101555 | 0.28 |
C11orf53 |
chromosome 11 open reading frame 53 |
25296 |
0.18 |
| chr3_7724702_7724983 | 0.28 |
RP11-157E16.1 |
|
75258 |
0.12 |
| chr9_33320777_33320971 | 0.28 |
NFX1 |
nuclear transcription factor, X-box binding 1 |
30331 |
0.12 |
| chr15_85873197_85873348 | 0.28 |
AKAP13 |
A kinase (PRKA) anchor protein 13 |
50546 |
0.11 |
| chr16_88521210_88521549 | 0.28 |
ZFPM1 |
zinc finger protein, FOG family member 1 |
1654 |
0.34 |
| chr6_154674292_154674443 | 0.27 |
IPCEF1 |
interaction protein for cytohesin exchange factors 1 |
3502 |
0.36 |
| chr18_33070553_33070704 | 0.27 |
INO80C |
INO80 complex subunit C |
3194 |
0.27 |
| chr20_52253425_52253583 | 0.27 |
ZNF217 |
zinc finger protein 217 |
28073 |
0.17 |
| chr3_4711265_4711416 | 0.27 |
ITPR1 |
inositol 1,4,5-trisphosphate receptor, type 1 |
153164 |
0.04 |
| chr2_86244884_86245048 | 0.27 |
POLR1A |
polymerase (RNA) I polypeptide A, 194kDa |
88312 |
0.06 |
| chr10_14996335_14996590 | 0.27 |
DCLRE1C |
DNA cross-link repair 1C |
31 |
0.98 |
| chr5_166410441_166410592 | 0.27 |
TENM2 |
teneurin transmembrane protein 2 |
301288 |
0.01 |
| chr14_76657346_76657538 | 0.27 |
GPATCH2L |
G patch domain containing 2-like |
4809 |
0.27 |
| chr3_194033211_194033441 | 0.27 |
CPN2 |
carboxypeptidase N, polypeptide 2 |
38721 |
0.15 |
| chr1_204452069_204452220 | 0.26 |
PIK3C2B |
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 beta |
7302 |
0.19 |
| chr12_127567917_127568068 | 0.26 |
ENSG00000239776 |
. |
82680 |
0.12 |
| chr3_69433628_69433794 | 0.26 |
FRMD4B |
FERM domain containing 4B |
1513 |
0.57 |
| chr19_19439561_19440144 | 0.26 |
MAU2 |
MAU2 sister chromatid cohesion factor |
8057 |
0.13 |
| chr12_9964006_9964157 | 0.26 |
KLRF1 |
killer cell lectin-like receptor subfamily F, member 1 |
15996 |
0.14 |
| chr2_27109662_27109955 | 0.26 |
DPYSL5 |
dihydropyrimidinase-like 5 |
38432 |
0.13 |
| chr6_11581693_11581844 | 0.26 |
TMEM170B |
transmembrane protein 170B |
43257 |
0.17 |
| chr1_203051666_203051922 | 0.26 |
MYOG |
myogenin (myogenic factor 4) |
3370 |
0.17 |
| chr3_128916268_128916630 | 0.26 |
CNBP |
CCHC-type zinc finger, nucleic acid binding protein |
13684 |
0.14 |
| chr3_100054819_100055039 | 0.26 |
NIT2 |
nitrilase family, member 2 |
1274 |
0.49 |
| chr9_7533037_7533188 | 0.25 |
TMEM261 |
transmembrane protein 261 |
266955 |
0.02 |
| chr3_187529226_187529377 | 0.25 |
BCL6 |
B-cell CLL/lymphoma 6 |
65786 |
0.12 |
| chr10_35784890_35785041 | 0.25 |
GJD4 |
gap junction protein, delta 4, 40.1kDa |
109373 |
0.06 |
| chr19_49127703_49127921 | 0.25 |
SPHK2 |
sphingosine kinase 2 |
127 |
0.77 |
| chr6_116833439_116833590 | 0.25 |
TRAPPC3L |
trafficking protein particle complex 3-like |
8 |
0.93 |
| chr12_54765807_54765981 | 0.25 |
GPR84 |
G protein-coupled receptor 84 |
7623 |
0.09 |
| chr19_54584658_54584809 | 0.25 |
TARM1 |
T cell-interacting, activating receptor on myeloid cells 1 |
99 |
0.93 |
| chr5_100114059_100114278 | 0.25 |
ENSG00000221263 |
. |
38101 |
0.19 |
| chr10_5987513_5987753 | 0.25 |
RP11-536K7.3 |
|
68 |
0.97 |
| chr8_109674261_109674444 | 0.25 |
TMEM74 |
transmembrane protein 74 |
125492 |
0.06 |
| chr5_68463336_68463586 | 0.25 |
CCNB1 |
cyclin B1 |
371 |
0.77 |
| chr12_26888585_26888736 | 0.25 |
ITPR2 |
inositol 1,4,5-trisphosphate receptor, type 2 |
97054 |
0.07 |
| chr16_4850786_4851082 | 0.25 |
ROGDI |
rogdi homolog (Drosophila) |
355 |
0.77 |
| chr9_77112278_77112453 | 0.25 |
RORB |
RAR-related orphan receptor B |
84 |
0.97 |
| chr4_173707980_173708237 | 0.25 |
ENSG00000241652 |
. |
354686 |
0.01 |
| chr15_50404931_50405220 | 0.25 |
ATP8B4 |
ATPase, class I, type 8B, member 4 |
1626 |
0.45 |
| chr5_150477986_150478137 | 0.25 |
TNIP1 |
TNFAIP3 interacting protein 1 |
4923 |
0.21 |
| chr1_197170977_197171128 | 0.24 |
CRB1 |
crumbs homolog 1 (Drosophila) |
460 |
0.77 |
| chr6_14744067_14744218 | 0.24 |
ENSG00000206960 |
. |
97376 |
0.09 |
| chr12_116757651_116758051 | 0.24 |
MED13L |
mediator complex subunit 13-like |
42708 |
0.16 |
| chr6_6857532_6857683 | 0.24 |
ENSG00000240936 |
. |
81523 |
0.11 |
| chr5_75992073_75992255 | 0.24 |
F2R |
coagulation factor II (thrombin) receptor |
19704 |
0.16 |
| chr4_60684323_60684624 | 0.24 |
ENSG00000201676 |
. |
14420 |
0.24 |
| chr7_18184166_18184423 | 0.24 |
ENSG00000221393 |
. |
17362 |
0.24 |
| chr4_105979647_105979990 | 0.24 |
ENSG00000252136 |
. |
47326 |
0.16 |
| chr20_44509523_44509687 | 0.24 |
ZSWIM1 |
zinc finger, SWIM-type containing 1 |
261 |
0.8 |
| chr1_180248686_180248990 | 0.24 |
RP5-1180C10.2 |
|
3479 |
0.3 |
| chr3_29194998_29195333 | 0.24 |
RBMS3 |
RNA binding motif, single stranded interacting protein 3 |
127308 |
0.05 |
| chr4_146540899_146541050 | 0.24 |
MMAA |
methylmalonic aciduria (cobalamin deficiency) cblA type |
1559 |
0.42 |
| chr12_121019480_121019828 | 0.24 |
POP5 |
processing of precursor 5, ribonuclease P/MRP subunit (S. cerevisiae) |
453 |
0.74 |
| chr3_169720111_169720354 | 0.24 |
SEC62-AS1 |
SEC62 antisense RNA 1 |
16729 |
0.13 |
| chr19_15054504_15054655 | 0.24 |
OR7C2 |
olfactory receptor, family 7, subfamily C, member 2 |
2278 |
0.26 |
| chrX_15448302_15448453 | 0.24 |
BMX |
BMX non-receptor tyrosine kinase |
33992 |
0.15 |
| chr1_45023108_45023259 | 0.24 |
ENSG00000263381 |
. |
12018 |
0.2 |
| chr2_145443692_145443848 | 0.24 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
165149 |
0.04 |
| chr10_125865907_125866246 | 0.24 |
CHST15 |
carbohydrate (N-acetylgalactosamine 4-sulfate 6-O) sulfotransferase 15 |
12870 |
0.28 |
| chr1_181056660_181057120 | 0.23 |
IER5 |
immediate early response 5 |
748 |
0.71 |
| chr2_148779503_148779985 | 0.23 |
ORC4 |
origin recognition complex, subunit 4 |
597 |
0.65 |
| chr2_118856015_118856215 | 0.23 |
INSIG2 |
insulin induced gene 2 |
10065 |
0.24 |
| chr3_128313216_128313433 | 0.23 |
C3orf27 |
chromosome 3 open reading frame 27 |
18395 |
0.19 |
| chr15_69075380_69075658 | 0.23 |
ENSG00000265195 |
. |
18745 |
0.22 |
| chr10_45874886_45875037 | 0.23 |
ALOX5 |
arachidonate 5-lipoxygenase |
5286 |
0.27 |
| chr6_21589300_21589525 | 0.23 |
SOX4 |
SRY (sex determining region Y)-box 4 |
4560 |
0.37 |
| chr8_102923998_102924149 | 0.23 |
NCALD |
neurocalcin delta |
63484 |
0.13 |
| chrX_13754467_13754844 | 0.23 |
OFD1 |
oral-facial-digital syndrome 1 |
1791 |
0.29 |
| chr7_100011522_100011687 | 0.23 |
ZCWPW1 |
zinc finger, CW type with PWWP domain 1 |
4436 |
0.09 |
| chr22_36556784_36556935 | 0.23 |
APOL3 |
apolipoprotein L, 3 |
28 |
0.98 |
| chr1_46052215_46052366 | 0.23 |
NASP |
nuclear autoantigenic sperm protein (histone-binding) |
2553 |
0.23 |
| chr2_176495143_176495353 | 0.23 |
AC016751.3 |
|
266816 |
0.01 |
| chr17_79360953_79361175 | 0.23 |
RP11-1055B8.6 |
Uncharacterized protein |
8211 |
0.12 |
| chr13_105829604_105829896 | 0.23 |
DAOA |
D-amino acid oxidase activator |
288842 |
0.01 |
| chr16_28518181_28518332 | 0.23 |
IL27 |
interleukin 27 |
101 |
0.95 |
| chr9_33759387_33759538 | 0.23 |
PRSS3 |
protease, serine, 3 |
8643 |
0.14 |
| chr19_14877311_14877462 | 0.23 |
EMR2 |
egf-like module containing, mucin-like, hormone receptor-like 2 |
5800 |
0.16 |
| chr11_66115431_66115582 | 0.23 |
RP11-867G23.8 |
Uncharacterized protein |
85 |
0.76 |
| chr4_85888329_85888497 | 0.23 |
WDFY3 |
WD repeat and FYVE domain containing 3 |
869 |
0.72 |
| chr12_57773877_57774028 | 0.23 |
ENSG00000201198 |
. |
35127 |
0.09 |
| chr1_85411015_85411201 | 0.23 |
MCOLN2 |
mucolipin 2 |
51515 |
0.12 |
| chr17_1992881_1993032 | 0.23 |
RP11-667K14.5 |
|
19 |
0.96 |
| chr17_59940538_59940689 | 0.22 |
BRIP1 |
BRCA1 interacting protein C-terminal helicase 1 |
92 |
0.97 |
| chr4_4292418_4292569 | 0.22 |
ZBTB49 |
zinc finger and BTB domain containing 49 |
497 |
0.52 |
| chr17_58754505_58754656 | 0.22 |
BCAS3 |
breast carcinoma amplified sequence 3 |
234 |
0.92 |
| chrX_13770949_13771164 | 0.22 |
OFD1 |
oral-facial-digital syndrome 1 |
18192 |
0.17 |
| chrX_16846441_16846592 | 0.22 |
RBBP7 |
retinoblastoma binding protein 7 |
23672 |
0.15 |
| chr17_30214979_30215206 | 0.22 |
UTP6 |
UTP6, small subunit (SSU) processome component, homolog (yeast) |
13692 |
0.18 |
| chr9_33221071_33221222 | 0.22 |
SPINK4 |
serine peptidase inhibitor, Kazal type 4 |
169 |
0.94 |
| chr11_108095836_108095987 | 0.22 |
ATM |
ataxia telangiectasia mutated |
1106 |
0.42 |
| chr15_52733385_52733536 | 0.22 |
MYO5A |
myosin VA (heavy chain 12, myoxin) |
5593 |
0.28 |
| chr12_53937677_53937943 | 0.22 |
RP11-793H13.3 |
|
30042 |
0.1 |
| chr11_128554939_128555157 | 0.22 |
RP11-744N12.3 |
|
1275 |
0.35 |
| chr1_20959712_20959863 | 0.22 |
PINK1 |
PTEN induced putative kinase 1 |
161 |
0.95 |
| chr6_611011_611162 | 0.22 |
HUS1B |
HUS1 checkpoint homolog b (S. pombe) |
45877 |
0.17 |
| chr1_228328808_228329114 | 0.22 |
GUK1 |
guanylate kinase 1 |
122 |
0.93 |
| chr6_5999961_6000211 | 0.22 |
NRN1 |
neuritin 1 |
7114 |
0.26 |
| chr1_89150817_89151520 | 0.22 |
RP11-76N22.2 |
|
281 |
0.84 |
| chr6_35697474_35697939 | 0.22 |
FKBP5 |
FK506 binding protein 5 |
1346 |
0.31 |
| chr11_47972062_47972213 | 0.22 |
PTPRJ |
protein tyrosine phosphatase, receptor type, J |
29976 |
0.17 |
| chr6_106546899_106547210 | 0.21 |
PRDM1 |
PR domain containing 1, with ZNF domain |
239 |
0.88 |
| chr5_137679824_137679975 | 0.21 |
RP11-256P1.1 |
|
5431 |
0.13 |
| chr18_20722941_20723198 | 0.21 |
ENSG00000222999 |
. |
6593 |
0.21 |
| chr4_169239823_169239974 | 0.21 |
DDX60 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 60 |
57 |
0.98 |
| chr10_126430141_126430442 | 0.21 |
FAM53B |
family with sequence similarity 53, member B |
1248 |
0.43 |
| chr22_19465272_19465744 | 0.21 |
UFD1L |
ubiquitin fusion degradation 1 like (yeast) |
946 |
0.35 |
| chr18_74162951_74163102 | 0.21 |
ZNF516 |
zinc finger protein 516 |
39709 |
0.14 |
| chr10_82215641_82216144 | 0.21 |
TSPAN14 |
tetraspanin 14 |
1781 |
0.38 |
| chr9_127005358_127005801 | 0.21 |
NEK6 |
NIMA-related kinase 6 |
14306 |
0.17 |
| chr3_135912382_135912568 | 0.21 |
MSL2 |
male-specific lethal 2 homolog (Drosophila) |
921 |
0.68 |
| chr4_125624499_125624650 | 0.21 |
ANKRD50 |
ankyrin repeat domain 50 |
9313 |
0.34 |
| chr12_22830855_22831006 | 0.21 |
ETNK1 |
ethanolamine kinase 1 |
52639 |
0.15 |
| chr3_176919574_176920023 | 0.21 |
TBL1XR1 |
transducin (beta)-like 1 X-linked receptor 1 |
4537 |
0.31 |
| chr1_26450910_26451453 | 0.21 |
PDIK1L |
PDLIM1 interacting kinase 1 like |
12841 |
0.11 |
| chr19_54559994_54560145 | 0.21 |
VSTM1 |
V-set and transmembrane domain containing 1 |
1608 |
0.22 |
| chr1_42195372_42195655 | 0.21 |
HIVEP3 |
human immunodeficiency virus type I enhancer binding protein 3 |
28841 |
0.2 |
| chr1_151668701_151668852 | 0.21 |
RP11-98D18.1 |
|
4726 |
0.1 |
| chr5_159698543_159698722 | 0.21 |
ENSG00000243654 |
. |
20163 |
0.16 |
| chr3_128583649_128583920 | 0.21 |
RP11-723O4.2 |
|
663 |
0.69 |
| chr17_59222293_59222572 | 0.21 |
RP11-136H19.1 |
|
9107 |
0.23 |
| chr16_48644531_48644682 | 0.20 |
N4BP1 |
NEDD4 binding protein 1 |
486 |
0.81 |
| chr3_71731013_71731282 | 0.20 |
EIF4E3 |
eukaryotic translation initiation factor 4E family member 3 |
43379 |
0.15 |
| chrX_109280296_109280497 | 0.20 |
ENSG00000208013 |
. |
18161 |
0.2 |
| chr9_92020971_92021287 | 0.20 |
SEMA4D |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D |
288 |
0.93 |
| chr7_116594399_116594611 | 0.20 |
ST7 |
suppression of tumorigenicity 7 |
60 |
0.54 |
| chr11_128089678_128089829 | 0.20 |
ETS1 |
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
285536 |
0.01 |
| chrX_9547118_9547269 | 0.20 |
TBL1X |
transducin (beta)-like 1X-linked |
44210 |
0.2 |
| chr15_50345522_50345833 | 0.20 |
ATP8B4 |
ATPase, class I, type 8B, member 4 |
6093 |
0.27 |
| chr1_21292036_21292187 | 0.20 |
EIF4G3 |
eukaryotic translation initiation factor 4 gamma, 3 |
7418 |
0.23 |
| chr2_145275679_145275945 | 0.20 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
16 |
0.51 |
| chr10_65388709_65389027 | 0.20 |
REEP3 |
receptor accessory protein 3 |
107745 |
0.07 |
| chr9_132682421_132682638 | 0.20 |
FNBP1 |
formin binding protein 1 |
940 |
0.56 |
| chr9_113096398_113096701 | 0.20 |
TXNDC8 |
thioredoxin domain containing 8 (spermatozoa) |
3539 |
0.28 |
| chr2_210852878_210853031 | 0.20 |
RPE |
ribulose-5-phosphate-3-epimerase |
14335 |
0.18 |
| chr16_54210277_54210428 | 0.20 |
RP11-324D17.1 |
HCG2045435; Uncharacterized protein |
70525 |
0.11 |
| chr8_124170086_124170384 | 0.20 |
FAM83A |
family with sequence similarity 83, member A |
21052 |
0.11 |
| chr22_25782542_25782779 | 0.20 |
LRP5L |
low density lipoprotein receptor-related protein 5-like |
5116 |
0.24 |
| chr2_169957115_169957325 | 0.20 |
AC007556.3 |
|
33 |
0.98 |
| chr7_150104070_150104221 | 0.20 |
RP4-584D14.6 |
|
23172 |
0.11 |
| chr1_212809981_212810132 | 0.20 |
RP11-338C15.5 |
|
9943 |
0.14 |
| chr9_35569725_35569931 | 0.20 |
FAM166B |
family with sequence similarity 166, member B |
5932 |
0.12 |
| chr2_237973979_237974130 | 0.20 |
AC105760.3 |
|
9244 |
0.19 |
| chr12_33047562_33047713 | 0.20 |
PKP2 |
plakophilin 2 |
2053 |
0.46 |
| chr6_2838201_2838352 | 0.20 |
SERPINB1 |
serpin peptidase inhibitor, clade B (ovalbumin), member 1 |
3818 |
0.22 |
| chr1_65522143_65522294 | 0.20 |
ENSG00000265996 |
. |
1307 |
0.39 |
| chr15_68502161_68502336 | 0.20 |
CALML4 |
calmodulin-like 4 |
3831 |
0.2 |
| chr10_7449534_7449830 | 0.20 |
SFMBT2 |
Scm-like with four mbt domains 2 |
1025 |
0.67 |
| chr14_106679120_106679271 | 0.19 |
IGHVII-20-1 |
immunoglobulin heavy variable (II)-20-1 (pseudogene) |
9244 |
0.05 |
| chr3_138954177_138954328 | 0.19 |
MRPS22 |
mitochondrial ribosomal protein S22 |
108609 |
0.06 |
| chr8_128887727_128887885 | 0.19 |
TMEM75 |
transmembrane protein 75 |
72785 |
0.09 |
| chr5_101622831_101622982 | 0.19 |
SLCO4C1 |
solute carrier organic anion transporter family, member 4C1 |
9347 |
0.21 |
| chr2_55747454_55747845 | 0.19 |
CCDC104 |
coiled-coil domain containing 104 |
782 |
0.58 |
| chr14_70172450_70172635 | 0.19 |
SRSF5 |
serine/arginine-rich splicing factor 5 |
21075 |
0.22 |
| chr19_14186633_14186784 | 0.19 |
hsa-mir-1199 |
hsa-mir-1199 |
2314 |
0.15 |
| chr7_48498957_48499108 | 0.19 |
ABCA13 |
ATP-binding cassette, sub-family A (ABC1), member 13 |
4372 |
0.37 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0031946 | regulation of glucocorticoid biosynthetic process(GO:0031946) |
| 0.1 | 0.2 | GO:0072110 | glomerular mesangial cell proliferation(GO:0072110) regulation of glomerular mesangial cell proliferation(GO:0072124) |
| 0.1 | 0.2 | GO:0033603 | positive regulation of dopamine secretion(GO:0033603) |
| 0.1 | 0.2 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.1 | 0.2 | GO:0072600 | protein targeting to Golgi(GO:0000042) establishment of protein localization to Golgi(GO:0072600) |
| 0.1 | 0.2 | GO:0002606 | positive regulation of dendritic cell antigen processing and presentation(GO:0002606) |
| 0.0 | 0.2 | GO:0045625 | regulation of T-helper 1 cell differentiation(GO:0045625) |
| 0.0 | 0.2 | GO:0045356 | positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.0 | 0.1 | GO:0015959 | diadenosine polyphosphate metabolic process(GO:0015959) |
| 0.0 | 0.1 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.0 | 0.2 | GO:0018101 | protein citrullination(GO:0018101) |
| 0.0 | 0.1 | GO:0070423 | cytoplasmic pattern recognition receptor signaling pathway(GO:0002753) nucleotide-binding domain, leucine rich repeat containing receptor signaling pathway(GO:0035872) nucleotide-binding oligomerization domain containing signaling pathway(GO:0070423) nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070431) |
| 0.0 | 0.1 | GO:2000143 | negative regulation of transcription initiation from RNA polymerase II promoter(GO:0060633) negative regulation of DNA-templated transcription, initiation(GO:2000143) |
| 0.0 | 0.1 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.0 | 0.1 | GO:0002282 | microglial cell activation involved in immune response(GO:0002282) |
| 0.0 | 0.1 | GO:0002328 | pro-B cell differentiation(GO:0002328) |
| 0.0 | 0.2 | GO:0033688 | regulation of osteoblast proliferation(GO:0033688) |
| 0.0 | 0.1 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.0 | 0.2 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.1 | GO:0046831 | regulation of RNA export from nucleus(GO:0046831) |
| 0.0 | 0.2 | GO:0002360 | T cell lineage commitment(GO:0002360) |
| 0.0 | 0.2 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.1 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.0 | 0.1 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.1 | GO:0021860 | pyramidal neuron differentiation(GO:0021859) pyramidal neuron development(GO:0021860) |
| 0.0 | 0.0 | GO:0010831 | positive regulation of myotube differentiation(GO:0010831) |
| 0.0 | 0.1 | GO:0071436 | sodium ion export(GO:0071436) |
| 0.0 | 0.1 | GO:0001306 | age-dependent response to oxidative stress(GO:0001306) age-dependent general metabolic decline(GO:0007571) |
| 0.0 | 0.0 | GO:0060061 | Spemann organizer formation(GO:0060061) |
| 0.0 | 0.1 | GO:0060287 | epithelial cilium movement(GO:0003351) epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.3 | GO:0006378 | mRNA polyadenylation(GO:0006378) |
| 0.0 | 0.4 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 0.1 | GO:0002713 | negative regulation of B cell mediated immunity(GO:0002713) negative regulation of immunoglobulin mediated immune response(GO:0002890) |
| 0.0 | 0.1 | GO:0034067 | protein localization to Golgi apparatus(GO:0034067) |
| 0.0 | 0.1 | GO:0008611 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.2 | GO:0006670 | sphingosine metabolic process(GO:0006670) |
| 0.0 | 0.1 | GO:0043374 | CD8-positive, alpha-beta T cell activation(GO:0036037) CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.1 | GO:0002538 | arachidonic acid metabolite production involved in inflammatory response(GO:0002538) leukotriene production involved in inflammatory response(GO:0002540) |
| 0.0 | 0.1 | GO:0045070 | positive regulation of viral genome replication(GO:0045070) |
| 0.0 | 0.1 | GO:0035461 | vitamin transmembrane transport(GO:0035461) |
| 0.0 | 0.1 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.0 | 0.1 | GO:1901798 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) positive regulation of signal transduction by p53 class mediator(GO:1901798) |
| 0.0 | 0.2 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.1 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.0 | 0.1 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 | 0.1 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 0.0 | GO:0060437 | lung growth(GO:0060437) |
| 0.0 | 0.1 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.0 | GO:0032239 | regulation of nucleobase-containing compound transport(GO:0032239) |
| 0.0 | 0.1 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.0 | 0.1 | GO:0035751 | lysosomal lumen acidification(GO:0007042) regulation of lysosomal lumen pH(GO:0035751) |
| 0.0 | 0.1 | GO:0045885 | obsolete positive regulation of survival gene product expression(GO:0045885) |
| 0.0 | 0.1 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.1 | GO:0018243 | protein O-linked glycosylation via serine(GO:0018242) protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.0 | GO:0060872 | semicircular canal development(GO:0060872) |
| 0.0 | 0.1 | GO:0000132 | establishment of mitotic spindle orientation(GO:0000132) establishment of spindle orientation(GO:0051294) |
| 0.0 | 0.1 | GO:0007262 | STAT protein import into nucleus(GO:0007262) |
| 0.0 | 0.0 | GO:0071803 | regulation of podosome assembly(GO:0071801) positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.0 | GO:0060737 | prostate gland morphogenetic growth(GO:0060737) |
| 0.0 | 0.0 | GO:0003078 | obsolete regulation of natriuresis(GO:0003078) |
| 0.0 | 0.1 | GO:0032727 | positive regulation of interferon-alpha production(GO:0032727) |
| 0.0 | 0.0 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.0 | 0.0 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.0 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.0 | 0.0 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.0 | 0.1 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 0.1 | GO:0071501 | SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.0 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.0 | GO:0006655 | phosphatidylglycerol biosynthetic process(GO:0006655) cardiolipin metabolic process(GO:0032048) cardiolipin biosynthetic process(GO:0032049) phosphatidylglycerol metabolic process(GO:0046471) |
| 0.0 | 0.1 | GO:0045579 | positive regulation of B cell differentiation(GO:0045579) |
| 0.0 | 0.1 | GO:0051570 | regulation of histone H3-K9 methylation(GO:0051570) |
| 0.0 | 0.1 | GO:0035608 | protein deglutamylation(GO:0035608) |
| 0.0 | 0.1 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.0 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 | 0.1 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.0 | 0.1 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 0.2 | GO:0007379 | segment specification(GO:0007379) |
| 0.0 | 0.0 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.1 | GO:0034405 | response to fluid shear stress(GO:0034405) |
| 0.0 | 0.0 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) phosphatidylethanolamine metabolic process(GO:0046337) |
| 0.0 | 0.1 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.0 | 0.2 | GO:0042255 | ribosome assembly(GO:0042255) |
| 0.0 | 0.0 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.0 | 0.0 | GO:1903429 | regulation of neuron maturation(GO:0014041) regulation of cell maturation(GO:1903429) |
| 0.0 | 0.0 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.0 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.0 | 0.1 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.1 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.0 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.0 | GO:0043320 | natural killer cell activation involved in immune response(GO:0002323) natural killer cell degranulation(GO:0043320) |
| 0.0 | 0.1 | GO:0008653 | lipopolysaccharide metabolic process(GO:0008653) |
| 0.0 | 0.2 | GO:0001893 | maternal placenta development(GO:0001893) |
| 0.0 | 0.0 | GO:0006991 | response to sterol depletion(GO:0006991) |
| 0.0 | 0.0 | GO:0003323 | type B pancreatic cell development(GO:0003323) |
| 0.0 | 0.1 | GO:0021903 | rostrocaudal neural tube patterning(GO:0021903) midbrain-hindbrain boundary development(GO:0030917) |
| 0.0 | 0.4 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
| 0.0 | 0.0 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) |
| 0.0 | 0.1 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 0.2 | GO:0015949 | nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.0 | 0.0 | GO:0010980 | regulation of vitamin D 24-hydroxylase activity(GO:0010979) positive regulation of vitamin D 24-hydroxylase activity(GO:0010980) |
| 0.0 | 0.1 | GO:0006824 | cobalt ion transport(GO:0006824) |
| 0.0 | 0.0 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.1 | GO:0018027 | peptidyl-lysine dimethylation(GO:0018027) |
| 0.0 | 0.1 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) |
| 0.0 | 0.0 | GO:0006533 | aspartate metabolic process(GO:0006531) aspartate catabolic process(GO:0006533) |
| 0.0 | 0.0 | GO:0007000 | nucleolus organization(GO:0007000) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.1 | 0.2 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.1 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.1 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.0 | 0.1 | GO:0044462 | external encapsulating structure part(GO:0044462) |
| 0.0 | 0.2 | GO:0030677 | nucleolar ribonuclease P complex(GO:0005655) ribonuclease P complex(GO:0030677) multimeric ribonuclease P complex(GO:0030681) |
| 0.0 | 0.1 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.0 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.0 | 0.4 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.1 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.2 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.1 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.2 | GO:0034518 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.0 | 0.1 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.0 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.1 | GO:0031094 | platelet dense tubular network(GO:0031094) |
| 0.0 | 0.0 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.1 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.1 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.5 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.0 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.3 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.0 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.1 | 0.2 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.1 | 0.3 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.3 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.3 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 0.4 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.2 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.1 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.2 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.1 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 0.1 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.0 | 0.1 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.0 | 0.3 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.2 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.0 | 0.1 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.1 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.1 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.0 | 0.2 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.1 | GO:0090482 | vitamin transmembrane transporter activity(GO:0090482) |
| 0.0 | 0.1 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.2 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.1 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.0 | 0.1 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.1 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.1 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.1 | GO:0016165 | linoleate 13S-lipoxygenase activity(GO:0016165) |
| 0.0 | 0.2 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.1 | GO:0022897 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 0.0 | 0.1 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.1 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.2 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.2 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.1 | GO:0008159 | obsolete positive transcription elongation factor activity(GO:0008159) |
| 0.0 | 0.1 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.0 | 0.1 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.1 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.1 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.1 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.3 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 0.0 | 0.0 | GO:0004525 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.0 | 0.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.1 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.1 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.0 | 0.1 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.0 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.0 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.0 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 0.1 | GO:0071553 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.0 | 0.0 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.0 | 0.0 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.0 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.0 | 0.0 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.0 | GO:0046980 | tapasin binding(GO:0046980) |
| 0.0 | 0.1 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.1 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.0 | 0.0 | GO:0045569 | TRAIL binding(GO:0045569) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.5 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 0.0 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.0 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.1 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 0.0 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.0 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 0.1 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.3 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.0 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 0.5 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.3 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.2 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.1 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.3 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.0 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.4 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.0 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.2 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 0.1 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.3 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.4 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.3 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.0 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.0 | 0.2 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.0 | 0.2 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.3 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.2 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.0 | 0.2 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 0.2 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.4 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.6 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.2 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.1 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.1 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.2 | REACTOME TAK1 ACTIVATES NFKB BY PHOSPHORYLATION AND ACTIVATION OF IKKS COMPLEX | Genes involved in TAK1 activates NFkB by phosphorylation and activation of IKKs complex |
| 0.0 | 0.3 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.1 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.6 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |