Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
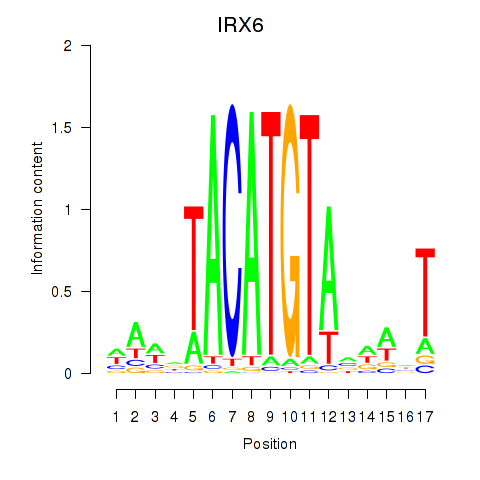
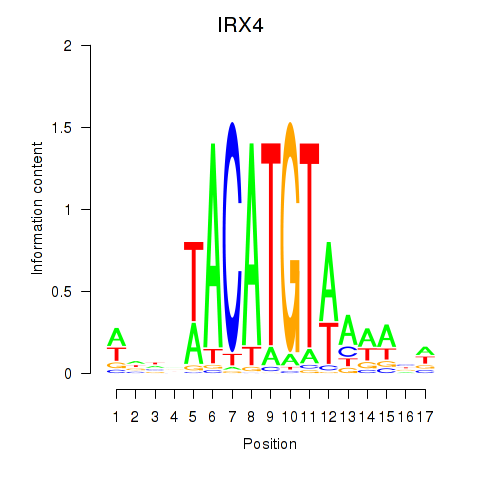
Results for IRX6_IRX4
Z-value: 0.62
Transcription factors associated with IRX6_IRX4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
IRX6
|
ENSG00000159387.7 | iroquois homeobox 6 |
|
IRX4
|
ENSG00000113430.5 | iroquois homeobox 4 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr5_1882240_1882502 | IRX4 | 487 | 0.715971 | 0.59 | 9.5e-02 | Click! |
| chr5_1881693_1881844 | IRX4 | 1090 | 0.418558 | 0.52 | 1.5e-01 | Click! |
| chr5_1882815_1883393 | IRX4 | 224 | 0.858060 | 0.43 | 2.5e-01 | Click! |
| chr5_1877685_1877836 | IRX4 | 5098 | 0.167410 | 0.30 | 4.3e-01 | Click! |
| chr5_1876111_1876262 | IRX4 | 6672 | 0.158586 | 0.29 | 4.5e-01 | Click! |
| chr16_55357840_55357991 | IRX6 | 243 | 0.928797 | 0.65 | 5.6e-02 | Click! |
| chr16_55359119_55359270 | IRX6 | 1522 | 0.403624 | -0.21 | 5.9e-01 | Click! |
Activity of the IRX6_IRX4 motif across conditions
Conditions sorted by the z-value of the IRX6_IRX4 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
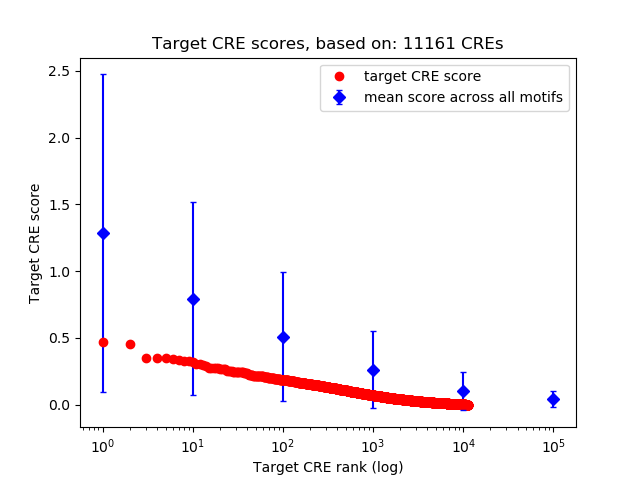
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr8_95037399_95037630 | 0.47 |
ENSG00000263855 |
. |
51432 |
0.15 |
| chr22_36945126_36945326 | 0.46 |
RP5-1119A7.10 |
|
2986 |
0.2 |
| chr3_125230252_125231290 | 0.35 |
ENSG00000201800 |
. |
5052 |
0.18 |
| chr7_100782348_100782499 | 0.35 |
SERPINE1 |
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 |
12044 |
0.1 |
| chr3_154512727_154512878 | 0.35 |
MME |
membrane metallo-endopeptidase |
229111 |
0.02 |
| chr6_14638853_14639371 | 0.34 |
ENSG00000206960 |
. |
7654 |
0.34 |
| chr9_128409096_128409536 | 0.34 |
MAPKAP1 |
mitogen-activated protein kinase associated protein 1 |
3380 |
0.33 |
| chr20_10538984_10539442 | 0.33 |
JAG1 |
jagged 1 |
103941 |
0.07 |
| chr15_93457053_93457941 | 0.33 |
CHD2 |
chromodomain helicase DNA binding protein 2 |
9794 |
0.2 |
| chr20_51111252_51111403 | 0.32 |
ZFP64 |
ZFP64 zinc finger protein |
302802 |
0.01 |
| chr6_140364803_140364954 | 0.31 |
ENSG00000252107 |
. |
114953 |
0.07 |
| chr8_122361324_122361475 | 0.30 |
ENSG00000221644 |
. |
162267 |
0.04 |
| chr11_122577004_122577557 | 0.30 |
ENSG00000239079 |
. |
19739 |
0.22 |
| chr1_150598433_150598584 | 0.29 |
ENSA |
endosulfine alpha |
3084 |
0.15 |
| chr3_185411880_185412031 | 0.28 |
C3orf65 |
chromosome 3 open reading frame 65 |
19125 |
0.22 |
| chr5_73529344_73529495 | 0.28 |
ENSG00000222551 |
. |
38171 |
0.21 |
| chr8_130709458_130709738 | 0.28 |
GSDMC |
gasdermin C |
89536 |
0.08 |
| chr4_127860021_127860172 | 0.27 |
ENSG00000199862 |
. |
89278 |
0.11 |
| chr19_9241411_9241562 | 0.27 |
OR7G3 |
olfactory receptor, family 7, subfamily G, member 3 |
3860 |
0.15 |
| chr2_3625268_3626011 | 0.27 |
ENSG00000252531 |
. |
2521 |
0.16 |
| chr1_210046746_210046897 | 0.27 |
DIEXF |
digestive organ expansion factor homolog (zebrafish) |
34671 |
0.15 |
| chr2_32936569_32936720 | 0.27 |
TTC27 |
tetratricopeptide repeat domain 27 |
32699 |
0.21 |
| chr5_171382567_171382718 | 0.27 |
FBXW11 |
F-box and WD repeat domain containing 11 |
22118 |
0.23 |
| chr20_52354290_52354570 | 0.26 |
ENSG00000238468 |
. |
69133 |
0.12 |
| chr7_12693175_12693326 | 0.25 |
ARL4A |
ADP-ribosylation factor-like 4A |
33231 |
0.17 |
| chr1_39652074_39652225 | 0.25 |
MACF1 |
microtubule-actin crosslinking factor 1 |
17769 |
0.15 |
| chr10_62186857_62187008 | 0.25 |
ANK3 |
ankyrin 3, node of Ranvier (ankyrin G) |
37444 |
0.23 |
| chr8_30278565_30278948 | 0.25 |
RBPMS |
RNA binding protein with multiple splicing |
21377 |
0.21 |
| chr4_169914702_169914853 | 0.25 |
ENSG00000201818 |
. |
11623 |
0.18 |
| chr1_27025127_27026141 | 0.25 |
ARID1A |
AT rich interactive domain 1A (SWI-like) |
2739 |
0.21 |
| chr2_230718614_230718765 | 0.24 |
TRIP12 |
thyroid hormone receptor interactor 12 |
5309 |
0.22 |
| chr6_80255213_80255459 | 0.24 |
LCA5 |
Leber congenital amaurosis 5 |
8161 |
0.3 |
| chr3_99565123_99565274 | 0.24 |
FILIP1L |
filamin A interacting protein 1-like |
2258 |
0.37 |
| chr3_142853615_142853766 | 0.24 |
CHST2 |
carbohydrate (N-acetylglucosamine-6-O) sulfotransferase 2 |
15517 |
0.21 |
| chr1_86020329_86020665 | 0.24 |
DDAH1 |
dimethylarginine dimethylaminohydrolase 1 |
23436 |
0.17 |
| chr1_243713303_243713454 | 0.24 |
RP11-269F20.1 |
|
4544 |
0.34 |
| chr6_131100314_131100854 | 0.24 |
SMLR1 |
small leucine-rich protein 1 |
47962 |
0.18 |
| chr2_217051631_217052157 | 0.24 |
AC012513.6 |
|
72424 |
0.09 |
| chr6_140204883_140205034 | 0.24 |
ENSG00000252107 |
. |
274873 |
0.02 |
| chr4_144273352_144273505 | 0.23 |
ENSG00000265623 |
. |
8815 |
0.21 |
| chr2_1746061_1746328 | 0.23 |
PXDN |
peroxidasin homolog (Drosophila) |
2020 |
0.44 |
| chr4_187597285_187597436 | 0.22 |
FAT1 |
FAT atypical cadherin 1 |
47649 |
0.17 |
| chr4_123703753_123703904 | 0.22 |
FGF2 |
fibroblast growth factor 2 (basic) |
44035 |
0.13 |
| chr1_232558087_232558238 | 0.22 |
SIPA1L2 |
signal-induced proliferation-associated 1 like 2 |
40001 |
0.22 |
| chr6_143722656_143722949 | 0.22 |
AL031320.1 |
|
36773 |
0.13 |
| chr15_63802358_63803207 | 0.22 |
USP3 |
ubiquitin specific peptidase 3 |
4863 |
0.26 |
| chr6_56555129_56555280 | 0.22 |
DST |
dystonin |
47410 |
0.18 |
| chr19_6103075_6103226 | 0.22 |
CTB-66B24.1 |
|
6183 |
0.15 |
| chr1_224972610_224972761 | 0.22 |
RP11-449J1.1 |
|
18083 |
0.21 |
| chr5_95430378_95430529 | 0.22 |
ENSG00000207578 |
. |
15611 |
0.25 |
| chr2_179106826_179106977 | 0.22 |
OSBPL6 |
oxysterol binding protein-like 6 |
42753 |
0.16 |
| chr7_44454046_44454197 | 0.21 |
NUDCD3 |
NudC domain containing 3 |
76358 |
0.08 |
| chr1_22047557_22047882 | 0.21 |
USP48 |
ubiquitin specific peptidase 48 |
3893 |
0.26 |
| chr12_25487343_25487617 | 0.21 |
ENSG00000201439 |
. |
69882 |
0.1 |
| chr2_216393689_216393840 | 0.21 |
AC012462.1 |
|
92788 |
0.08 |
| chr7_41727697_41728013 | 0.21 |
INHBA-AS1 |
INHBA antisense RNA 1 |
5659 |
0.22 |
| chr9_35497818_35497969 | 0.21 |
RUSC2 |
RUN and SH3 domain containing 2 |
7769 |
0.18 |
| chrX_71494679_71495271 | 0.21 |
RPS4X |
ribosomal protein S4, X-linked |
2123 |
0.24 |
| chr18_51804642_51804943 | 0.21 |
POLI |
polymerase (DNA directed) iota |
4484 |
0.25 |
| chr1_63187029_63187180 | 0.21 |
RP11-230B22.1 |
|
32951 |
0.16 |
| chr11_103396303_103396486 | 0.21 |
DYNC2H1 |
dynein, cytoplasmic 2, heavy chain 1 |
167423 |
0.04 |
| chr5_33306659_33307137 | 0.21 |
CTD-2203K17.1 |
|
133827 |
0.05 |
| chr2_238106047_238106302 | 0.21 |
AC112715.2 |
Uncharacterized protein |
59560 |
0.13 |
| chr3_57963869_57964020 | 0.21 |
FLNB |
filamin B, beta |
30183 |
0.2 |
| chr2_198836723_198836874 | 0.21 |
PLCL1 |
phospholipase C-like 1 |
111738 |
0.07 |
| chr2_210290098_210290249 | 0.21 |
MAP2 |
microtubule-associated protein 2 |
1391 |
0.61 |
| chr19_34724547_34724719 | 0.21 |
KIAA0355 |
KIAA0355 |
20809 |
0.21 |
| chr12_91522053_91522204 | 0.20 |
LUM |
lumican |
16520 |
0.21 |
| chr1_178065326_178065477 | 0.20 |
RASAL2 |
RAS protein activator like 2 |
2125 |
0.47 |
| chr18_65027187_65027338 | 0.20 |
ENSG00000265580 |
. |
63743 |
0.14 |
| chr11_117184410_117184561 | 0.20 |
CEP164 |
centrosomal protein 164kDa |
788 |
0.54 |
| chr19_10731127_10731278 | 0.20 |
SLC44A2 |
solute carrier family 44 (choline transporter), member 2 |
4732 |
0.12 |
| chr17_60904793_60905165 | 0.20 |
MARCH10 |
membrane-associated ring finger (C3HC4) 10, E3 ubiquitin protein ligase |
19274 |
0.23 |
| chr1_186157290_186157441 | 0.20 |
GS1-174L6.4 |
|
10931 |
0.22 |
| chr7_20259929_20260378 | 0.20 |
MACC1 |
metastasis associated in colon cancer 1 |
3126 |
0.32 |
| chr3_100008069_100008220 | 0.20 |
TBC1D23 |
TBC1 domain family, member 23 |
9860 |
0.2 |
| chr1_205715034_205715185 | 0.20 |
NUCKS1 |
nuclear casein kinase and cyclin-dependent kinase substrate 1 |
4295 |
0.17 |
| chr12_9100275_9100426 | 0.20 |
M6PR |
mannose-6-phosphate receptor (cation dependent) |
906 |
0.49 |
| chr9_97663549_97663700 | 0.20 |
RP11-49O14.2 |
|
913 |
0.6 |
| chr16_89467841_89467992 | 0.20 |
RP1-168P16.2 |
|
11056 |
0.14 |
| chr3_11427274_11427546 | 0.20 |
ATG7 |
autophagy related 7 |
21277 |
0.26 |
| chr14_24834583_24834734 | 0.20 |
NFATC4 |
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4 |
1459 |
0.21 |
| chr12_122430745_122430896 | 0.20 |
BCL7A |
B-cell CLL/lymphoma 7A |
26508 |
0.17 |
| chr2_46542258_46542445 | 0.19 |
EPAS1 |
endothelial PAS domain protein 1 |
17810 |
0.25 |
| chr15_101299874_101300025 | 0.19 |
ALDH1A3 |
aldehyde dehydrogenase 1 family, member A3 |
117970 |
0.05 |
| chr11_46974144_46974439 | 0.19 |
C11orf49 |
chromosome 11 open reading frame 49 |
15960 |
0.18 |
| chr4_81186341_81186492 | 0.19 |
FGF5 |
fibroblast growth factor 5 |
1337 |
0.53 |
| chr10_112062798_112063458 | 0.19 |
SMNDC1 |
survival motor neuron domain containing 1 |
1327 |
0.53 |
| chr19_16179828_16179979 | 0.19 |
TPM4 |
tropomyosin 4 |
1393 |
0.4 |
| chr1_100817300_100817579 | 0.19 |
CDC14A |
cell division cycle 14A |
149 |
0.96 |
| chr11_33714452_33714603 | 0.19 |
RP4-541C22.5 |
|
3720 |
0.2 |
| chr7_7744988_7745289 | 0.19 |
RPA3 |
replication protein A3, 14kDa |
13100 |
0.25 |
| chrX_55743062_55743213 | 0.19 |
RRAGB |
Ras-related GTP binding B |
1035 |
0.62 |
| chr2_167993645_167993796 | 0.19 |
XIRP2-AS1 |
XIRP2 antisense RNA 1 |
3745 |
0.34 |
| chr6_138976214_138976649 | 0.19 |
RP11-390P2.4 |
|
37254 |
0.17 |
| chr3_183563662_183564003 | 0.19 |
PARL |
presenilin associated, rhomboid-like |
1770 |
0.27 |
| chr5_57158791_57158942 | 0.19 |
ENSG00000266864 |
. |
10007 |
0.31 |
| chr12_52348007_52348213 | 0.19 |
ACVR1B |
activin A receptor, type IB |
946 |
0.5 |
| chr15_72525332_72525483 | 0.19 |
PKM |
pyruvate kinase, muscle |
1243 |
0.4 |
| chr3_141579230_141579381 | 0.19 |
ATP1B3 |
ATPase, Na+/K+ transporting, beta 3 polypeptide |
15661 |
0.18 |
| chr2_165752799_165752950 | 0.19 |
ENSG00000223318 |
. |
587 |
0.74 |
| chr20_43181909_43182060 | 0.19 |
PKIG |
protein kinase (cAMP-dependent, catalytic) inhibitor gamma |
21414 |
0.13 |
| chr3_149301783_149301934 | 0.19 |
WWTR1 |
WW domain containing transcription regulator 1 |
7819 |
0.21 |
| chr3_177246534_177246685 | 0.18 |
ENSG00000252028 |
. |
25269 |
0.26 |
| chr13_33847390_33847541 | 0.18 |
ENSG00000236581 |
. |
1878 |
0.38 |
| chr11_117020661_117020812 | 0.18 |
PAFAH1B2 |
platelet-activating factor acetylhydrolase 1b, catalytic subunit 2 (30kDa) |
5676 |
0.16 |
| chr3_99614299_99614817 | 0.18 |
FILIP1L |
filamin A interacting protein 1-like |
19512 |
0.23 |
| chr1_246150119_246150270 | 0.18 |
RP11-83A16.1 |
|
46659 |
0.18 |
| chr1_178061988_178062215 | 0.18 |
RASAL2 |
RAS protein activator like 2 |
763 |
0.79 |
| chr16_18970727_18970878 | 0.18 |
TMC7 |
transmembrane channel-like 7 |
24454 |
0.12 |
| chr2_67628597_67628898 | 0.18 |
ETAA1 |
Ewing tumor-associated antigen 1 |
4296 |
0.38 |
| chr20_36429033_36429188 | 0.18 |
CTNNBL1 |
catenin, beta like 1 |
23435 |
0.24 |
| chr10_17066021_17066526 | 0.18 |
CUBN |
cubilin (intrinsic factor-cobalamin receptor) |
40079 |
0.2 |
| chr17_11939642_11939793 | 0.18 |
ENSG00000252707 |
. |
237 |
0.92 |
| chr2_40079075_40079226 | 0.18 |
SLC8A1-AS1 |
SLC8A1 antisense RNA 1 |
65557 |
0.13 |
| chr2_174985605_174985756 | 0.18 |
OLA1 |
Obg-like ATPase 1 |
96994 |
0.07 |
| chr4_26621376_26621527 | 0.18 |
ENSG00000265627 |
. |
17661 |
0.22 |
| chr5_105728908_105729059 | 0.18 |
ENSG00000252337 |
. |
153383 |
0.05 |
| chr3_187663809_187663960 | 0.18 |
BCL6 |
B-cell CLL/lymphoma 6 |
200369 |
0.02 |
| chr18_22566959_22567110 | 0.18 |
ZNF521 |
zinc finger protein 521 |
237719 |
0.02 |
| chr7_129599031_129599182 | 0.18 |
ENSG00000207184 |
. |
2810 |
0.17 |
| chr3_45052908_45053120 | 0.18 |
CLEC3B |
C-type lectin domain family 3, member B |
14661 |
0.15 |
| chr5_125786033_125786184 | 0.18 |
GRAMD3 |
GRAM domain containing 3 |
11929 |
0.22 |
| chr13_98036472_98036623 | 0.18 |
ENSG00000202290 |
. |
21192 |
0.2 |
| chr1_41449008_41449159 | 0.18 |
CTPS1 |
CTP synthase 1 |
250 |
0.94 |
| chr15_76348972_76349262 | 0.18 |
NRG4 |
neuregulin 4 |
2952 |
0.25 |
| chr8_103252757_103253104 | 0.18 |
KB-431C1.4 |
|
1308 |
0.34 |
| chr5_14264414_14265476 | 0.18 |
TRIO |
trio Rho guanine nucleotide exchange factor |
26141 |
0.27 |
| chr11_10322719_10322870 | 0.18 |
RP11-351I24.1 |
|
2457 |
0.22 |
| chr14_68698275_68698426 | 0.18 |
ENSG00000243546 |
. |
4906 |
0.3 |
| chr15_41958458_41958609 | 0.18 |
MGA |
MGA, MAX dimerization protein |
2493 |
0.26 |
| chr4_57915542_57915693 | 0.18 |
ENSG00000251703 |
. |
46759 |
0.1 |
| chr15_71341987_71342266 | 0.18 |
THSD4 |
thrombospondin, type I, domain containing 4 |
47165 |
0.15 |
| chr1_234752857_234753272 | 0.17 |
IRF2BP2 |
interferon regulatory factor 2 binding protein 2 |
7793 |
0.19 |
| chr17_58439093_58439244 | 0.17 |
USP32 |
ubiquitin specific peptidase 32 |
30168 |
0.17 |
| chr9_84386849_84387000 | 0.17 |
ENSG00000263404 |
. |
34072 |
0.17 |
| chr2_73207406_73207767 | 0.17 |
EMX1 |
empty spiracles homeobox 1 |
56469 |
0.12 |
| chr1_8065443_8065723 | 0.17 |
ERRFI1 |
ERBB receptor feedback inhibitor 1 |
10110 |
0.18 |
| chr12_19174986_19175137 | 0.17 |
PLEKHA5 |
pleckstrin homology domain containing, family A member 5 |
107587 |
0.07 |
| chr16_85470909_85471082 | 0.17 |
ENSG00000264203 |
. |
4103 |
0.3 |
| chr8_122931309_122931460 | 0.17 |
HAS2-AS1 |
HAS2 antisense RNA 1 |
277708 |
0.01 |
| chr12_50940008_50940333 | 0.17 |
DIP2B |
DIP2 disco-interacting protein 2 homolog B (Drosophila) |
41280 |
0.15 |
| chr9_12718941_12719092 | 0.17 |
TYRP1 |
tyrosinase-related protein 1 |
23296 |
0.2 |
| chr5_131750080_131750814 | 0.17 |
C5orf56 |
chromosome 5 open reading frame 56 |
3766 |
0.16 |
| chr14_69535729_69536049 | 0.17 |
ENSG00000206768 |
. |
74781 |
0.09 |
| chr15_33494490_33494641 | 0.17 |
FMN1 |
formin 1 |
7668 |
0.29 |
| chr1_154152302_154152968 | 0.17 |
TPM3 |
tropomyosin 3 |
1976 |
0.18 |
| chr15_51203212_51203412 | 0.17 |
AP4E1 |
adaptor-related protein complex 4, epsilon 1 subunit |
2320 |
0.4 |
| chr4_169540123_169540274 | 0.17 |
PALLD |
palladin, cytoskeletal associated protein |
12568 |
0.21 |
| chr4_114880757_114880908 | 0.17 |
RP11-26P13.2 |
|
16420 |
0.24 |
| chr8_104293348_104293656 | 0.17 |
RP11-318M2.2 |
|
4628 |
0.2 |
| chr1_117601173_117601556 | 0.17 |
RP11-27K13.3 |
|
748 |
0.55 |
| chr8_89288165_89288316 | 0.17 |
RP11-586K2.1 |
|
50825 |
0.16 |
| chr13_33836579_33836730 | 0.16 |
ENSG00000236581 |
. |
8862 |
0.23 |
| chr6_143050462_143050730 | 0.16 |
RP1-67K17.3 |
|
18984 |
0.27 |
| chr7_65760902_65761053 | 0.16 |
ENSG00000252126 |
. |
48314 |
0.13 |
| chr6_145276841_145276992 | 0.16 |
UTRN |
utrophin |
156399 |
0.04 |
| chr4_170174772_170174923 | 0.16 |
SH3RF1 |
SH3 domain containing ring finger 1 |
16261 |
0.25 |
| chr8_28982615_28982843 | 0.16 |
CTD-2647L4.1 |
|
14639 |
0.15 |
| chr7_130539521_130539776 | 0.16 |
ENSG00000226380 |
. |
21932 |
0.23 |
| chr7_77496191_77496692 | 0.16 |
PHTF2 |
putative homeodomain transcription factor 2 |
26944 |
0.18 |
| chr1_178207539_178207690 | 0.16 |
RASAL2 |
RAS protein activator like 2 |
102992 |
0.08 |
| chr15_39288838_39288989 | 0.16 |
RP11-624L4.1 |
|
122001 |
0.06 |
| chr3_71085056_71085207 | 0.16 |
FOXP1 |
forkhead box P1 |
28946 |
0.25 |
| chr1_7283919_7284070 | 0.16 |
ENSG00000207056 |
. |
4574 |
0.33 |
| chr4_107506143_107506294 | 0.16 |
ENSG00000222333 |
. |
169276 |
0.04 |
| chr8_13124406_13124699 | 0.16 |
DLC1 |
deleted in liver cancer 1 |
9503 |
0.27 |
| chr2_171787186_171787958 | 0.16 |
GORASP2 |
golgi reassembly stacking protein 2, 55kDa |
1741 |
0.39 |
| chr4_174411815_174412157 | 0.16 |
HAND2-AS1 |
HAND2 antisense RNA 1 (head to head) |
36435 |
0.14 |
| chr6_81258118_81258393 | 0.16 |
BCKDHB |
branched chain keto acid dehydrogenase E1, beta polypeptide |
441872 |
0.01 |
| chr6_16682722_16682873 | 0.16 |
RP1-151F17.1 |
|
78572 |
0.11 |
| chr6_157223655_157223806 | 0.16 |
ARID1B |
AT rich interactive domain 1B (SWI1-like) |
1223 |
0.62 |
| chr3_149469081_149469660 | 0.16 |
COMMD2 |
COMM domain containing 2 |
916 |
0.55 |
| chrX_13450415_13450566 | 0.16 |
ATXN3L |
ataxin 3-like |
111972 |
0.06 |
| chr11_88288822_88288973 | 0.16 |
GRM5-AS1 |
GRM5 antisense RNA 1 |
43981 |
0.18 |
| chr7_129551821_129551972 | 0.16 |
UBE2H |
ubiquitin-conjugating enzyme E2H |
39271 |
0.11 |
| chr3_25834915_25835066 | 0.16 |
OXSM |
3-oxoacyl-ACP synthase, mitochondrial |
2713 |
0.25 |
| chr6_37212230_37212381 | 0.16 |
ENSG00000238375 |
. |
6777 |
0.17 |
| chr6_51167339_51167725 | 0.16 |
ENSG00000212532 |
. |
161956 |
0.04 |
| chr4_55529764_55530043 | 0.16 |
KIT |
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog |
5818 |
0.34 |
| chr4_90741767_90741918 | 0.16 |
SNCA |
synuclein, alpha (non A4 component of amyloid precursor) |
14989 |
0.21 |
| chr5_77973860_77974011 | 0.16 |
LHFPL2 |
lipoma HMGIC fusion partner-like 2 |
29287 |
0.25 |
| chr4_41146468_41146619 | 0.16 |
ENSG00000207198 |
. |
30584 |
0.17 |
| chr12_49664011_49664162 | 0.16 |
RP11-161H23.5 |
|
3003 |
0.15 |
| chr5_139555723_139556039 | 0.16 |
CYSTM1 |
cysteine-rich transmembrane module containing 1 |
1654 |
0.3 |
| chr11_130819091_130819698 | 0.16 |
SNX19 |
sorting nexin 19 |
32990 |
0.21 |
| chr7_21583848_21583999 | 0.16 |
DNAH11 |
dynein, axonemal, heavy chain 11 |
1090 |
0.63 |
| chr3_187980787_187980938 | 0.16 |
LPP |
LIM domain containing preferred translocation partner in lipoma |
23210 |
0.25 |
| chr3_71635193_71635783 | 0.16 |
FOXP1 |
forkhead box P1 |
2348 |
0.32 |
| chrX_4628432_4628686 | 0.15 |
ENSG00000222834 |
. |
8396 |
0.33 |
| chr7_75696719_75696870 | 0.15 |
MDH2 |
malate dehydrogenase 2, NAD (mitochondrial) |
19315 |
0.12 |
| chr2_211340830_211341097 | 0.15 |
LANCL1 |
LanC lantibiotic synthetase component C-like 1 (bacterial) |
466 |
0.76 |
| chr4_177927532_177927683 | 0.15 |
ENSG00000222859 |
. |
174989 |
0.03 |
| chr12_76271999_76272150 | 0.15 |
ENSG00000243420 |
. |
79836 |
0.1 |
| chr16_50229405_50229706 | 0.15 |
PAPD5 |
PAP associated domain containing 5 |
41776 |
0.12 |
| chr7_120938645_120938796 | 0.15 |
WNT16 |
wingless-type MMTV integration site family, member 16 |
26701 |
0.23 |
| chr2_159950478_159950629 | 0.15 |
ENSG00000202029 |
. |
66899 |
0.12 |
| chr20_39686412_39686563 | 0.15 |
TOP1 |
topoisomerase (DNA) I |
29029 |
0.17 |
| chr4_19833823_19833974 | 0.15 |
SLIT2 |
slit homolog 2 (Drosophila) |
420985 |
0.01 |
| chr13_20699448_20699599 | 0.15 |
GJA3 |
gap junction protein, alpha 3, 46kDa |
35665 |
0.16 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.0 | 0.1 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.1 | GO:0035385 | Roundabout signaling pathway(GO:0035385) |
| 0.0 | 0.1 | GO:0046881 | positive regulation of gonadotropin secretion(GO:0032278) positive regulation of follicle-stimulating hormone secretion(GO:0046881) |
| 0.0 | 0.1 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.1 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.0 | 0.0 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.0 | 0.1 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.0 | 0.1 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.0 | 0.1 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.1 | GO:0009128 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.0 | 0.0 | GO:0051832 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.0 | 0.1 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.0 | 0.1 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.1 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.0 | 0.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.1 | GO:1901532 | regulation of megakaryocyte differentiation(GO:0045652) regulation of hematopoietic progenitor cell differentiation(GO:1901532) |
| 0.0 | 0.0 | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.0 | 0.1 | GO:0045542 | positive regulation of cholesterol biosynthetic process(GO:0045542) |
| 0.0 | 0.1 | GO:0010586 | miRNA metabolic process(GO:0010586) miRNA catabolic process(GO:0010587) |
| 0.0 | 0.1 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 0.1 | GO:0045112 | integrin biosynthetic process(GO:0045112) |
| 0.0 | 0.1 | GO:2000178 | negative regulation of neuroblast proliferation(GO:0007406) negative regulation of neural precursor cell proliferation(GO:2000178) |
| 0.0 | 0.1 | GO:0042663 | endodermal cell fate specification(GO:0001714) regulation of endodermal cell fate specification(GO:0042663) regulation of endodermal cell differentiation(GO:1903224) |
| 0.0 | 0.1 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.1 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.0 | 0.0 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.1 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.0 | 0.0 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.1 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 | 0.1 | GO:0060839 | endothelial cell fate commitment(GO:0060839) |
| 0.0 | 0.0 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.0 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.0 | 0.1 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.1 | GO:0060710 | chorio-allantoic fusion(GO:0060710) |
| 0.0 | 0.0 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.0 | 0.1 | GO:0031657 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G1/S transition of mitotic cell cycle(GO:0031657) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G1/S transition of mitotic cell cycle(GO:0031659) |
| 0.0 | 0.1 | GO:0000052 | citrulline metabolic process(GO:0000052) |
| 0.0 | 0.1 | GO:0061162 | establishment of apical/basal cell polarity(GO:0035089) establishment of monopolar cell polarity(GO:0061162) establishment or maintenance of monopolar cell polarity(GO:0061339) |
| 0.0 | 0.1 | GO:0006264 | mitochondrial DNA replication(GO:0006264) |
| 0.0 | 0.0 | GO:0014874 | response to stimulus involved in regulation of muscle adaptation(GO:0014874) |
| 0.0 | 0.1 | GO:0060390 | regulation of SMAD protein import into nucleus(GO:0060390) |
| 0.0 | 0.0 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.0 | 0.1 | GO:0051124 | synaptic growth at neuromuscular junction(GO:0051124) |
| 0.0 | 0.0 | GO:0002072 | optic cup morphogenesis involved in camera-type eye development(GO:0002072) |
| 0.0 | 0.1 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.1 | GO:0090322 | regulation of superoxide metabolic process(GO:0090322) |
| 0.0 | 0.0 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.0 | 0.1 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.1 | GO:0022038 | corpus callosum development(GO:0022038) |
| 0.0 | 0.0 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.0 | 0.1 | GO:0032776 | DNA methylation on cytosine(GO:0032776) C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 0.0 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.1 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.1 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.0 | GO:0050703 | interleukin-1 alpha secretion(GO:0050703) |
| 0.0 | 0.1 | GO:0043312 | neutrophil activation involved in immune response(GO:0002283) neutrophil degranulation(GO:0043312) |
| 0.0 | 0.1 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.0 | 0.1 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 0.1 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.1 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.0 | GO:0061072 | iris morphogenesis(GO:0061072) |
| 0.0 | 0.1 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.0 | 0.0 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.0 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) positive regulation of signal transduction by p53 class mediator(GO:1901798) |
| 0.0 | 0.0 | GO:0008300 | isoprenoid catabolic process(GO:0008300) |
| 0.0 | 0.0 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.0 | GO:0070272 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.0 | 0.0 | GO:0070933 | histone H3 deacetylation(GO:0070932) histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.0 | GO:0035929 | steroid hormone secretion(GO:0035929) |
| 0.0 | 0.1 | GO:0006266 | DNA ligation(GO:0006266) |
| 0.0 | 0.0 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.1 | GO:0090399 | replicative senescence(GO:0090399) |
| 0.0 | 0.0 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.0 | GO:0061029 | eyelid development in camera-type eye(GO:0061029) |
| 0.0 | 0.2 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.1 | GO:0060261 | positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 0.0 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.1 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.0 | 0.0 | GO:0002138 | retinoic acid biosynthetic process(GO:0002138) diterpenoid biosynthetic process(GO:0016102) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.1 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.1 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.1 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.0 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.1 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.0 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 0.0 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.2 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.0 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.1 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.0 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.1 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.0 | 0.1 | GO:0043256 | laminin complex(GO:0043256) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.1 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.0 | 0.1 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.1 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.1 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.2 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.1 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.1 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.1 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.2 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.1 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.1 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.1 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.1 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.1 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.2 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.1 | GO:0016972 | thiol oxidase activity(GO:0016972) |
| 0.0 | 0.1 | GO:1900750 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.0 | 0.0 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.0 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.1 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.0 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.0 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.0 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 0.1 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.0 | 0.1 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.1 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.1 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.2 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.0 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.0 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.0 | 0.0 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.0 | 0.1 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.1 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 0.1 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.1 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.1 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.1 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.0 | 0.0 | GO:0033765 | steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) |
| 0.0 | 0.1 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.1 | GO:0070717 | poly-purine tract binding(GO:0070717) |
| 0.0 | 0.0 | GO:0016901 | oxidoreductase activity, acting on the CH-OH group of donors, quinone or similar compound as acceptor(GO:0016901) |
| 0.0 | 0.0 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.0 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.0 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.0 | 0.0 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.0 | 0.1 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.0 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.2 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 0.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.2 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.3 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.0 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.1 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.1 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |