Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
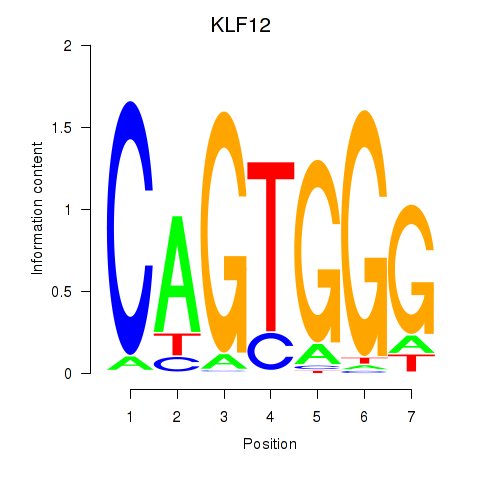
Results for KLF12
Z-value: 1.53
Transcription factors associated with KLF12
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
KLF12
|
ENSG00000118922.12 | Kruppel like factor 12 |
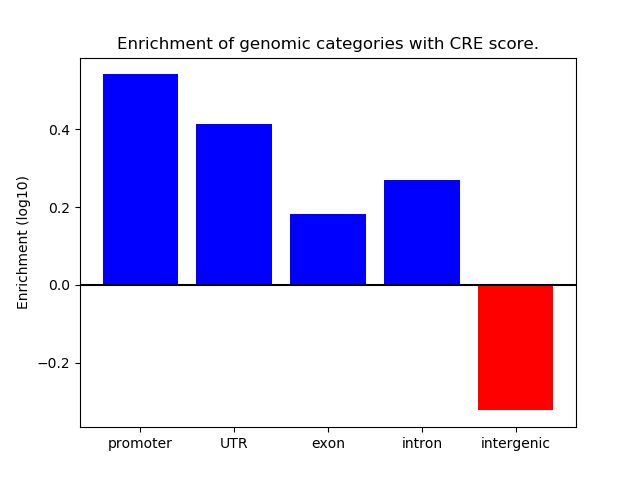
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr13_74655507_74655658 | KLF12 | 52812 | 0.183538 | -0.95 | 6.9e-05 | Click! |
| chr13_74655680_74655831 | KLF12 | 52639 | 0.184049 | -0.93 | 2.6e-04 | Click! |
| chr13_74332901_74333052 | KLF12 | 236210 | 0.020378 | -0.89 | 1.3e-03 | Click! |
| chr13_74610738_74610889 | KLF12 | 41627 | 0.221304 | -0.89 | 1.3e-03 | Click! |
| chr13_74610274_74610425 | KLF12 | 41163 | 0.222906 | -0.87 | 2.0e-03 | Click! |
Activity of the KLF12 motif across conditions
Conditions sorted by the z-value of the KLF12 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
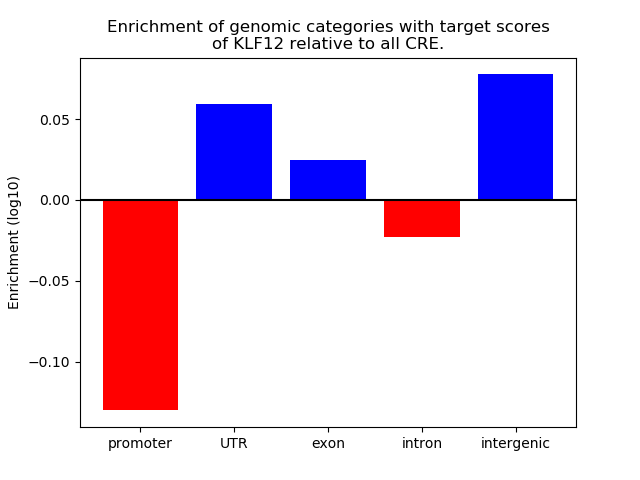
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr15_101741938_101742937 | 0.56 |
CHSY1 |
chondroitin sulfate synthase 1 |
49700 |
0.12 |
| chr1_235053615_235054079 | 0.52 |
ENSG00000239690 |
. |
13914 |
0.26 |
| chr3_197829897_197830311 | 0.52 |
AC073135.3 |
|
6973 |
0.18 |
| chr16_27253790_27254558 | 0.48 |
NSMCE1 |
non-SMC element 1 homolog (S. cerevisiae) |
9775 |
0.17 |
| chr10_81006599_81006910 | 0.47 |
ZMIZ1 |
zinc finger, MIZ-type containing 1 |
59221 |
0.14 |
| chr7_75592437_75593112 | 0.37 |
POR |
P450 (cytochrome) oxidoreductase |
4387 |
0.19 |
| chr18_43588789_43589171 | 0.37 |
ENSG00000222179 |
. |
19069 |
0.17 |
| chr3_148425752_148425954 | 0.35 |
AGTR1 |
angiotensin II receptor, type 1 |
10148 |
0.28 |
| chr9_116376964_116377197 | 0.34 |
RGS3 |
regulator of G-protein signaling 3 |
21314 |
0.2 |
| chr2_112133274_112133686 | 0.33 |
ENSG00000266139 |
. |
54812 |
0.17 |
| chr15_67055010_67055873 | 0.30 |
SMAD6 |
SMAD family member 6 |
51406 |
0.15 |
| chr16_85110269_85110512 | 0.29 |
KIAA0513 |
KIAA0513 |
676 |
0.74 |
| chr10_80009477_80009629 | 0.29 |
ENSG00000201393 |
. |
117711 |
0.06 |
| chr19_19083853_19084052 | 0.29 |
ENSG00000241172 |
. |
14885 |
0.1 |
| chr12_57621019_57621403 | 0.29 |
ENSG00000202067 |
. |
546 |
0.57 |
| chr11_131697224_131697416 | 0.27 |
AP004372.1 |
|
69682 |
0.12 |
| chr7_47267280_47267469 | 0.27 |
TNS3 |
tensin 3 |
52542 |
0.19 |
| chr15_75418307_75418680 | 0.27 |
ENSG00000252722 |
. |
4486 |
0.2 |
| chr1_31870163_31870721 | 0.27 |
SERINC2 |
serine incorporator 2 |
11970 |
0.15 |
| chr15_100051927_100052783 | 0.27 |
AC015660.1 |
HCG1993240; Serologically defined breast cancer antigen NY-BR-40; Uncharacterized protein |
13777 |
0.21 |
| chr7_31039847_31040026 | 0.26 |
GHRHR |
growth hormone releasing hormone receptor |
30788 |
0.19 |
| chr9_139963617_139963929 | 0.26 |
SAPCD2 |
suppressor APC domain containing 2 |
1267 |
0.17 |
| chr1_201642000_201642151 | 0.25 |
RP11-25B7.1 |
|
1197 |
0.41 |
| chr3_123140534_123140685 | 0.25 |
ADCY5 |
adenylate cyclase 5 |
1488 |
0.52 |
| chr13_98066168_98066319 | 0.25 |
RAP2A |
RAP2A, member of RAS oncogene family |
20233 |
0.22 |
| chr18_11149717_11149948 | 0.25 |
PIEZO2 |
piezo-type mechanosensitive ion channel component 2 |
1245 |
0.64 |
| chr4_8284667_8284834 | 0.25 |
HTRA3 |
HtrA serine peptidase 3 |
13246 |
0.22 |
| chr6_138752975_138753135 | 0.25 |
ENSG00000266555 |
. |
3376 |
0.26 |
| chr7_116774717_116775343 | 0.25 |
ST7-AS2 |
ST7 antisense RNA 2 |
10582 |
0.23 |
| chrX_138285001_138285220 | 0.24 |
FGF13 |
fibroblast growth factor 13 |
577 |
0.86 |
| chr12_13132371_13132545 | 0.24 |
RP11-392P7.6 |
|
313 |
0.84 |
| chr10_35853810_35854235 | 0.24 |
GJD4 |
gap junction protein, delta 4, 40.1kDa |
40316 |
0.15 |
| chr10_134389363_134389791 | 0.24 |
INPP5A |
inositol polyphosphate-5-phosphatase, 40kDa |
31853 |
0.19 |
| chr6_108786068_108786353 | 0.24 |
ENSG00000200799 |
. |
72933 |
0.1 |
| chr11_70261497_70261648 | 0.24 |
CTTN |
cortactin |
4659 |
0.16 |
| chr19_50284218_50285014 | 0.24 |
AP2A1 |
adaptor-related protein complex 2, alpha 1 subunit |
14225 |
0.07 |
| chr14_24837858_24838637 | 0.24 |
NFATC4 |
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4 |
41 |
0.94 |
| chr10_44475712_44476330 | 0.24 |
ENSG00000238957 |
. |
53967 |
0.17 |
| chr14_59213631_59214204 | 0.23 |
ENSG00000221427 |
. |
24047 |
0.25 |
| chr18_74175634_74175889 | 0.23 |
ZNF516 |
zinc finger protein 516 |
26974 |
0.16 |
| chr17_77806355_77806637 | 0.23 |
CBX4 |
chromobox homolog 4 |
5184 |
0.16 |
| chr2_55364453_55364798 | 0.23 |
RTN4 |
reticulon 4 |
24868 |
0.16 |
| chrX_135241873_135242024 | 0.23 |
FHL1 |
four and a half LIM domains 1 |
7215 |
0.22 |
| chr9_93536564_93536934 | 0.22 |
SYK |
spleen tyrosine kinase |
27320 |
0.27 |
| chr11_61484218_61484613 | 0.22 |
MYRF |
myelin regulatory factor |
35706 |
0.11 |
| chr5_134691363_134691514 | 0.22 |
C5orf66 |
chromosome 5 open reading frame 66 |
17809 |
0.18 |
| chr17_79396387_79396545 | 0.22 |
RP11-1055B8.7 |
BAH and coiled-coil domain-containing protein 1 |
8930 |
0.11 |
| chr17_32574614_32574944 | 0.22 |
CCL2 |
chemokine (C-C motif) ligand 2 |
7525 |
0.13 |
| chr1_150244620_150245106 | 0.22 |
C1orf54 |
chromosome 1 open reading frame 54 |
239 |
0.83 |
| chr2_223656927_223657285 | 0.22 |
ACSL3 |
acyl-CoA synthetase long-chain family member 3 |
68546 |
0.11 |
| chr6_158735535_158735730 | 0.22 |
TULP4 |
tubby like protein 4 |
1940 |
0.42 |
| chr15_64278058_64278452 | 0.22 |
DAPK2 |
death-associated protein kinase 2 |
54211 |
0.11 |
| chr17_25866922_25867277 | 0.22 |
KSR1 |
kinase suppressor of ras 1 |
42848 |
0.14 |
| chr22_44423046_44423480 | 0.22 |
PARVB |
parvin, beta |
3106 |
0.31 |
| chr5_139035453_139035988 | 0.22 |
CXXC5 |
CXXC finger protein 5 |
3420 |
0.27 |
| chr16_88220538_88220993 | 0.22 |
BANP |
BTG3 associated nuclear protein |
217141 |
0.02 |
| chr11_36112461_36112922 | 0.22 |
ENSG00000263389 |
. |
81043 |
0.09 |
| chr20_60819766_60820011 | 0.21 |
OSBPL2 |
oxysterol binding protein-like 2 |
6308 |
0.16 |
| chr10_60336642_60336940 | 0.21 |
BICC1 |
bicaudal C homolog 1 (Drosophila) |
63891 |
0.15 |
| chr19_1513130_1513873 | 0.21 |
ADAMTSL5 |
ADAMTS-like 5 |
313 |
0.73 |
| chr15_89565634_89565983 | 0.21 |
ENSG00000265866 |
. |
50125 |
0.12 |
| chr2_85940079_85940420 | 0.21 |
ENSG00000199687 |
. |
15211 |
0.13 |
| chr15_66909608_66909886 | 0.21 |
RP11-321F6.1 |
HCG2003567; Uncharacterized protein |
35219 |
0.12 |
| chr11_111385155_111385345 | 0.21 |
C11orf88 |
chromosome 11 open reading frame 88 |
260 |
0.77 |
| chr22_46518789_46519087 | 0.21 |
ENSG00000207875 |
. |
9372 |
0.11 |
| chr1_246345460_246345709 | 0.21 |
ENSG00000202184 |
. |
6836 |
0.23 |
| chr20_3745410_3745658 | 0.21 |
C20orf27 |
chromosome 20 open reading frame 27 |
2841 |
0.17 |
| chr8_72087638_72087789 | 0.21 |
RP11-326E22.1 |
|
19956 |
0.28 |
| chr17_78746879_78747269 | 0.21 |
RP11-28G8.1 |
|
32358 |
0.19 |
| chr13_40468804_40469100 | 0.21 |
ENSG00000212553 |
. |
37588 |
0.21 |
| chr15_101678714_101678865 | 0.21 |
RP11-505E24.2 |
|
52518 |
0.13 |
| chr6_170089803_170089954 | 0.21 |
ENSG00000265654 |
. |
7466 |
0.13 |
| chr22_38438083_38438604 | 0.21 |
RP5-1039K5.16 |
|
8108 |
0.11 |
| chr1_110165621_110165808 | 0.21 |
AMPD2 |
adenosine monophosphate deaminase 2 |
722 |
0.46 |
| chr4_14531121_14531355 | 0.21 |
ENSG00000202449 |
. |
161096 |
0.04 |
| chr19_17790699_17790876 | 0.21 |
UNC13A |
unc-13 homolog A (C. elegans) |
3809 |
0.18 |
| chr12_12016346_12017014 | 0.21 |
ETV6 |
ets variant 6 |
22191 |
0.26 |
| chr20_56482827_56482978 | 0.21 |
ENSG00000221007 |
. |
11038 |
0.21 |
| chr15_101656629_101657100 | 0.21 |
RP11-505E24.2 |
|
30593 |
0.19 |
| chr14_65813869_65814020 | 0.21 |
ENSG00000266740 |
. |
12043 |
0.25 |
| chr11_73032066_73032515 | 0.21 |
ARHGEF17 |
Rho guanine nucleotide exchange factor (GEF) 17 |
9680 |
0.14 |
| chr11_33982202_33982416 | 0.21 |
LMO2 |
LIM domain only 2 (rhombotin-like 1) |
68473 |
0.1 |
| chr12_58846810_58846966 | 0.20 |
RP11-362K2.2 |
Protein LOC100506869 |
91019 |
0.1 |
| chr9_35553068_35553219 | 0.20 |
FAM166B |
family with sequence similarity 166, member B |
10753 |
0.12 |
| chr19_50310955_50311176 | 0.20 |
FUZ |
fuzzy planar cell polarity protein |
831 |
0.3 |
| chr19_17221488_17221977 | 0.20 |
MYO9B |
myosin IXB |
9204 |
0.12 |
| chr10_100108984_100109336 | 0.20 |
ENSG00000221419 |
. |
45904 |
0.13 |
| chr1_149377811_149378198 | 0.20 |
FCGR1C |
Fc fragment of IgG, high affinity Ic, receptor (CD64), pseudogene |
8647 |
0.14 |
| chr16_85084815_85085194 | 0.20 |
KIAA0513 |
KIAA0513 |
11814 |
0.21 |
| chr12_125253324_125253475 | 0.20 |
SCARB1 |
scavenger receptor class B, member 1 |
48854 |
0.17 |
| chr2_206595324_206595734 | 0.20 |
AC007362.3 |
|
33148 |
0.2 |
| chr8_6149074_6149420 | 0.20 |
RP11-115C21.2 |
|
114816 |
0.06 |
| chr3_128690038_128690295 | 0.20 |
RP11-723O4.6 |
Uncharacterized protein FLJ43738 |
7 |
0.97 |
| chr7_73634593_73634808 | 0.20 |
LAT2 |
linker for activation of T cells family, member 2 |
10347 |
0.15 |
| chr22_38135230_38135506 | 0.20 |
TRIOBP |
TRIO and F-actin binding protein |
6867 |
0.12 |
| chr10_126916868_126917019 | 0.20 |
CTBP2 |
C-terminal binding protein 2 |
67313 |
0.13 |
| chr19_42470438_42470824 | 0.19 |
RABAC1 |
Rab acceptor 1 (prenylated) |
7089 |
0.14 |
| chr19_39156030_39156719 | 0.19 |
ACTN4 |
actinin, alpha 4 |
17996 |
0.11 |
| chr4_3495752_3496051 | 0.19 |
RP11-529E10.6 |
|
10283 |
0.16 |
| chr10_104436480_104437015 | 0.19 |
TRIM8 |
tripartite motif containing 8 |
32103 |
0.12 |
| chr10_60338881_60339032 | 0.19 |
BICC1 |
bicaudal C homolog 1 (Drosophila) |
66056 |
0.14 |
| chr15_40389876_40390209 | 0.19 |
BMF |
Bcl2 modifying factor |
8245 |
0.15 |
| chr22_39865304_39865652 | 0.19 |
MGAT3 |
mannosyl (beta-1,4-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase |
3315 |
0.18 |
| chr16_81868234_81868705 | 0.19 |
PLCG2 |
phospholipase C, gamma 2 (phosphatidylinositol-specific) |
48405 |
0.17 |
| chr10_73358221_73358464 | 0.19 |
CDH23 |
cadherin-related 23 |
27790 |
0.22 |
| chr8_144479919_144480248 | 0.19 |
RP11-909N17.2 |
|
14836 |
0.09 |
| chr16_31138954_31139105 | 0.19 |
KAT8 |
K(lysine) acetyltransferase 8 |
436 |
0.58 |
| chr6_611191_611411 | 0.19 |
HUS1B |
HUS1 checkpoint homolog b (S. pombe) |
45662 |
0.17 |
| chr11_69171319_69171574 | 0.19 |
MYEOV |
myeloma overexpressed |
109821 |
0.07 |
| chr3_10396103_10396350 | 0.19 |
ENSG00000264534 |
. |
24313 |
0.12 |
| chr10_89790579_89790730 | 0.19 |
ENSG00000200891 |
. |
36202 |
0.2 |
| chr3_69813958_69814283 | 0.19 |
MITF |
microphthalmia-associated transcription factor |
1158 |
0.56 |
| chr2_218776748_218776899 | 0.19 |
TNS1 |
tensin 1 |
6639 |
0.26 |
| chr15_89680763_89681294 | 0.19 |
ENSG00000239151 |
. |
17018 |
0.17 |
| chr10_440686_440837 | 0.18 |
DIP2C |
DIP2 disco-interacting protein 2 homolog C (Drosophila) |
7012 |
0.21 |
| chr17_6269742_6269893 | 0.18 |
AIPL1 |
aryl hydrocarbon receptor interacting protein-like 1 |
68582 |
0.1 |
| chr16_89622328_89622613 | 0.18 |
SPG7 |
spastic paraplegia 7 (pure and complicated autosomal recessive) |
2998 |
0.14 |
| chr18_10059619_10060204 | 0.18 |
ENSG00000263630 |
. |
54717 |
0.15 |
| chr9_137216593_137216789 | 0.18 |
RXRA |
retinoid X receptor, alpha |
1735 |
0.46 |
| chr3_45687605_45688015 | 0.18 |
LIMD1-AS1 |
LIMD1 antisense RNA 1 |
42564 |
0.12 |
| chr11_73041779_73041997 | 0.18 |
ARHGEF17 |
Rho guanine nucleotide exchange factor (GEF) 17 |
19278 |
0.12 |
| chr3_151122388_151122772 | 0.18 |
P2RY12 |
purinergic receptor P2Y, G-protein coupled, 12 |
19980 |
0.18 |
| chr3_122803672_122804475 | 0.18 |
PDIA5 |
protein disulfide isomerase family A, member 5 |
17878 |
0.23 |
| chr8_37660288_37660608 | 0.18 |
GPR124 |
G protein-coupled receptor 124 |
5674 |
0.16 |
| chr3_129666488_129666817 | 0.18 |
RP11-93K22.6 |
|
21485 |
0.15 |
| chr16_16168105_16168644 | 0.18 |
ABCC1 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 1 |
64661 |
0.11 |
| chr8_135033119_135033449 | 0.18 |
ENSG00000212273 |
. |
376316 |
0.01 |
| chr10_115753755_115753984 | 0.18 |
ADRB1 |
adrenoceptor beta 1 |
49937 |
0.13 |
| chr2_27301986_27302370 | 0.18 |
EMILIN1 |
elastin microfibril interfacer 1 |
743 |
0.39 |
| chr9_38038631_38039019 | 0.18 |
SHB |
Src homology 2 domain containing adaptor protein B |
30383 |
0.2 |
| chr10_29297172_29297323 | 0.18 |
ENSG00000199402 |
. |
133811 |
0.05 |
| chr15_82307610_82307761 | 0.18 |
ENSG00000222521 |
. |
28950 |
0.18 |
| chr9_130624750_130625121 | 0.18 |
ENSG00000263979 |
. |
6839 |
0.08 |
| chr1_158604437_158604588 | 0.18 |
OR10Z1 |
olfactory receptor, family 10, subfamily Z, member 1 |
28283 |
0.13 |
| chr16_16165629_16165805 | 0.18 |
ABCC1 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 1 |
62004 |
0.12 |
| chr1_36831464_36831995 | 0.18 |
STK40 |
serine/threonine kinase 40 |
4788 |
0.15 |
| chr13_108102676_108103085 | 0.18 |
ENSG00000221650 |
. |
80716 |
0.11 |
| chr2_127912815_127913111 | 0.18 |
BIN1 |
bridging integrator 1 |
48032 |
0.14 |
| chr3_129280215_129280484 | 0.18 |
PLXND1 |
plexin D1 |
450 |
0.78 |
| chr7_102530812_102531077 | 0.18 |
LRRC17 |
leucine rich repeat containing 17 |
22494 |
0.16 |
| chr5_177869992_177870288 | 0.18 |
CTB-26E19.1 |
|
45 |
0.98 |
| chr7_101360762_101360997 | 0.18 |
MYL10 |
myosin, light chain 10, regulatory |
88303 |
0.08 |
| chr17_47905442_47905593 | 0.18 |
RP11-304F15.3 |
|
17755 |
0.14 |
| chr1_17621907_17622242 | 0.18 |
PADI4 |
peptidyl arginine deiminase, type IV |
12616 |
0.15 |
| chr19_14486268_14486999 | 0.18 |
CD97 |
CD97 molecule |
5335 |
0.16 |
| chr22_33158107_33158258 | 0.18 |
LL22NC01-116C6.1 |
|
20981 |
0.22 |
| chr3_4518637_4518947 | 0.18 |
SUMF1 |
sulfatase modifying factor 1 |
9827 |
0.2 |
| chr14_71276457_71276666 | 0.17 |
MAP3K9 |
mitogen-activated protein kinase kinase kinase 9 |
310 |
0.94 |
| chr11_2763311_2763710 | 0.17 |
KCNQ1OT1 |
KCNQ1 opposite strand/antisense transcript 1 (non-protein coding) |
42286 |
0.15 |
| chr12_111873433_111874308 | 0.17 |
SH2B3 |
SH2B adaptor protein 3 |
1204 |
0.47 |
| chr6_114071112_114071516 | 0.17 |
ENSG00000253091 |
. |
31353 |
0.18 |
| chr6_54453109_54453260 | 0.17 |
ENSG00000251946 |
. |
198001 |
0.03 |
| chr10_14778907_14779278 | 0.17 |
FAM107B |
family with sequence similarity 107, member B |
37804 |
0.16 |
| chr11_118023633_118024140 | 0.17 |
SCN4B |
sodium channel, voltage-gated, type IV, beta subunit |
283 |
0.88 |
| chr6_89271446_89271597 | 0.17 |
ENSG00000222145 |
. |
152628 |
0.04 |
| chr6_39771799_39772005 | 0.17 |
DAAM2 |
dishevelled associated activator of morphogenesis 2 |
11108 |
0.26 |
| chr20_30657043_30657343 | 0.17 |
HCK |
hemopoietic cell kinase |
17129 |
0.12 |
| chr11_130253658_130253916 | 0.17 |
ADAMTS8 |
ADAM metallopeptidase with thrombospondin type 1 motif, 8 |
45101 |
0.15 |
| chr11_66368217_66368514 | 0.17 |
CCS |
copper chaperone for superoxide dismutase |
860 |
0.38 |
| chr11_17877616_17877767 | 0.17 |
KCNC1 |
potassium voltage-gated channel, Shaw-related subfamily, member 1 |
120171 |
0.05 |
| chr2_189540640_189540876 | 0.17 |
GULP1 |
GULP, engulfment adaptor PTB domain containing 1 |
105948 |
0.07 |
| chr5_76598141_76598292 | 0.17 |
PDE8B |
phosphodiesterase 8B |
23302 |
0.24 |
| chr6_20028504_20028655 | 0.17 |
RP11-239H6.2 |
|
183739 |
0.03 |
| chr2_216764593_216764988 | 0.17 |
ENSG00000212055 |
. |
21148 |
0.25 |
| chrX_135291994_135292627 | 0.17 |
FHL1 |
four and a half LIM domains 1 |
5430 |
0.24 |
| chr16_31037072_31037504 | 0.17 |
STX4 |
syntaxin 4 |
6922 |
0.08 |
| chr19_16556596_16556763 | 0.17 |
CTD-2013N17.4 |
|
4922 |
0.15 |
| chr21_41004853_41005004 | 0.17 |
AF064860.5 |
|
2730 |
0.27 |
| chr11_10347895_10348118 | 0.17 |
AMPD3 |
adenosine monophosphate deaminase 3 |
8788 |
0.17 |
| chr9_132096828_132097338 | 0.17 |
ENSG00000242281 |
. |
35657 |
0.16 |
| chr17_59311556_59311803 | 0.17 |
RP11-136H19.1 |
|
98354 |
0.06 |
| chr7_5649047_5649443 | 0.17 |
FSCN1 |
fascin homolog 1, actin-bundling protein (Strongylocentrotus purpuratus) |
14928 |
0.16 |
| chr5_77796055_77796354 | 0.17 |
LHFPL2 |
lipoma HMGIC fusion partner-like 2 |
48770 |
0.18 |
| chr15_78375318_78375576 | 0.17 |
SH2D7 |
SH2 domain containing 7 |
5297 |
0.14 |
| chr4_146702479_146702874 | 0.17 |
ZNF827 |
zinc finger protein 827 |
6857 |
0.25 |
| chr9_127025264_127025533 | 0.17 |
RP11-121A14.3 |
|
243 |
0.88 |
| chr20_48843010_48843216 | 0.17 |
CEBPB |
CCAAT/enhancer binding protein (C/EBP), beta |
35737 |
0.17 |
| chr4_86853987_86854157 | 0.17 |
ARHGAP24 |
Rho GTPase activating protein 24 |
2646 |
0.31 |
| chr12_2468276_2468593 | 0.17 |
CACNA1C-IT3 |
CACNA1C intronic transcript 3 (non-protein coding) |
89492 |
0.09 |
| chr3_183025161_183025367 | 0.17 |
B3GNT5 |
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5 |
42132 |
0.14 |
| chr8_37660040_37660273 | 0.17 |
GPR124 |
G protein-coupled receptor 124 |
5382 |
0.16 |
| chr11_114006986_114007170 | 0.17 |
ENSG00000221112 |
. |
49427 |
0.14 |
| chr18_54126758_54127059 | 0.17 |
TXNL1 |
thioredoxin-like 1 |
154814 |
0.04 |
| chr12_19727679_19727830 | 0.17 |
AEBP2 |
AE binding protein 2 |
74691 |
0.12 |
| chr9_79206598_79206788 | 0.17 |
ENSG00000241781 |
. |
19962 |
0.22 |
| chr2_127862116_127862267 | 0.17 |
BIN1 |
bridging integrator 1 |
2386 |
0.38 |
| chr4_54552846_54552997 | 0.17 |
RP11-317M11.1 |
|
9147 |
0.23 |
| chr14_100240950_100241315 | 0.17 |
EML1 |
echinoderm microtubule associated protein like 1 |
1091 |
0.59 |
| chr10_75727518_75727836 | 0.17 |
VCL |
vinculin |
30195 |
0.12 |
| chr21_36277232_36277480 | 0.17 |
RUNX1 |
runt-related transcription factor 1 |
15269 |
0.29 |
| chr1_184120290_184120490 | 0.17 |
ENSG00000199840 |
. |
20567 |
0.24 |
| chr1_21991980_21992494 | 0.17 |
RAP1GAP |
RAP1 GTPase activating protein |
3557 |
0.26 |
| chr1_39995674_39995906 | 0.17 |
RP11-69E11.4 |
|
4433 |
0.16 |
| chr10_134755869_134756456 | 0.17 |
TTC40 |
tetratricopeptide repeat domain 40 |
73 |
0.98 |
| chr15_63073163_63073722 | 0.17 |
ENSG00000265317 |
. |
5564 |
0.2 |
| chr5_139041444_139041605 | 0.17 |
CXXC5 |
CXXC finger protein 5 |
2384 |
0.32 |
| chr17_40111299_40111636 | 0.17 |
CNP |
2',3'-cyclic nucleotide 3' phosphodiesterase |
7292 |
0.12 |
| chr19_18476603_18476927 | 0.17 |
PGPEP1 |
pyroglutamyl-peptidase I |
8449 |
0.09 |
| chr6_37668651_37668857 | 0.17 |
MDGA1 |
MAM domain containing glycosylphosphatidylinositol anchor 1 |
1672 |
0.48 |
| chr6_43805886_43806228 | 0.17 |
VEGFA |
vascular endothelial growth factor A |
63967 |
0.1 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0045425 | positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) |
| 0.1 | 0.3 | GO:0007028 | cytoplasm organization(GO:0007028) |
| 0.1 | 0.3 | GO:0071731 | response to nitric oxide(GO:0071731) |
| 0.1 | 0.3 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.1 | 0.2 | GO:0033622 | integrin activation(GO:0033622) |
| 0.1 | 0.2 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.1 | 0.2 | GO:0032455 | nerve growth factor processing(GO:0032455) |
| 0.1 | 0.2 | GO:0060083 | smooth muscle contraction involved in micturition(GO:0060083) |
| 0.1 | 0.2 | GO:0051852 | disruption by host of symbiont cells(GO:0051852) killing by host of symbiont cells(GO:0051873) |
| 0.1 | 0.2 | GO:1901534 | positive regulation of megakaryocyte differentiation(GO:0045654) positive regulation of hematopoietic progenitor cell differentiation(GO:1901534) |
| 0.1 | 0.1 | GO:2000095 | regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.1 | 0.2 | GO:0003078 | obsolete regulation of natriuresis(GO:0003078) |
| 0.0 | 0.1 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.0 | 0.0 | GO:0060831 | smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:0060831) |
| 0.0 | 0.2 | GO:0070874 | negative regulation of glycogen metabolic process(GO:0070874) |
| 0.0 | 0.1 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.1 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.0 | 0.1 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.0 | 0.1 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.0 | 0.1 | GO:0032959 | inositol trisphosphate biosynthetic process(GO:0032959) |
| 0.0 | 0.1 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.1 | GO:0018101 | protein citrullination(GO:0018101) |
| 0.0 | 0.0 | GO:0051883 | disruption of cells of other organism involved in symbiotic interaction(GO:0051818) killing of cells in other organism involved in symbiotic interaction(GO:0051883) |
| 0.0 | 0.1 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.0 | 0.1 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.0 | 0.0 | GO:0072338 | allantoin metabolic process(GO:0000255) creatinine metabolic process(GO:0046449) cellular lactam metabolic process(GO:0072338) |
| 0.0 | 0.1 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) semicircular canal development(GO:0060872) |
| 0.0 | 0.2 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.1 | GO:0002138 | retinoic acid biosynthetic process(GO:0002138) diterpenoid biosynthetic process(GO:0016102) |
| 0.0 | 0.2 | GO:0045634 | regulation of melanocyte differentiation(GO:0045634) regulation of pigment cell differentiation(GO:0050932) |
| 0.0 | 0.2 | GO:0043116 | negative regulation of vascular permeability(GO:0043116) |
| 0.0 | 0.2 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.1 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) |
| 0.0 | 0.1 | GO:0034139 | regulation of toll-like receptor 3 signaling pathway(GO:0034139) |
| 0.0 | 0.3 | GO:0033866 | nucleoside bisphosphate biosynthetic process(GO:0033866) ribonucleoside bisphosphate biosynthetic process(GO:0034030) purine nucleoside bisphosphate biosynthetic process(GO:0034033) |
| 0.0 | 0.0 | GO:0002538 | arachidonic acid metabolite production involved in inflammatory response(GO:0002538) leukotriene production involved in inflammatory response(GO:0002540) |
| 0.0 | 0.1 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 | 0.1 | GO:0046010 | positive regulation of circadian sleep/wake cycle, non-REM sleep(GO:0046010) |
| 0.0 | 0.1 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.0 | 0.1 | GO:0061299 | retina vasculature development in camera-type eye(GO:0061298) retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 | 0.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.1 | GO:0061030 | epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.0 | 0.2 | GO:0006896 | Golgi to endosome transport(GO:0006895) Golgi to vacuole transport(GO:0006896) |
| 0.0 | 0.1 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.1 | GO:0045992 | negative regulation of embryonic development(GO:0045992) |
| 0.0 | 0.1 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.0 | 0.1 | GO:0002679 | respiratory burst involved in defense response(GO:0002679) |
| 0.0 | 0.1 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.2 | GO:0051957 | positive regulation of amino acid transport(GO:0051957) |
| 0.0 | 0.1 | GO:0002689 | negative regulation of leukocyte chemotaxis(GO:0002689) |
| 0.0 | 0.1 | GO:0015864 | pyrimidine nucleoside transport(GO:0015864) |
| 0.0 | 0.1 | GO:0001820 | serotonin secretion(GO:0001820) |
| 0.0 | 0.1 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 0.0 | GO:0060406 | positive regulation of penile erection(GO:0060406) |
| 0.0 | 0.1 | GO:0031558 | obsolete induction of apoptosis in response to chemical stimulus(GO:0031558) |
| 0.0 | 0.1 | GO:0021631 | optic nerve morphogenesis(GO:0021631) |
| 0.0 | 0.0 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.0 | 0.1 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.0 | 0.1 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.0 | 0.1 | GO:0016264 | gap junction assembly(GO:0016264) |
| 0.0 | 0.1 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.0 | 0.1 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.0 | 0.1 | GO:0001967 | suckling behavior(GO:0001967) |
| 0.0 | 0.1 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.0 | 0.0 | GO:0010193 | response to ozone(GO:0010193) |
| 0.0 | 0.0 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
| 0.0 | 0.1 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.2 | GO:0002040 | sprouting angiogenesis(GO:0002040) |
| 0.0 | 0.2 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.1 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.1 | GO:0046618 | drug export(GO:0046618) |
| 0.0 | 0.2 | GO:0070373 | negative regulation of ERK1 and ERK2 cascade(GO:0070373) |
| 0.0 | 0.1 | GO:0060638 | mesenchymal-epithelial cell signaling(GO:0060638) |
| 0.0 | 0.1 | GO:0010891 | negative regulation of sequestering of triglyceride(GO:0010891) |
| 0.0 | 0.1 | GO:0046532 | regulation of photoreceptor cell differentiation(GO:0046532) |
| 0.0 | 0.0 | GO:0046877 | regulation of saliva secretion(GO:0046877) |
| 0.0 | 0.1 | GO:0043277 | apoptotic cell clearance(GO:0043277) |
| 0.0 | 0.1 | GO:0042339 | keratan sulfate metabolic process(GO:0042339) |
| 0.0 | 0.0 | GO:0010759 | regulation of macrophage chemotaxis(GO:0010758) positive regulation of macrophage chemotaxis(GO:0010759) mononuclear cell migration(GO:0071674) regulation of mononuclear cell migration(GO:0071675) |
| 0.0 | 0.0 | GO:0042494 | detection of bacterial lipoprotein(GO:0042494) detection of bacterial lipopeptide(GO:0070340) |
| 0.0 | 0.3 | GO:0044364 | killing of cells of other organism(GO:0031640) disruption of cells of other organism(GO:0044364) |
| 0.0 | 0.1 | GO:0043030 | regulation of macrophage activation(GO:0043030) |
| 0.0 | 0.2 | GO:0000085 | mitotic G2 phase(GO:0000085) G2 phase(GO:0051319) |
| 0.0 | 0.0 | GO:0048245 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.0 | 0.2 | GO:0006911 | phagocytosis, engulfment(GO:0006911) membrane invagination(GO:0010324) |
| 0.0 | 0.1 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) positive regulation of neutrophil migration(GO:1902624) |
| 0.0 | 0.1 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 0.0 | 0.1 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 0.2 | GO:0060644 | mammary gland epithelial cell differentiation(GO:0060644) |
| 0.0 | 0.2 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.0 | 0.1 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.0 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.0 | 0.1 | GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0043619) |
| 0.0 | 0.1 | GO:0001553 | luteinization(GO:0001553) |
| 0.0 | 0.1 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.1 | GO:0060180 | regulation of female receptivity(GO:0045924) female mating behavior(GO:0060180) |
| 0.0 | 0.1 | GO:0032303 | regulation of icosanoid secretion(GO:0032303) positive regulation of icosanoid secretion(GO:0032305) regulation of prostaglandin secretion(GO:0032306) positive regulation of prostaglandin secretion(GO:0032308) prostaglandin secretion(GO:0032310) |
| 0.0 | 0.1 | GO:0061318 | renal filtration cell differentiation(GO:0061318) glomerular epithelium development(GO:0072010) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.0 | 0.0 | GO:0072179 | nephric duct formation(GO:0072179) |
| 0.0 | 0.1 | GO:1900115 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.1 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.0 | 0.1 | GO:0045652 | regulation of megakaryocyte differentiation(GO:0045652) regulation of hematopoietic progenitor cell differentiation(GO:1901532) |
| 0.0 | 0.1 | GO:0034331 | cell junction maintenance(GO:0034331) cell-cell junction maintenance(GO:0045217) |
| 0.0 | 0.1 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.0 | 0.1 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.0 | 0.1 | GO:0055064 | cellular chloride ion homeostasis(GO:0030644) chloride ion homeostasis(GO:0055064) |
| 0.0 | 0.0 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.0 | GO:0070254 | mucus secretion(GO:0070254) |
| 0.0 | 0.1 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.0 | 0.1 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.1 | GO:0032373 | positive regulation of sterol transport(GO:0032373) positive regulation of cholesterol transport(GO:0032376) |
| 0.0 | 0.1 | GO:0090231 | regulation of spindle checkpoint(GO:0090231) |
| 0.0 | 0.1 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.1 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.0 | 0.1 | GO:0052805 | histidine catabolic process(GO:0006548) imidazole-containing compound catabolic process(GO:0052805) |
| 0.0 | 0.0 | GO:0039656 | modulation by virus of host gene expression(GO:0039656) |
| 0.0 | 0.1 | GO:0015936 | coenzyme A metabolic process(GO:0015936) |
| 0.0 | 0.1 | GO:0002052 | positive regulation of neuroblast proliferation(GO:0002052) |
| 0.0 | 0.0 | GO:0060014 | granulosa cell differentiation(GO:0060014) |
| 0.0 | 0.0 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.0 | 0.3 | GO:0050926 | regulation of positive chemotaxis(GO:0050926) |
| 0.0 | 0.1 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) phosphatidylethanolamine metabolic process(GO:0046337) |
| 0.0 | 0.0 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.0 | 0.1 | GO:0006533 | aspartate metabolic process(GO:0006531) aspartate catabolic process(GO:0006533) |
| 0.0 | 0.0 | GO:0048875 | surfactant homeostasis(GO:0043129) chemical homeostasis within a tissue(GO:0048875) |
| 0.0 | 0.0 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.0 | 0.0 | GO:0031223 | response to auditory stimulus(GO:0010996) auditory behavior(GO:0031223) |
| 0.0 | 0.1 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.2 | GO:0048286 | lung alveolus development(GO:0048286) |
| 0.0 | 0.1 | GO:0009129 | pyrimidine nucleoside monophosphate metabolic process(GO:0009129) |
| 0.0 | 0.1 | GO:0042119 | neutrophil activation(GO:0042119) |
| 0.0 | 0.0 | GO:0048311 | mitochondrion distribution(GO:0048311) |
| 0.0 | 0.1 | GO:0034505 | tooth mineralization(GO:0034505) |
| 0.0 | 0.0 | GO:0051386 | regulation of neurotrophin TRK receptor signaling pathway(GO:0051386) |
| 0.0 | 0.0 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.1 | GO:0006551 | leucine metabolic process(GO:0006551) |
| 0.0 | 0.0 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.0 | 0.1 | GO:0070498 | interleukin-1-mediated signaling pathway(GO:0070498) |
| 0.0 | 0.0 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.1 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.0 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.0 | 0.1 | GO:0016265 | obsolete death(GO:0016265) |
| 0.0 | 0.0 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.0 | 0.0 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.1 | GO:0031033 | myosin filament organization(GO:0031033) |
| 0.0 | 0.1 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 0.0 | GO:0021910 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) |
| 0.0 | 0.0 | GO:0048865 | stem cell fate commitment(GO:0048865) stem cell fate determination(GO:0048867) |
| 0.0 | 0.1 | GO:0090025 | regulation of monocyte chemotaxis(GO:0090025) positive regulation of monocyte chemotaxis(GO:0090026) |
| 0.0 | 0.0 | GO:0003085 | negative regulation of systemic arterial blood pressure(GO:0003085) |
| 0.0 | 0.1 | GO:0032206 | positive regulation of telomere maintenance(GO:0032206) |
| 0.0 | 0.0 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.0 | 0.0 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.0 | GO:0003032 | detection of oxygen(GO:0003032) |
| 0.0 | 0.1 | GO:0015824 | proline transport(GO:0015824) |
| 0.0 | 0.0 | GO:0014052 | regulation of gamma-aminobutyric acid secretion(GO:0014052) |
| 0.0 | 0.1 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.1 | GO:0033962 | cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.0 | 0.1 | GO:0090398 | cellular senescence(GO:0090398) |
| 0.0 | 0.1 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.0 | 0.0 | GO:0006547 | histidine metabolic process(GO:0006547) |
| 0.0 | 0.1 | GO:0042693 | muscle cell fate commitment(GO:0042693) |
| 0.0 | 0.2 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.1 | GO:0046835 | carbohydrate phosphorylation(GO:0046835) |
| 0.0 | 0.0 | GO:0021938 | smoothened signaling pathway involved in regulation of cerebellar granule cell precursor cell proliferation(GO:0021938) |
| 0.0 | 0.0 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.1 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.1 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 0.0 | GO:0051305 | chromosome movement towards spindle pole(GO:0051305) |
| 0.0 | 0.1 | GO:0014912 | negative regulation of smooth muscle cell migration(GO:0014912) |
| 0.0 | 0.0 | GO:0045901 | translational frameshifting(GO:0006452) positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.0 | GO:0051901 | positive regulation of mitochondrial depolarization(GO:0051901) positive regulation of membrane depolarization(GO:1904181) |
| 0.0 | 0.0 | GO:0072526 | pyridine-containing compound catabolic process(GO:0072526) |
| 0.0 | 0.0 | GO:0060916 | mesenchymal cell proliferation involved in lung development(GO:0060916) |
| 0.0 | 0.0 | GO:0045348 | positive regulation of MHC class II biosynthetic process(GO:0045348) |
| 0.0 | 0.0 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.0 | 0.1 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.0 | GO:0043045 | DNA methylation involved in embryo development(GO:0043045) changes to DNA methylation involved in embryo development(GO:1901538) |
| 0.0 | 0.0 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.0 | GO:0010874 | regulation of cholesterol efflux(GO:0010874) |
| 0.0 | 0.1 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.1 | GO:0007263 | nitric oxide mediated signal transduction(GO:0007263) |
| 0.0 | 0.0 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 0.1 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.0 | 0.0 | GO:1903392 | negative regulation of focal adhesion assembly(GO:0051895) negative regulation of cell junction assembly(GO:1901889) negative regulation of adherens junction organization(GO:1903392) |
| 0.0 | 0.0 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.1 | GO:0071545 | inositol phosphate catabolic process(GO:0071545) |
| 0.0 | 0.0 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.0 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 0.0 | GO:0006893 | Golgi to plasma membrane transport(GO:0006893) vesicle-mediated transport to the plasma membrane(GO:0098876) |
| 0.0 | 0.0 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.0 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 | 0.0 | GO:0032026 | response to magnesium ion(GO:0032026) |
| 0.0 | 0.1 | GO:0071347 | cellular response to interleukin-1(GO:0071347) |
| 0.0 | 0.1 | GO:0007199 | G-protein coupled receptor signaling pathway coupled to cGMP nucleotide second messenger(GO:0007199) |
| 0.0 | 0.1 | GO:0007549 | dosage compensation(GO:0007549) |
| 0.0 | 0.1 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 0.1 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.0 | GO:0035511 | oxidative DNA demethylation(GO:0035511) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.2 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 0.2 | GO:0070083 | clathrin-sculpted monoamine transport vesicle(GO:0070081) clathrin-sculpted monoamine transport vesicle membrane(GO:0070083) |
| 0.0 | 0.1 | GO:0005954 | calcium- and calmodulin-dependent protein kinase complex(GO:0005954) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.1 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.1 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.1 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.1 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.1 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.1 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.1 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.1 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.1 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.1 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.1 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 0.0 | GO:0002141 | stereocilia coupling link(GO:0002139) stereocilia ankle link(GO:0002141) stereocilia ankle link complex(GO:0002142) |
| 0.0 | 0.2 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.3 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.1 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.1 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.1 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.2 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.1 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.1 | GO:0001740 | Barr body(GO:0001740) |
| 0.0 | 0.0 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.1 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.0 | 0.0 | GO:0044462 | external encapsulating structure part(GO:0044462) |
| 0.0 | 0.1 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.1 | GO:0098642 | network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.0 | 0.0 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.0 | 0.1 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.0 | 0.0 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.1 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.0 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.0 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.0 | GO:0098651 | basement membrane collagen trimer(GO:0098651) |
| 0.0 | 0.1 | GO:0043205 | fibril(GO:0043205) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0001098 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.1 | 0.2 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 0.2 | GO:0050542 | icosanoid binding(GO:0050542) fatty acid derivative binding(GO:1901567) |
| 0.1 | 0.2 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.3 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.1 | GO:0042808 | obsolete neuronal Cdc2-like kinase binding(GO:0042808) |
| 0.0 | 0.2 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.2 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.1 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.0 | 0.0 | GO:0050543 | icosatetraenoic acid binding(GO:0050543) |
| 0.0 | 0.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.1 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.2 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.1 | GO:0071553 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.0 | 0.3 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 0.1 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.1 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.1 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.0 | 0.1 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.0 | 0.1 | GO:0052659 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) inositol-1,3,4,5-tetrakisphosphate 5-phosphatase activity(GO:0052659) |
| 0.0 | 0.3 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.0 | 0.1 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.3 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.0 | 0.2 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.1 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.1 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.1 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.1 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.1 | GO:0016717 | oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water(GO:0016717) |
| 0.0 | 0.1 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.2 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.0 | 0.1 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.0 | 0.0 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.1 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.1 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.1 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.1 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.0 | 0.1 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.0 | 0.1 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.1 | GO:0070696 | transmembrane receptor protein serine/threonine kinase binding(GO:0070696) |
| 0.0 | 0.0 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.1 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.1 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.1 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.1 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.1 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.0 | 0.1 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.1 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.2 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.1 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.1 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.1 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.1 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.1 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.0 | 0.1 | GO:0019959 | C-X-C chemokine binding(GO:0019958) interleukin-8 binding(GO:0019959) |
| 0.0 | 0.2 | GO:0051635 | obsolete bacterial cell surface binding(GO:0051635) |
| 0.0 | 0.1 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.1 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.0 | 0.8 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 0.1 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.0 | GO:0099602 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.0 | 0.1 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 0.0 | GO:0019863 | IgE binding(GO:0019863) |
| 0.0 | 0.1 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.1 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.4 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.0 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.0 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.1 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.0 | GO:0005350 | pyrimidine nucleobase transmembrane transporter activity(GO:0005350) |
| 0.0 | 0.0 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.0 | 0.2 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.0 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.0 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.0 | 0.0 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.1 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.0 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.0 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.1 | GO:0001846 | opsonin binding(GO:0001846) |
| 0.0 | 0.0 | GO:0005497 | androgen binding(GO:0005497) |
| 0.0 | 0.1 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.0 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.0 | GO:0047115 | trans-1,2-dihydrobenzene-1,2-diol dehydrogenase activity(GO:0047115) |
| 0.0 | 0.0 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.1 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.0 | 0.0 | GO:0036041 | long-chain fatty acid binding(GO:0036041) |
| 0.0 | 0.1 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.0 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.0 | GO:0002054 | nucleobase binding(GO:0002054) purine nucleobase binding(GO:0002060) |
| 0.0 | 0.1 | GO:0016595 | glutamate binding(GO:0016595) |
| 0.0 | 0.1 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 0.0 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.0 | 0.2 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.0 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.0 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.1 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.0 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.1 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.0 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.0 | 0.0 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.0 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.0 | GO:0001537 | N-acetylgalactosamine 4-O-sulfotransferase activity(GO:0001537) |
| 0.0 | 0.1 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.0 | 0.0 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.0 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.0 | 0.0 | GO:0035514 | DNA demethylase activity(GO:0035514) DNA-N1-methyladenine dioxygenase activity(GO:0043734) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.1 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 0.0 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.1 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.1 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.3 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.0 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.0 | 0.1 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | REACTOME PHOSPHOLIPASE C MEDIATED CASCADE | Genes involved in Phospholipase C-mediated cascade |
| 0.0 | 0.3 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.0 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.2 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.3 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.0 | REACTOME HEMOSTASIS | Genes involved in Hemostasis |
| 0.0 | 0.2 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.1 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.2 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.0 | REACTOME ACTIVATION OF NMDA RECEPTOR UPON GLUTAMATE BINDING AND POSTSYNAPTIC EVENTS | Genes involved in Activation of NMDA receptor upon glutamate binding and postsynaptic events |
| 0.0 | 0.0 | REACTOME APC C CDC20 MEDIATED DEGRADATION OF MITOTIC PROTEINS | Genes involved in APC/C:Cdc20 mediated degradation of mitotic proteins |
| 0.0 | 0.0 | REACTOME NUCLEAR EVENTS KINASE AND TRANSCRIPTION FACTOR ACTIVATION | Genes involved in Nuclear Events (kinase and transcription factor activation) |
| 0.0 | 0.1 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 0.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.1 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.2 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.0 | REACTOME SIGNALING BY ERBB2 | Genes involved in Signaling by ERBB2 |
| 0.0 | 0.2 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.1 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.0 | 0.0 | REACTOME FRS2 MEDIATED CASCADE | Genes involved in FRS2-mediated cascade |
| 0.0 | 0.1 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.1 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |