Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
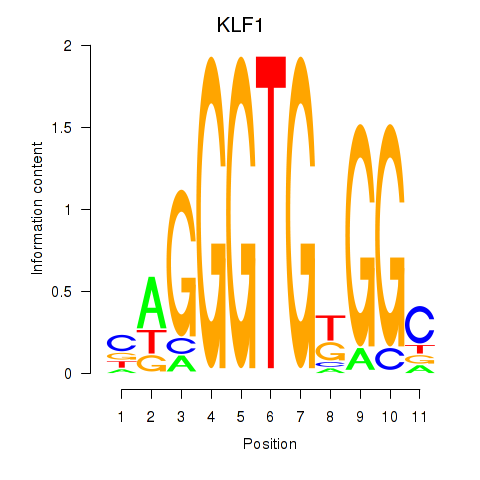
Results for KLF1
Z-value: 2.06
Transcription factors associated with KLF1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
KLF1
|
ENSG00000105610.4 | Kruppel like factor 1 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr19_12996557_12997061 | KLF1 | 1186 | 0.201684 | 0.66 | 5.4e-02 | Click! |
| chr19_12996010_12996205 | KLF1 | 1888 | 0.120145 | -0.48 | 1.9e-01 | Click! |
| chr19_12995388_12995986 | KLF1 | 2308 | 0.099733 | 0.32 | 4.0e-01 | Click! |
| chr19_12997876_12998027 | KLF1 | 44 | 0.918445 | -0.23 | 5.6e-01 | Click! |
| chr19_12996291_12996442 | KLF1 | 1629 | 0.140044 | -0.13 | 7.3e-01 | Click! |
Activity of the KLF1 motif across conditions
Conditions sorted by the z-value of the KLF1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
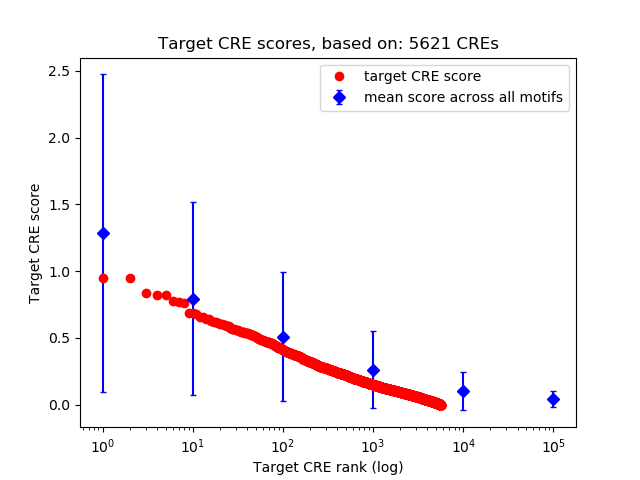
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr7_158656716_158656867 | 0.95 |
WDR60 |
WD repeat domain 60 |
5346 |
0.27 |
| chr2_237657369_237658039 | 0.95 |
ACKR3 |
atypical chemokine receptor 3 |
179420 |
0.03 |
| chr1_154979701_154979987 | 0.84 |
ZBTB7B |
zinc finger and BTB domain containing 7B |
4549 |
0.08 |
| chr19_13113558_13113812 | 0.82 |
NFIX |
nuclear factor I/X (CCAAT-binding transcription factor) |
7033 |
0.09 |
| chr1_21629611_21629762 | 0.82 |
ECE1 |
endothelin converting enzyme 1 |
4194 |
0.22 |
| chr2_86055197_86055348 | 0.77 |
ENSG00000252321 |
. |
7537 |
0.17 |
| chr9_137444423_137444866 | 0.77 |
COL5A1 |
collagen, type V, alpha 1 |
88976 |
0.08 |
| chr3_47447809_47448062 | 0.76 |
PTPN23 |
protein tyrosine phosphatase, non-receptor type 23 |
25425 |
0.13 |
| chr11_7686997_7687148 | 0.69 |
CYB5R2 |
cytochrome b5 reductase 2 |
7631 |
0.17 |
| chr20_35504605_35504756 | 0.69 |
TLDC2 |
TBC/LysM-associated domain containing 2 |
146 |
0.95 |
| chr1_225925038_225925189 | 0.68 |
RP11-145A3.2 |
|
1137 |
0.42 |
| chr17_79064346_79064714 | 0.66 |
BAIAP2 |
BAI1-associated protein 2 |
4269 |
0.14 |
| chr1_6238952_6239103 | 0.65 |
CHD5 |
chromodomain helicase DNA binding protein 5 |
1156 |
0.38 |
| chr6_36684328_36684602 | 0.64 |
RAB44 |
RAB44, member RAS oncogene family |
1209 |
0.43 |
| chr11_119560225_119560376 | 0.64 |
ENSG00000199217 |
. |
33279 |
0.14 |
| chr8_22847555_22847827 | 0.63 |
RHOBTB2 |
Rho-related BTB domain containing 2 |
2696 |
0.18 |
| chr8_143758469_143758620 | 0.62 |
PSCA |
prostate stem cell antigen |
3330 |
0.15 |
| chr5_149887043_149887237 | 0.62 |
NDST1 |
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
534 |
0.77 |
| chr22_18433540_18433818 | 0.61 |
ENSG00000207780 |
. |
30048 |
0.14 |
| chr22_37905149_37905300 | 0.61 |
CARD10 |
caspase recruitment domain family, member 10 |
68 |
0.96 |
| chr10_79321861_79322012 | 0.60 |
ENSG00000199592 |
. |
24871 |
0.23 |
| chr10_126029743_126029894 | 0.60 |
OAT |
ornithine aminotransferase |
77687 |
0.1 |
| chr1_94204227_94204378 | 0.59 |
RP11-488P3.1 |
|
36698 |
0.17 |
| chr19_9957964_9958210 | 0.59 |
ENSG00000266247 |
. |
4017 |
0.11 |
| chr2_241391028_241391179 | 0.59 |
GPC1 |
glypican 1 |
1125 |
0.42 |
| chr9_130298577_130298728 | 0.58 |
FAM129B |
family with sequence similarity 129, member B |
32715 |
0.13 |
| chr3_48538894_48539045 | 0.57 |
SHISA5 |
shisa family member 5 |
2593 |
0.15 |
| chr5_16820384_16820535 | 0.57 |
ENSG00000253093 |
. |
33644 |
0.2 |
| chr1_94511054_94511205 | 0.57 |
ABCA4 |
ATP-binding cassette, sub-family A (ABC1), member 4 |
26901 |
0.2 |
| chr1_32155176_32155569 | 0.56 |
COL16A1 |
collagen, type XVI, alpha 1 |
6023 |
0.15 |
| chr16_89436687_89437018 | 0.56 |
ANKRD11 |
ankyrin repeat domain 11 |
41458 |
0.09 |
| chr21_44465729_44466640 | 0.56 |
CBS |
cystathionine-beta-synthase |
12867 |
0.18 |
| chr2_20412540_20412691 | 0.55 |
SDC1 |
syndecan 1 |
11426 |
0.17 |
| chr17_26439314_26439465 | 0.55 |
ENSG00000252283 |
. |
50396 |
0.1 |
| chr17_76405529_76405680 | 0.55 |
PGS1 |
phosphatidylglycerophosphate synthase 1 |
5559 |
0.15 |
| chr1_2431468_2431763 | 0.55 |
RP3-395M20.3 |
|
5697 |
0.11 |
| chr2_119967496_119967647 | 0.54 |
STEAP3 |
STEAP family member 3, metalloreductase |
13813 |
0.19 |
| chr17_72364891_72365060 | 0.54 |
GPR142 |
G protein-coupled receptor 142 |
1378 |
0.3 |
| chr1_205249223_205249374 | 0.54 |
TMCC2 |
transmembrane and coiled-coil domain family 2 |
23969 |
0.14 |
| chr6_33546571_33546722 | 0.54 |
BAK1 |
BCL2-antagonist/killer 1 |
1335 |
0.33 |
| chr10_50016038_50016189 | 0.53 |
WDFY4 |
WDFY family member 4 |
5330 |
0.26 |
| chr17_16977412_16977763 | 0.53 |
MPRIP-AS1 |
MPRIP antisense RNA 1 |
3479 |
0.23 |
| chr10_50398424_50398575 | 0.53 |
C10orf128 |
chromosome 10 open reading frame 128 |
2063 |
0.33 |
| chr11_67190760_67190911 | 0.52 |
PPP1CA |
protein phosphatase 1, catalytic subunit, alpha isozyme |
2181 |
0.11 |
| chr8_144657419_144657635 | 0.52 |
RP11-661A12.9 |
|
1867 |
0.12 |
| chr3_195949731_195949882 | 0.52 |
SLC51A |
solute carrier family 51, alpha subunit |
5043 |
0.14 |
| chr20_62317786_62317937 | 0.52 |
RTEL1 |
regulator of telomere elongation helicase 1 |
1831 |
0.18 |
| chr12_122235279_122235721 | 0.51 |
RHOF |
ras homolog family member F (in filopodia) |
3083 |
0.21 |
| chr10_82245535_82245824 | 0.51 |
TSPAN14 |
tetraspanin 14 |
26621 |
0.17 |
| chr2_144480307_144480458 | 0.51 |
RP11-434H14.1 |
|
18481 |
0.21 |
| chr13_27883554_27883724 | 0.51 |
ENSG00000221756 |
. |
10511 |
0.14 |
| chr9_124396687_124396838 | 0.50 |
DAB2IP |
DAB2 interacting protein |
17111 |
0.22 |
| chr16_89699348_89699594 | 0.50 |
DPEP1 |
dipeptidase 1 (renal) |
2774 |
0.15 |
| chr6_138213965_138214116 | 0.49 |
RP11-356I2.4 |
|
24670 |
0.2 |
| chr12_122468255_122468406 | 0.49 |
BCL7A |
B-cell CLL/lymphoma 7A |
8538 |
0.22 |
| chr8_144302255_144302406 | 0.49 |
GPIHBP1 |
glycosylphosphatidylinositol anchored high density lipoprotein binding protein 1 |
7262 |
0.14 |
| chr8_129789388_129789539 | 0.49 |
ENSG00000221351 |
. |
42577 |
0.22 |
| chr17_79386350_79386501 | 0.48 |
ENSG00000266392 |
. |
11847 |
0.11 |
| chr9_132465308_132465459 | 0.48 |
RP11-483H20.6 |
|
15531 |
0.14 |
| chr2_217501804_217502154 | 0.48 |
IGFBP2 |
insulin-like growth factor binding protein 2, 36kDa |
3861 |
0.22 |
| chr12_111831727_111831878 | 0.48 |
SH2B3 |
SH2B adaptor protein 3 |
11950 |
0.18 |
| chr11_2734182_2734333 | 0.48 |
KCNQ1OT1 |
KCNQ1 opposite strand/antisense transcript 1 (non-protein coding) |
13033 |
0.24 |
| chr11_72902349_72902500 | 0.47 |
RP11-800A3.2 |
|
24534 |
0.14 |
| chr19_40440819_40441084 | 0.47 |
FCGBP |
Fc fragment of IgG binding protein |
418 |
0.82 |
| chr3_124880829_124880980 | 0.47 |
ENSG00000264986 |
. |
10508 |
0.19 |
| chr18_32989920_32990179 | 0.47 |
ZNF396 |
zinc finger protein 396 |
32748 |
0.17 |
| chr16_3041774_3041925 | 0.47 |
PKMYT1 |
protein kinase, membrane associated tyrosine/threonine 1 |
11309 |
0.06 |
| chr11_2411400_2411766 | 0.47 |
CD81 |
CD81 molecule |
258 |
0.86 |
| chr9_130906009_130906346 | 0.47 |
LCN2 |
lipocalin 2 |
5173 |
0.09 |
| chr17_2115110_2115261 | 0.47 |
SMG6 |
SMG6 nonsense mediated mRNA decay factor |
846 |
0.42 |
| chr7_1164785_1164936 | 0.47 |
C7orf50 |
chromosome 7 open reading frame 50 |
13016 |
0.12 |
| chr9_137247824_137248108 | 0.46 |
ENSG00000263897 |
. |
23291 |
0.21 |
| chr10_49196316_49196467 | 0.46 |
FAM25C |
family with sequence similarity 25, member C |
11427 |
0.23 |
| chr9_131398817_131398968 | 0.46 |
WDR34 |
WD repeat domain 34 |
20068 |
0.11 |
| chr2_64558219_64558458 | 0.46 |
ENSG00000264297 |
. |
9555 |
0.26 |
| chr10_45476036_45476374 | 0.46 |
C10orf10 |
chromosome 10 open reading frame 10 |
1947 |
0.22 |
| chr17_76247695_76247857 | 0.45 |
TMEM235 |
transmembrane protein 235 |
19654 |
0.13 |
| chr11_12112374_12112662 | 0.45 |
MICAL2 |
microtubule associated monooxygenase, calponin and LIM domain containing 2 |
3025 |
0.36 |
| chr1_36771499_36771945 | 0.45 |
SH3D21 |
SH3 domain containing 21 |
266 |
0.89 |
| chr1_64179739_64179890 | 0.44 |
ENSG00000244256 |
. |
58400 |
0.12 |
| chr15_86315521_86315672 | 0.44 |
ENSG00000221634 |
. |
1787 |
0.23 |
| chr7_158750678_158750919 | 0.44 |
ENSG00000231419 |
. |
50314 |
0.16 |
| chr8_67030629_67031150 | 0.44 |
TRIM55 |
tripartite motif containing 55 |
8242 |
0.26 |
| chr9_136926080_136926374 | 0.44 |
BRD3 |
bromodomain containing 3 |
5093 |
0.18 |
| chr10_125846427_125846578 | 0.44 |
CHST15 |
carbohydrate (N-acetylgalactosamine 4-sulfate 6-O) sulfotransferase 15 |
5468 |
0.31 |
| chr6_35460175_35460326 | 0.43 |
TEAD3 |
TEA domain family member 3 |
4477 |
0.19 |
| chr5_149903159_149903581 | 0.43 |
NDST1 |
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
15696 |
0.16 |
| chr1_183221071_183221222 | 0.43 |
NMNAT2 |
nicotinamide nucleotide adenylyltransferase 2 |
52863 |
0.14 |
| chr10_82264968_82265119 | 0.43 |
RP11-137H2.4 |
|
30655 |
0.16 |
| chr2_28600761_28600914 | 0.43 |
FOSL2 |
FOS-like antigen 2 |
14832 |
0.16 |
| chr18_74779636_74780174 | 0.43 |
MBP |
myelin basic protein |
37312 |
0.2 |
| chr8_25088407_25088558 | 0.43 |
DOCK5 |
dedicator of cytokinesis 5 |
46102 |
0.16 |
| chr17_71537670_71537821 | 0.43 |
RP11-449L23.3 |
|
21760 |
0.2 |
| chr20_60926120_60926271 | 0.42 |
RP11-157P1.5 |
|
1856 |
0.23 |
| chr12_6321121_6321344 | 0.42 |
CD9 |
CD9 molecule |
11269 |
0.18 |
| chr5_130869288_130869613 | 0.42 |
RAPGEF6 |
Rap guanine nucleotide exchange factor (GEF) 6 |
724 |
0.8 |
| chr11_65257627_65258285 | 0.42 |
SCYL1 |
SCY1-like 1 (S. cerevisiae) |
34592 |
0.08 |
| chr12_123619407_123619701 | 0.42 |
PITPNM2 |
phosphatidylinositol transfer protein, membrane-associated 2 |
15008 |
0.17 |
| chr2_68942118_68942311 | 0.41 |
ARHGAP25 |
Rho GTPase activating protein 25 |
4580 |
0.29 |
| chr2_134938887_134939038 | 0.41 |
ENSG00000222921 |
. |
49804 |
0.15 |
| chr14_35839032_35839183 | 0.41 |
NFKBIA |
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha |
34239 |
0.18 |
| chr1_43472890_43473041 | 0.41 |
ENSG00000207256 |
. |
15856 |
0.16 |
| chr16_57418424_57418638 | 0.41 |
CX3CL1 |
chemokine (C-X3-C motif) ligand 1 |
7647 |
0.14 |
| chr2_102603897_102604048 | 0.41 |
IL1R2 |
interleukin 1 receptor, type II |
4334 |
0.3 |
| chr10_80908519_80908825 | 0.40 |
ZMIZ1 |
zinc finger, MIZ-type containing 1 |
79880 |
0.1 |
| chr14_77512064_77512283 | 0.40 |
IRF2BPL |
interferon regulatory factor 2 binding protein-like |
17139 |
0.17 |
| chr11_64951007_64951158 | 0.40 |
CAPN1 |
calpain 1, (mu/I) large subunit |
275 |
0.8 |
| chr10_129843874_129844131 | 0.40 |
PTPRE |
protein tyrosine phosphatase, receptor type, E |
1668 |
0.5 |
| chr17_7348880_7349137 | 0.40 |
CHRNB1 |
cholinergic receptor, nicotinic, beta 1 (muscle) |
158 |
0.84 |
| chr20_4878879_4879030 | 0.40 |
SLC23A2 |
solute carrier family 23 (ascorbic acid transporter), member 2 |
1339 |
0.42 |
| chr17_201097_201248 | 0.39 |
RP11-1260E13.2 |
|
1257 |
0.28 |
| chr9_137248525_137248692 | 0.39 |
ENSG00000263897 |
. |
22649 |
0.21 |
| chr8_103757441_103757701 | 0.39 |
ENSG00000266799 |
. |
11624 |
0.23 |
| chr16_2173270_2173421 | 0.39 |
RP11-304L19.2 |
|
4137 |
0.06 |
| chr10_81915562_81915788 | 0.39 |
ANXA11 |
annexin A11 |
6093 |
0.21 |
| chr16_11654458_11655352 | 0.39 |
LITAF |
lipopolysaccharide-induced TNF factor |
25324 |
0.15 |
| chr3_45062662_45062813 | 0.39 |
CLEC3B |
C-type lectin domain family 3, member B |
4938 |
0.19 |
| chr15_100016511_100016662 | 0.39 |
MEF2A |
myocyte enhancer factor 2A |
784 |
0.68 |
| chr3_48636605_48636768 | 0.39 |
COL7A1 |
collagen, type VII, alpha 1 |
3986 |
0.11 |
| chr2_15829588_15829739 | 0.39 |
AC008278.3 |
|
9055 |
0.22 |
| chr4_143321954_143322429 | 0.38 |
INPP4B |
inositol polyphosphate-4-phosphatase, type II, 105kDa |
30221 |
0.27 |
| chr8_22439472_22439623 | 0.38 |
PDLIM2 |
PDZ and LIM domain 2 (mystique) |
1505 |
0.23 |
| chr20_18688451_18688602 | 0.38 |
RP11-379J5.5 |
|
9173 |
0.23 |
| chr9_114614005_114614347 | 0.38 |
ENSG00000206907 |
. |
12068 |
0.2 |
| chr6_39171595_39171746 | 0.38 |
KCNK5 |
potassium channel, subfamily K, member 5 |
25556 |
0.22 |
| chr22_39486625_39486957 | 0.38 |
RP4-742C19.12 |
|
800 |
0.52 |
| chr8_141146867_141147018 | 0.38 |
C8orf17 |
chromosome 8 open reading frame 17 |
203526 |
0.03 |
| chr22_38291291_38291442 | 0.38 |
ENSG00000207227 |
. |
4012 |
0.12 |
| chr8_28246819_28246970 | 0.38 |
ZNF395 |
zinc finger protein 395 |
2911 |
0.2 |
| chr18_43232666_43232907 | 0.38 |
RP11-116O18.1 |
|
6668 |
0.21 |
| chr9_124497789_124497940 | 0.38 |
DAB2IP |
DAB2 interacting protein |
7200 |
0.28 |
| chr5_131590562_131590713 | 0.38 |
PDLIM4 |
PDZ and LIM domain 4 |
2727 |
0.23 |
| chr1_161170632_161171366 | 0.38 |
NDUFS2 |
NADH dehydrogenase (ubiquinone) Fe-S protein 2, 49kDa (NADH-coenzyme Q reductase) |
938 |
0.27 |
| chr10_47170047_47170291 | 0.37 |
ANXA8L1 |
annexin A8-like 1 |
3825 |
0.18 |
| chrX_153632756_153632907 | 0.37 |
DNASE1L1 |
deoxyribonuclease I-like 1 |
1120 |
0.25 |
| chr10_105210970_105211151 | 0.37 |
CALHM2 |
calcium homeostasis modulator 2 |
1015 |
0.26 |
| chr3_49943488_49944003 | 0.37 |
CTD-2330K9.3 |
Uncharacterized protein |
2325 |
0.15 |
| chr19_14608235_14608386 | 0.37 |
GIPC1 |
GIPC PDZ domain containing family, member 1 |
1366 |
0.27 |
| chr1_46668774_46668925 | 0.37 |
LURAP1 |
leucine rich adaptor protein 1 |
157 |
0.93 |
| chr11_119584963_119585114 | 0.37 |
PVRL1 |
poliovirus receptor-related 1 (herpesvirus entry mediator C) |
14225 |
0.19 |
| chr19_16994459_16994610 | 0.37 |
F2RL3 |
coagulation factor II (thrombin) receptor-like 3 |
5137 |
0.13 |
| chr14_65142113_65142264 | 0.37 |
PLEKHG3 |
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
28632 |
0.17 |
| chr19_15947461_15947612 | 0.37 |
OR10H1 |
olfactory receptor, family 10, subfamily H, member 1 |
28600 |
0.14 |
| chr11_57125235_57125386 | 0.37 |
ENSG00000266018 |
. |
1886 |
0.18 |
| chr17_74486749_74486900 | 0.37 |
RHBDF2 |
rhomboid 5 homolog 2 (Drosophila) |
2412 |
0.17 |
| chr7_2549413_2549701 | 0.37 |
LFNG |
LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
2606 |
0.23 |
| chr3_182792190_182792488 | 0.37 |
MCCC1 |
methylcrotonoyl-CoA carboxylase 1 (alpha) |
24967 |
0.19 |
| chr14_24783456_24784225 | 0.37 |
LTB4R |
leukotriene B4 receptor |
66 |
0.91 |
| chr12_110278481_110278632 | 0.36 |
TRPV4 |
transient receptor potential cation channel, subfamily V, member 4 |
7344 |
0.15 |
| chr10_48259069_48259363 | 0.36 |
ANXA8 |
annexin A8 |
3863 |
0.17 |
| chr8_103741782_103741933 | 0.36 |
ENSG00000266799 |
. |
4090 |
0.28 |
| chr15_42102101_42102252 | 0.36 |
RP11-23P13.4 |
|
17877 |
0.09 |
| chr16_89368420_89368663 | 0.36 |
AC137932.5 |
|
4277 |
0.15 |
| chr16_11292647_11292983 | 0.36 |
RMI2 |
RecQ mediated genome instability 2 |
50691 |
0.08 |
| chr1_111745643_111746099 | 0.36 |
DENND2D |
DENN/MADD domain containing 2D |
1160 |
0.35 |
| chr20_3782789_3782940 | 0.36 |
CDC25B |
cell division cycle 25B |
5786 |
0.13 |
| chr7_128006485_128006636 | 0.36 |
PRRT4 |
proline-rich transmembrane protein 4 |
4821 |
0.16 |
| chr14_81894135_81894460 | 0.36 |
STON2 |
stonin 2 |
549 |
0.86 |
| chr6_36630670_36630821 | 0.35 |
ENSG00000251864 |
. |
9870 |
0.14 |
| chr16_10711037_10711188 | 0.35 |
EMP2 |
epithelial membrane protein 2 |
36557 |
0.13 |
| chr8_146015607_146015758 | 0.35 |
RPL8 |
ribosomal protein L8 |
1856 |
0.19 |
| chr2_31476447_31476598 | 0.35 |
EHD3 |
EH-domain containing 3 |
19319 |
0.18 |
| chr11_116645513_116645664 | 0.35 |
AP006216.10 |
|
1483 |
0.25 |
| chr11_75243357_75243645 | 0.35 |
GDPD5 |
glycerophosphodiester phosphodiesterase domain containing 5 |
6553 |
0.15 |
| chr1_233462761_233462977 | 0.34 |
MLK4 |
Mitogen-activated protein kinase kinase kinase MLK4 |
645 |
0.79 |
| chr1_9781158_9781391 | 0.34 |
PIK3CD |
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta |
3147 |
0.23 |
| chr15_63189996_63190147 | 0.34 |
RP11-1069G10.1 |
|
13838 |
0.23 |
| chr17_73841947_73842104 | 0.34 |
UNC13D |
unc-13 homolog D (C. elegans) |
1227 |
0.25 |
| chr15_101733144_101733684 | 0.34 |
CHSY1 |
chondroitin sulfate synthase 1 |
58723 |
0.11 |
| chr4_185601941_185602092 | 0.34 |
PRIMPOL |
primase and polymerase (DNA-directed) |
2585 |
0.29 |
| chr17_1520967_1521246 | 0.34 |
ENSG00000238946 |
. |
1517 |
0.25 |
| chr9_136858320_136859229 | 0.34 |
VAV2 |
vav 2 guanine nucleotide exchange factor |
1048 |
0.54 |
| chr17_48998396_48998613 | 0.34 |
TOB1 |
transducer of ERBB2, 1 |
53165 |
0.1 |
| chr4_1754587_1754779 | 0.34 |
TACC3 |
transforming, acidic coiled-coil containing protein 3 |
24552 |
0.13 |
| chr7_58045_58196 | 0.34 |
FAM20C |
family with sequence similarity 20, member C |
134849 |
0.05 |
| chr7_157145222_157145792 | 0.34 |
DNAJB6 |
DnaJ (Hsp40) homolog, subfamily B, member 6 |
12992 |
0.24 |
| chr22_18268291_18268442 | 0.33 |
BID |
BH3 interacting domain death agonist |
10935 |
0.18 |
| chr5_169950070_169950221 | 0.33 |
KCNIP1 |
Kv channel interacting protein 1 |
18568 |
0.21 |
| chr5_176864330_176864481 | 0.33 |
PRR7-AS1 |
PRR7 antisense RNA 1 |
7073 |
0.09 |
| chrX_47432713_47432906 | 0.33 |
TIMP1 |
TIMP metallopeptidase inhibitor 1 |
8903 |
0.13 |
| chr5_146257186_146257400 | 0.33 |
PPP2R2B |
protein phosphatase 2, regulatory subunit B, beta |
912 |
0.68 |
| chr15_77286567_77286718 | 0.33 |
PSTPIP1 |
proline-serine-threonine phosphatase interacting protein 1 |
784 |
0.65 |
| chr1_16277969_16279055 | 0.33 |
ZBTB17 |
zinc finger and BTB domain containing 17 |
7589 |
0.15 |
| chr12_69864753_69865036 | 0.33 |
FRS2 |
fibroblast growth factor receptor substrate 2 |
680 |
0.74 |
| chr4_74751671_74751822 | 0.33 |
CXCL1 |
chemokine (C-X-C motif) ligand 1 (melanoma growth stimulating activity, alpha) |
16625 |
0.15 |
| chr1_154405311_154405652 | 0.33 |
IL6R |
interleukin 6 receptor |
1648 |
0.28 |
| chr5_56013223_56013374 | 0.33 |
MAP3K1 |
mitogen-activated protein kinase kinase kinase 1, E3 ubiquitin protein ligase |
98103 |
0.07 |
| chr20_24914059_24914210 | 0.33 |
CST7 |
cystatin F (leukocystatin) |
15732 |
0.21 |
| chr11_47093587_47093738 | 0.33 |
C11orf49 |
chromosome 11 open reading frame 49 |
19650 |
0.17 |
| chr7_27993630_27993781 | 0.33 |
JAZF1 |
JAZF zinc finger 1 |
37970 |
0.19 |
| chr19_17907446_17907815 | 0.33 |
B3GNT3 |
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 3 |
1664 |
0.26 |
| chrX_153666332_153666497 | 0.33 |
GDI1 |
GDP dissociation inhibitor 1 |
1148 |
0.24 |
| chr14_69441554_69441705 | 0.32 |
ACTN1 |
actinin, alpha 1 |
2654 |
0.3 |
| chr12_1780268_1780419 | 0.32 |
ENSG00000266043 |
. |
10797 |
0.19 |
| chr12_111483776_111483927 | 0.32 |
CUX2 |
cut-like homeobox 2 |
12023 |
0.23 |
| chr4_177195681_177195832 | 0.32 |
ASB5 |
ankyrin repeat and SOCS box containing 5 |
2966 |
0.29 |
| chr14_105433817_105433968 | 0.32 |
AHNAK2 |
AHNAK nucleoprotein 2 |
10802 |
0.16 |
| chr11_3216794_3216945 | 0.32 |
AC108448.3 |
|
6093 |
0.14 |
| chr5_53968473_53968624 | 0.32 |
SNX18 |
sorting nexin 18 |
154955 |
0.04 |
| chr12_52629846_52629997 | 0.32 |
KRT7 |
keratin 7 |
3023 |
0.14 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.2 | 0.6 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.2 | 0.5 | GO:0046813 | receptor-mediated virion attachment to host cell(GO:0046813) |
| 0.1 | 0.3 | GO:0044413 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.1 | 0.4 | GO:0071731 | response to nitric oxide(GO:0071731) |
| 0.1 | 0.3 | GO:0002323 | natural killer cell activation involved in immune response(GO:0002323) natural killer cell degranulation(GO:0043320) |
| 0.1 | 0.4 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.1 | 0.2 | GO:0060340 | positive regulation of type I interferon-mediated signaling pathway(GO:0060340) |
| 0.1 | 0.3 | GO:0071034 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.1 | 0.3 | GO:0001743 | optic placode formation(GO:0001743) |
| 0.1 | 0.3 | GO:0060633 | negative regulation of transcription initiation from RNA polymerase II promoter(GO:0060633) negative regulation of DNA-templated transcription, initiation(GO:2000143) |
| 0.1 | 0.3 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 0.3 | GO:0060068 | vagina development(GO:0060068) |
| 0.1 | 0.4 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.1 | 0.1 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.1 | 0.3 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.1 | 0.4 | GO:0051642 | centrosome localization(GO:0051642) |
| 0.1 | 0.1 | GO:0021819 | layer formation in cerebral cortex(GO:0021819) |
| 0.1 | 0.2 | GO:0034139 | regulation of toll-like receptor 3 signaling pathway(GO:0034139) |
| 0.1 | 0.2 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.1 | 0.2 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.1 | 0.2 | GO:0070669 | response to interleukin-2(GO:0070669) |
| 0.1 | 0.2 | GO:0032353 | negative regulation of hormone metabolic process(GO:0032351) negative regulation of hormone biosynthetic process(GO:0032353) |
| 0.1 | 0.1 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.0 | 0.2 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.3 | GO:0036296 | response to increased oxygen levels(GO:0036296) response to hyperoxia(GO:0055093) |
| 0.0 | 0.2 | GO:0051873 | disruption by host of symbiont cells(GO:0051852) killing by host of symbiont cells(GO:0051873) |
| 0.0 | 0.2 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) regulation of store-operated calcium channel activity(GO:1901339) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.1 | GO:0030812 | negative regulation of nucleotide catabolic process(GO:0030812) |
| 0.0 | 0.2 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.2 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.0 | 0.1 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.0 | 0.1 | GO:0046185 | aldehyde catabolic process(GO:0046185) |
| 0.0 | 0.1 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.0 | GO:0022030 | cerebral cortex radial glia guided migration(GO:0021801) telencephalon glial cell migration(GO:0022030) |
| 0.0 | 0.6 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) NIK/NF-kappaB signaling(GO:0038061) regulation of NIK/NF-kappaB signaling(GO:1901222) positive regulation of NIK/NF-kappaB signaling(GO:1901224) |
| 0.0 | 0.2 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 0.1 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.1 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.1 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) |
| 0.0 | 0.2 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.0 | 0.1 | GO:0015864 | pyrimidine nucleoside transport(GO:0015864) |
| 0.0 | 0.1 | GO:0019605 | butyrate metabolic process(GO:0019605) |
| 0.0 | 0.1 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.0 | 0.1 | GO:0021798 | forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.0 | 0.2 | GO:0090218 | positive regulation of lipid kinase activity(GO:0090218) |
| 0.0 | 0.1 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 0.1 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.0 | 0.1 | GO:0006565 | L-serine catabolic process(GO:0006565) |
| 0.0 | 0.2 | GO:0032288 | myelin assembly(GO:0032288) |
| 0.0 | 0.1 | GO:0060585 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.0 | 0.1 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 0.1 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.0 | 0.1 | GO:0043276 | anoikis(GO:0043276) |
| 0.0 | 0.2 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 0.1 | GO:0043441 | acetoacetic acid biosynthetic process(GO:0043441) |
| 0.0 | 0.3 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.1 | GO:0002679 | respiratory burst involved in defense response(GO:0002679) |
| 0.0 | 0.1 | GO:0051124 | synaptic growth at neuromuscular junction(GO:0051124) |
| 0.0 | 0.1 | GO:0015816 | glycine transport(GO:0015816) |
| 0.0 | 0.1 | GO:0070977 | organ maturation(GO:0048799) bone maturation(GO:0070977) |
| 0.0 | 0.1 | GO:0070561 | vitamin D receptor signaling pathway(GO:0070561) |
| 0.0 | 0.1 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.1 | GO:1902803 | regulation of synaptic vesicle transport(GO:1902803) regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.0 | 0.1 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.0 | 0.1 | GO:0042268 | regulation of cytolysis(GO:0042268) |
| 0.0 | 0.1 | GO:0045112 | integrin biosynthetic process(GO:0045112) |
| 0.0 | 0.2 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.1 | GO:0048537 | mucosal-associated lymphoid tissue development(GO:0048537) |
| 0.0 | 0.1 | GO:0014832 | urinary bladder smooth muscle contraction(GO:0014832) |
| 0.0 | 0.0 | GO:0072393 | microtubule anchoring at microtubule organizing center(GO:0072393) |
| 0.0 | 0.1 | GO:0072367 | regulation of lipid transport by regulation of transcription from RNA polymerase II promoter(GO:0072367) |
| 0.0 | 0.1 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.2 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) regulation of dendritic spine morphogenesis(GO:0061001) dendritic spine organization(GO:0097061) |
| 0.0 | 0.1 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 0.1 | GO:0045657 | positive regulation of monocyte differentiation(GO:0045657) |
| 0.0 | 0.0 | GO:0043173 | nucleotide salvage(GO:0043173) |
| 0.0 | 0.1 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.0 | 0.1 | GO:0016188 | synaptic vesicle maturation(GO:0016188) |
| 0.0 | 0.1 | GO:0030187 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.0 | 0.1 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.0 | 0.2 | GO:0015884 | folic acid transport(GO:0015884) |
| 0.0 | 0.1 | GO:0032075 | positive regulation of nuclease activity(GO:0032075) |
| 0.0 | 0.1 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.0 | 0.2 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.3 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.2 | GO:0014911 | positive regulation of smooth muscle cell migration(GO:0014911) |
| 0.0 | 0.1 | GO:0060897 | neural plate anterior/posterior regionalization(GO:0021999) neural plate regionalization(GO:0060897) |
| 0.0 | 0.1 | GO:0070120 | ciliary neurotrophic factor-mediated signaling pathway(GO:0070120) |
| 0.0 | 0.0 | GO:0014848 | urinary tract smooth muscle contraction(GO:0014848) |
| 0.0 | 0.2 | GO:0046718 | viral entry into host cell(GO:0046718) |
| 0.0 | 0.1 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.0 | GO:0034201 | response to oleic acid(GO:0034201) |
| 0.0 | 0.0 | GO:0003179 | heart valve development(GO:0003170) heart valve morphogenesis(GO:0003179) |
| 0.0 | 0.1 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.1 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.1 | GO:0050965 | detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.0 | 0.0 | GO:0072216 | positive regulation of metanephros development(GO:0072216) |
| 0.0 | 0.1 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.1 | GO:0032691 | negative regulation of interleukin-1 beta production(GO:0032691) |
| 0.0 | 0.1 | GO:0046137 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.0 | 0.2 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 0.1 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 | 0.1 | GO:0097502 | protein mannosylation(GO:0035268) protein O-linked mannosylation(GO:0035269) mannosylation(GO:0097502) |
| 0.0 | 0.1 | GO:0014912 | negative regulation of smooth muscle cell migration(GO:0014912) |
| 0.0 | 0.1 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 | 0.0 | GO:0042754 | negative regulation of circadian sleep/wake cycle, sleep(GO:0042321) negative regulation of circadian rhythm(GO:0042754) negative regulation of behavior(GO:0048521) |
| 0.0 | 0.1 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.0 | 0.0 | GO:0051409 | response to nitrosative stress(GO:0051409) |
| 0.0 | 0.1 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.0 | 0.0 | GO:0021860 | pyramidal neuron differentiation(GO:0021859) pyramidal neuron development(GO:0021860) |
| 0.0 | 0.1 | GO:0042492 | gamma-delta T cell differentiation(GO:0042492) |
| 0.0 | 0.2 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.0 | GO:2000542 | negative regulation of gastrulation(GO:2000542) |
| 0.0 | 0.1 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.1 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 0.0 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.0 | 0.1 | GO:0014819 | regulation of skeletal muscle contraction(GO:0014819) |
| 0.0 | 0.1 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.0 | 0.1 | GO:0032515 | negative regulation of phosphatase activity(GO:0010923) negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.0 | 0.1 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.0 | 0.2 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.4 | GO:0009267 | cellular response to starvation(GO:0009267) |
| 0.0 | 0.1 | GO:0042415 | norepinephrine metabolic process(GO:0042415) |
| 0.0 | 0.1 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.0 | 0.1 | GO:0009128 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.0 | 0.1 | GO:0045714 | regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045714) |
| 0.0 | 0.0 | GO:0042320 | regulation of circadian sleep/wake cycle, REM sleep(GO:0042320) positive regulation of circadian sleep/wake cycle, sleep(GO:0045938) positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.0 | 0.2 | GO:0007143 | female meiotic division(GO:0007143) |
| 0.0 | 0.1 | GO:0032277 | negative regulation of gonadotropin secretion(GO:0032277) |
| 0.0 | 0.0 | GO:0034113 | heterotypic cell-cell adhesion(GO:0034113) regulation of heterotypic cell-cell adhesion(GO:0034114) |
| 0.0 | 0.0 | GO:0046101 | hypoxanthine biosynthetic process(GO:0046101) |
| 0.0 | 0.3 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.0 | 0.0 | GO:0045906 | negative regulation of vasoconstriction(GO:0045906) |
| 0.0 | 0.1 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.1 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.0 | 0.1 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.0 | 0.1 | GO:0042501 | serine phosphorylation of STAT protein(GO:0042501) |
| 0.0 | 0.2 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.1 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.0 | 0.0 | GO:0036336 | dendritic cell chemotaxis(GO:0002407) dendritic cell migration(GO:0036336) |
| 0.0 | 0.0 | GO:0032898 | neurotrophin production(GO:0032898) |
| 0.0 | 0.0 | GO:0002227 | innate immune response in mucosa(GO:0002227) mucosal immune response(GO:0002385) |
| 0.0 | 0.1 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.0 | 0.1 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.0 | GO:0060596 | mammary placode formation(GO:0060596) |
| 0.0 | 0.3 | GO:0046676 | negative regulation of peptide secretion(GO:0002792) negative regulation of insulin secretion(GO:0046676) negative regulation of peptide hormone secretion(GO:0090278) |
| 0.0 | 0.1 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.1 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 0.1 | GO:0050872 | white fat cell differentiation(GO:0050872) |
| 0.0 | 0.0 | GO:0010871 | negative regulation of receptor biosynthetic process(GO:0010871) |
| 0.0 | 0.1 | GO:0019511 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 0.1 | GO:0007494 | midgut development(GO:0007494) |
| 0.0 | 0.1 | GO:0060055 | angiogenesis involved in wound healing(GO:0060055) |
| 0.0 | 0.1 | GO:0045990 | carbon catabolite regulation of transcription(GO:0045990) regulation of transcription by glucose(GO:0046015) |
| 0.0 | 0.0 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.0 | 0.1 | GO:0019240 | protein citrullination(GO:0018101) citrulline biosynthetic process(GO:0019240) |
| 0.0 | 0.1 | GO:0035583 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.0 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) |
| 0.0 | 0.4 | GO:0006691 | leukotriene metabolic process(GO:0006691) |
| 0.0 | 0.1 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 0.1 | GO:0046337 | phosphatidylethanolamine biosynthetic process(GO:0006646) phosphatidylethanolamine metabolic process(GO:0046337) |
| 0.0 | 0.1 | GO:0043545 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.0 | 0.1 | GO:0075713 | establishment of viral latency(GO:0019043) establishment of integrated proviral latency(GO:0075713) |
| 0.0 | 0.0 | GO:0046471 | phosphatidylglycerol biosynthetic process(GO:0006655) cardiolipin metabolic process(GO:0032048) cardiolipin biosynthetic process(GO:0032049) phosphatidylglycerol metabolic process(GO:0046471) |
| 0.0 | 0.1 | GO:0006498 | N-terminal protein lipidation(GO:0006498) |
| 0.0 | 0.1 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 | 0.1 | GO:0032988 | ribonucleoprotein complex disassembly(GO:0032988) |
| 0.0 | 0.0 | GO:0032823 | regulation of natural killer cell differentiation(GO:0032823) positive regulation of natural killer cell differentiation(GO:0032825) |
| 0.0 | 0.1 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.1 | GO:0006621 | protein retention in ER lumen(GO:0006621) maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.0 | 0.1 | GO:0019255 | glucose 1-phosphate metabolic process(GO:0019255) |
| 0.0 | 0.1 | GO:0016115 | diterpenoid catabolic process(GO:0016103) terpenoid catabolic process(GO:0016115) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.0 | GO:0070572 | positive regulation of axon regeneration(GO:0048680) positive regulation of neuron projection regeneration(GO:0070572) |
| 0.0 | 0.2 | GO:0001914 | regulation of T cell mediated cytotoxicity(GO:0001914) |
| 0.0 | 0.1 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.0 | 0.1 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 0.1 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.0 | 0.0 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.0 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.1 | GO:0009629 | response to gravity(GO:0009629) |
| 0.0 | 0.1 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.1 | GO:0042117 | monocyte activation(GO:0042117) |
| 0.0 | 0.1 | GO:0001921 | positive regulation of receptor recycling(GO:0001921) |
| 0.0 | 0.0 | GO:0045007 | depurination(GO:0045007) |
| 0.0 | 0.2 | GO:0051205 | protein insertion into membrane(GO:0051205) |
| 0.0 | 0.2 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 | 0.0 | GO:0000966 | RNA 5'-end processing(GO:0000966) |
| 0.0 | 0.1 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.0 | 0.0 | GO:0021548 | pons development(GO:0021548) |
| 0.0 | 0.0 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.0 | 0.0 | GO:2000008 | regulation of protein localization to cell surface(GO:2000008) |
| 0.0 | 0.0 | GO:0014742 | positive regulation of cardiac muscle hypertrophy(GO:0010613) positive regulation of muscle hypertrophy(GO:0014742) |
| 0.0 | 0.1 | GO:0048265 | response to pain(GO:0048265) |
| 0.0 | 0.0 | GO:0032328 | alanine transport(GO:0032328) |
| 0.0 | 0.0 | GO:0017014 | protein nitrosylation(GO:0017014) peptidyl-cysteine S-nitrosylation(GO:0018119) |
| 0.0 | 0.0 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.0 | 0.0 | GO:0010224 | response to UV-B(GO:0010224) |
| 0.0 | 0.1 | GO:0009180 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) ribonucleoside diphosphate biosynthetic process(GO:0009188) |
| 0.0 | 0.2 | GO:0031102 | neuron projection regeneration(GO:0031102) |
| 0.0 | 0.0 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.2 | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.0 | 0.1 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.1 | GO:0009200 | deoxyribonucleoside triphosphate metabolic process(GO:0009200) |
| 0.0 | 0.0 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.0 | 0.0 | GO:0000492 | small nucleolar ribonucleoprotein complex assembly(GO:0000491) box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.1 | GO:0060678 | dichotomous subdivision of terminal units involved in ureteric bud branching(GO:0060678) |
| 0.0 | 0.4 | GO:0051149 | positive regulation of muscle cell differentiation(GO:0051149) |
| 0.0 | 0.0 | GO:0018126 | protein hydroxylation(GO:0018126) |
| 0.0 | 0.0 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.0 | 0.0 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 0.1 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.0 | 0.1 | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.0 | 0.1 | GO:0098876 | Golgi to plasma membrane transport(GO:0006893) vesicle-mediated transport to the plasma membrane(GO:0098876) |
| 0.0 | 0.0 | GO:0071503 | response to heparin(GO:0071503) |
| 0.0 | 0.0 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 | 0.1 | GO:0006558 | L-phenylalanine metabolic process(GO:0006558) L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.0 | 0.0 | GO:0045618 | positive regulation of keratinocyte differentiation(GO:0045618) |
| 0.0 | 0.1 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.0 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 0.1 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.1 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.0 | GO:0090037 | regulation of protein kinase C signaling(GO:0090036) positive regulation of protein kinase C signaling(GO:0090037) |
| 0.0 | 0.0 | GO:0032495 | response to muramyl dipeptide(GO:0032495) |
| 0.0 | 0.1 | GO:0046500 | S-adenosylmethionine metabolic process(GO:0046500) |
| 0.0 | 0.1 | GO:0046689 | response to mercury ion(GO:0046689) |
| 0.0 | 0.1 | GO:0042487 | regulation of odontogenesis of dentin-containing tooth(GO:0042487) |
| 0.0 | 0.0 | GO:0006290 | pyrimidine dimer repair(GO:0006290) |
| 0.0 | 0.0 | GO:0000281 | mitotic cytokinesis(GO:0000281) |
| 0.0 | 0.1 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.0 | 0.0 | GO:0006562 | proline catabolic process(GO:0006562) |
| 0.0 | 0.1 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.1 | GO:0030225 | macrophage differentiation(GO:0030225) |
| 0.0 | 0.0 | GO:0050765 | negative regulation of phagocytosis(GO:0050765) |
| 0.0 | 0.0 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.0 | 0.1 | GO:0045040 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.0 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.0 | 0.0 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.1 | GO:0015840 | urea transport(GO:0015840) |
| 0.0 | 0.1 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.0 | 0.0 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.0 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.0 | 0.0 | GO:0048289 | isotype switching to IgE isotypes(GO:0048289) regulation of isotype switching to IgE isotypes(GO:0048293) |
| 0.0 | 0.0 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 0.0 | 0.1 | GO:0042401 | cellular biogenic amine biosynthetic process(GO:0042401) |
| 0.0 | 0.0 | GO:0060317 | cardiac epithelial to mesenchymal transition(GO:0060317) |
| 0.0 | 0.2 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.0 | 0.2 | GO:0006692 | prostanoid metabolic process(GO:0006692) |
| 0.0 | 0.0 | GO:0001866 | NK T cell proliferation(GO:0001866) NK T cell activation(GO:0051132) |
| 0.0 | 0.5 | GO:0050434 | positive regulation of viral transcription(GO:0050434) |
| 0.0 | 0.2 | GO:0015949 | nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.0 | 0.0 | GO:0048840 | otolith development(GO:0048840) |
| 0.0 | 0.0 | GO:0031659 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G1/S transition of mitotic cell cycle(GO:0031657) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G1/S transition of mitotic cell cycle(GO:0031659) |
| 0.0 | 0.0 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.0 | 0.0 | GO:0019336 | phenol-containing compound catabolic process(GO:0019336) catechol-containing compound catabolic process(GO:0019614) catecholamine catabolic process(GO:0042424) |
| 0.0 | 0.1 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 | 0.1 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) |
| 0.0 | 0.1 | GO:0071459 | protein localization to chromosome, centromeric region(GO:0071459) |
| 0.0 | 0.2 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 0.2 | GO:0050771 | negative regulation of axonogenesis(GO:0050771) |
| 0.0 | 0.0 | GO:0006216 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.0 | 0.1 | GO:0040020 | regulation of meiotic nuclear division(GO:0040020) |
| 0.0 | 0.1 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.1 | GO:0003337 | mesenchymal to epithelial transition involved in metanephros morphogenesis(GO:0003337) mesenchymal to epithelial transition(GO:0060231) metanephric renal vesicle morphogenesis(GO:0072283) |
| 0.0 | 0.0 | GO:0051882 | mitochondrial depolarization(GO:0051882) |
| 0.0 | 0.1 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.1 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.0 | GO:0044803 | multi-organism membrane organization(GO:0044803) |
| 0.0 | 0.0 | GO:0019856 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.0 | 0.0 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 0.1 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.0 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.0 | 0.0 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.1 | GO:0019682 | pentose-phosphate shunt(GO:0006098) glyceraldehyde-3-phosphate metabolic process(GO:0019682) |
| 0.0 | 0.0 | GO:0036473 | intrinsic apoptotic signaling pathway in response to oxidative stress(GO:0008631) cell death in response to oxidative stress(GO:0036473) |
| 0.0 | 0.0 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.0 | 0.1 | GO:1903846 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.0 | 0.2 | GO:0035303 | regulation of dephosphorylation(GO:0035303) |
| 0.0 | 0.0 | GO:0003230 | cardiac atrium morphogenesis(GO:0003209) cardiac atrium development(GO:0003230) |
| 0.0 | 0.1 | GO:0000085 | mitotic G2 phase(GO:0000085) G2 phase(GO:0051319) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0032449 | CBM complex(GO:0032449) |
| 0.1 | 0.3 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.1 | 0.3 | GO:0043218 | compact myelin(GO:0043218) |
| 0.1 | 0.3 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.1 | 0.2 | GO:0033646 | host(GO:0018995) host cell part(GO:0033643) host intracellular part(GO:0033646) intracellular region of host(GO:0043656) host cell(GO:0043657) other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) |
| 0.1 | 0.3 | GO:0036454 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.1 | 0.2 | GO:0043260 | laminin-11 complex(GO:0043260) |
| 0.0 | 0.1 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.2 | GO:0060203 | clathrin-sculpted glutamate transport vesicle(GO:0060199) clathrin-sculpted glutamate transport vesicle membrane(GO:0060203) |
| 0.0 | 0.3 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.1 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.1 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.1 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.3 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.1 | GO:0005593 | FACIT collagen trimer(GO:0005593) |
| 0.0 | 0.6 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.2 | GO:0042581 | specific granule(GO:0042581) |
| 0.0 | 0.1 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.5 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.1 | GO:0002178 | palmitoyltransferase complex(GO:0002178) serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.1 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 0.1 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 0.1 | GO:0033185 | mannosyltransferase complex(GO:0031501) dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 0.5 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.1 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.0 | 0.1 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 0.1 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.1 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.2 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.1 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.0 | 0.1 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.1 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.1 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.1 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.1 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.4 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.1 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.1 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.5 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 0.1 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.4 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.0 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 0.1 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.1 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.0 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.0 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.0 | 0.2 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.2 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 0.1 | GO:0000800 | lateral element(GO:0000800) |
| 0.0 | 0.3 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 0.1 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.1 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.4 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.1 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.0 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.2 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.1 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.2 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.3 | GO:0008287 | protein serine/threonine phosphatase complex(GO:0008287) phosphatase complex(GO:1903293) |
| 0.0 | 0.0 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.0 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.0 | 0.1 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.0 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.1 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.6 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.1 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.3 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.0 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.1 | GO:0031332 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 0.0 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.1 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 0.1 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.0 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.7 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 0.5 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.1 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.1 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.0 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.1 | GO:0002080 | acrosomal membrane(GO:0002080) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.1 | 0.4 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.1 | 0.5 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.1 | 0.3 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.1 | 0.1 | GO:0008518 | reduced folate carrier activity(GO:0008518) |
| 0.1 | 0.4 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.1 | 0.2 | GO:0099602 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.1 | 0.2 | GO:0032554 | purine deoxyribonucleotide binding(GO:0032554) adenyl deoxyribonucleotide binding(GO:0032558) |
| 0.1 | 0.3 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.1 | 0.2 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.1 | 0.2 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.1 | 0.3 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) |
| 0.1 | 0.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 0.3 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 0.2 | GO:0017060 | 3-galactosyl-N-acetylglucosaminide 4-alpha-L-fucosyltransferase activity(GO:0017060) |
| 0.1 | 0.2 | GO:0046980 | tapasin binding(GO:0046980) |
| 0.1 | 0.2 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.1 | 0.4 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.1 | 0.2 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.1 | 0.2 | GO:0015065 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.0 | 0.2 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.3 | GO:0050656 | 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.0 | 0.2 | GO:0090482 | vitamin transmembrane transporter activity(GO:0090482) |
| 0.0 | 0.2 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.1 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.6 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.4 | GO:0042166 | acetylcholine binding(GO:0042166) |
| 0.0 | 0.2 | GO:0015168 | glycerol transmembrane transporter activity(GO:0015168) |
| 0.0 | 0.1 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.0 | 0.1 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.3 | GO:0016944 | obsolete RNA polymerase II transcription elongation factor activity(GO:0016944) |
| 0.0 | 0.1 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.2 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.1 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.1 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.0 | 0.4 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.1 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.2 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.2 | GO:0016717 | oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water(GO:0016717) |
| 0.0 | 0.1 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.1 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.0 | 0.1 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.0 | 0.1 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.0 | 0.1 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.0 | 0.1 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.2 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.3 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.1 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.1 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.0 | 0.2 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.1 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 0.1 | GO:0004904 | interferon receptor activity(GO:0004904) |
| 0.0 | 0.4 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.5 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 0.0 | 0.2 | GO:0042153 | obsolete RPTP-like protein binding(GO:0042153) |
| 0.0 | 0.1 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.2 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.0 | 0.1 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.1 | GO:0000977 | RNA polymerase II regulatory region sequence-specific DNA binding(GO:0000977) RNA polymerase II regulatory region DNA binding(GO:0001012) |
| 0.0 | 0.0 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.5 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.1 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.0 | 0.6 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.0 | 0.0 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.0 | 0.1 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.1 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.0 | 0.2 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.1 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.0 | GO:0003964 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.0 | 0.1 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.0 | 0.1 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.1 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.3 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.1 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.1 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) |
| 0.0 | 0.1 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
| 0.0 | 0.5 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.0 | GO:0015199 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.0 | 0.1 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.1 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.0 | 0.1 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.1 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.3 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.1 | GO:0004607 | phosphatidylcholine-sterol O-acyltransferase activity(GO:0004607) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.1 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.1 | GO:0008035 | high-density lipoprotein particle binding(GO:0008035) |
| 0.0 | 0.1 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.0 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.0 | 0.1 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.2 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.1 | GO:0030151 | molybdenum ion binding(GO:0030151) |
| 0.0 | 0.2 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.1 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.1 | GO:0019863 | IgE binding(GO:0019863) |
| 0.0 | 0.2 | GO:0043176 | amine binding(GO:0043176) |
| 0.0 | 0.1 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.1 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.0 | 0.1 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.1 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.0 | 0.3 | GO:0004532 | exoribonuclease activity(GO:0004532) |
| 0.0 | 0.0 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.0 | 0.0 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.1 | GO:0043295 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.0 | 0.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.1 | GO:0015333 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 0.0 | 0.1 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.1 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.1 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.1 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.0 | 0.1 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.0 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.0 | 0.1 | GO:0097506 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.0 | 0.0 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.0 | 0.1 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.1 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.2 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.0 | GO:0019798 | procollagen-proline dioxygenase activity(GO:0019798) |
| 0.0 | 0.1 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.0 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.1 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.2 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.0 | GO:0031708 | bombesin receptor binding(GO:0031705) endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.1 | GO:0015556 | C4-dicarboxylate transmembrane transporter activity(GO:0015556) |
| 0.0 | 0.0 | GO:0034648 | histone demethylase activity (H3-K4 specific)(GO:0032453) histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.0 | 0.2 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.0 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.0 | 0.1 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.1 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.1 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.1 | GO:0045569 | TRAIL binding(GO:0045569) |
| 0.0 | 0.1 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.2 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.1 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.0 | 0.3 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.1 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 0.0 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.1 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.3 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.0 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.0 | 0.1 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.0 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.1 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.2 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.1 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.1 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.1 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.2 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.0 | 0.1 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 0.2 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.0 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.0 | GO:0052659 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) inositol-1,3,4,5-tetrakisphosphate 5-phosphatase activity(GO:0052659) |
| 0.0 | 0.0 | GO:0016803 | ether hydrolase activity(GO:0016803) |
| 0.0 | 0.1 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.0 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.1 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.3 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.0 | 0.1 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.0 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.0 | 0.0 | GO:0004738 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.3 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.1 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.1 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.3 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.0 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.0 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.0 | 0.1 | GO:0005542 | folic acid binding(GO:0005542) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.1 | GO:0016847 | 1-aminocyclopropane-1-carboxylate synthase activity(GO:0016847) |
| 0.0 | 0.0 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.0 | 0.0 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.0 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.0 | 0.0 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.0 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) |
| 0.0 | 0.1 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) histone deacetylase activity (H4-K16 specific)(GO:0034739) NAD-dependent histone deacetylase activity (H4-K16 specific)(GO:0046970) |
| 0.0 | 0.0 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) |
| 0.0 | 0.0 | GO:0052834 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.0 | GO:0032139 | dinucleotide insertion or deletion binding(GO:0032139) |
| 0.0 | 0.0 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 0.0 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.1 | GO:0016421 | CoA carboxylase activity(GO:0016421) |
| 0.0 | 0.1 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.0 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.0 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.0 | 0.6 | GO:0005178 | integrin binding(GO:0005178) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 0.4 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.4 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.9 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 1.1 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.4 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.6 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.4 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 0.7 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.2 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.1 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.0 | 0.7 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.5 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.3 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.2 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.2 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.3 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.0 | 0.4 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.0 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.2 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.2 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 0.2 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.0 | 0.3 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.2 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.2 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.4 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.3 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.4 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.1 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.0 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.1 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.2 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.0 | 0.6 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 1.3 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.1 | REACTOME HEMOSTASIS | Genes involved in Hemostasis |
| 0.0 | 0.6 | REACTOME RORA ACTIVATES CIRCADIAN EXPRESSION | Genes involved in RORA Activates Circadian Expression |
| 0.0 | 0.5 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.5 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.3 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.0 | REACTOME SIGNALING BY FGFR IN DISEASE | Genes involved in Signaling by FGFR in disease |
| 0.0 | 0.4 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.4 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.3 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.1 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 0.5 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.2 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.2 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.0 | REACTOME APC CDC20 MEDIATED DEGRADATION OF NEK2A | Genes involved in APC-Cdc20 mediated degradation of Nek2A |
| 0.0 | 0.0 | REACTOME PLATELET AGGREGATION PLUG FORMATION | Genes involved in Platelet Aggregation (Plug Formation) |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.1 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 0.5 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 0.1 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.2 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.2 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.1 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 0.2 | REACTOME PROLONGED ERK ACTIVATION EVENTS | Genes involved in Prolonged ERK activation events |
| 0.0 | 0.2 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.2 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.1 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.5 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.1 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.0 | 0.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.2 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.2 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.2 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.0 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.1 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 0.6 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.1 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.1 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.2 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.0 | 0.4 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 0.0 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 0.0 | 0.2 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.0 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 0.2 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.1 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.1 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.1 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 0.1 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.1 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.3 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.3 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.1 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.1 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.3 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.0 | REACTOME SCFSKP2 MEDIATED DEGRADATION OF P27 P21 | Genes involved in SCF(Skp2)-mediated degradation of p27/p21 |
| 0.0 | 0.1 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.2 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.1 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.1 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.0 | 0.1 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.1 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.2 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.1 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.2 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.1 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.0 | 0.1 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.1 | REACTOME RESOLUTION OF AP SITES VIA THE MULTIPLE NUCLEOTIDE PATCH REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the multiple-nucleotide patch replacement pathway |
| 0.0 | 0.2 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.1 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |