Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
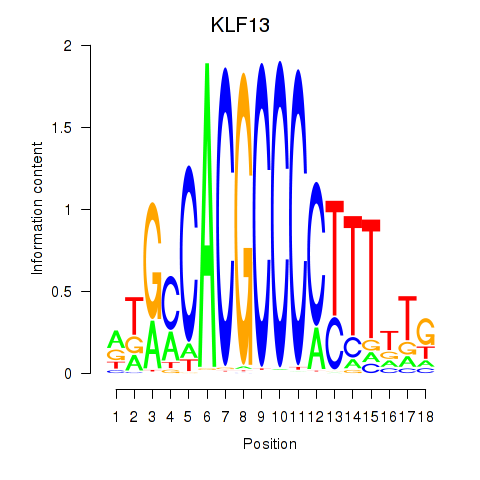
Results for KLF13
Z-value: 1.02
Transcription factors associated with KLF13
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
KLF13
|
ENSG00000169926.5 | Kruppel like factor 13 |
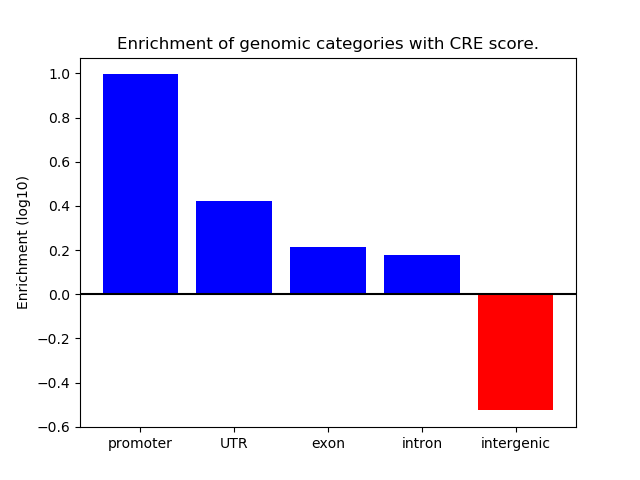
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr15_31619464_31619615 | KLF13 | 481 | 0.887517 | 0.90 | 8.1e-04 | Click! |
| chr15_31621843_31621994 | KLF13 | 2860 | 0.395501 | 0.86 | 3.1e-03 | Click! |
| chr15_31645569_31645720 | KLF13 | 12713 | 0.294014 | 0.86 | 3.2e-03 | Click! |
| chr15_31600904_31601060 | KLF13 | 18076 | 0.272251 | 0.85 | 3.7e-03 | Click! |
| chr15_31644683_31644849 | KLF13 | 12630 | 0.294151 | 0.83 | 5.2e-03 | Click! |
Activity of the KLF13 motif across conditions
Conditions sorted by the z-value of the KLF13 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
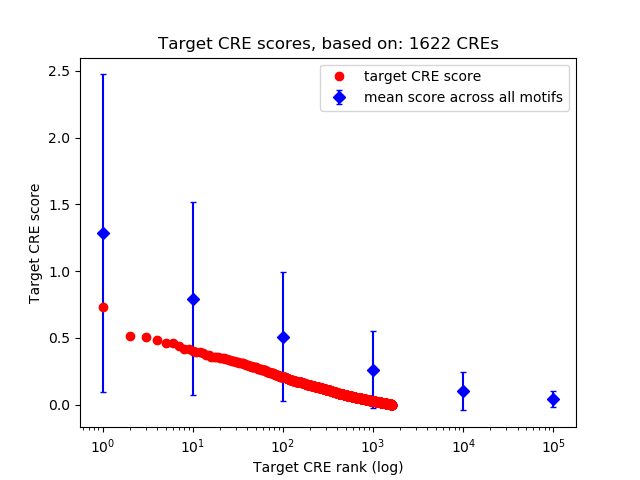
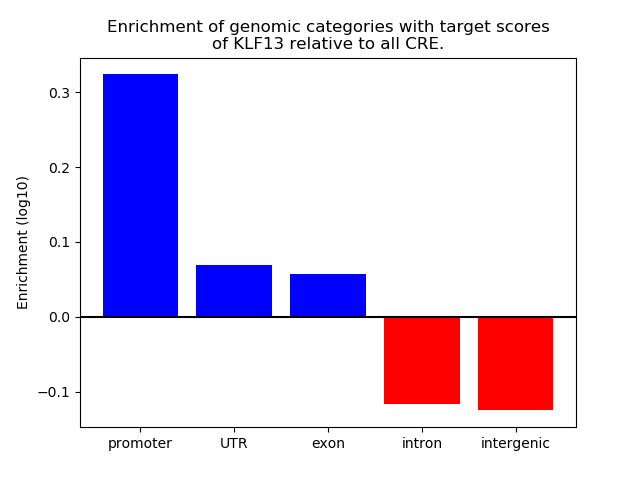
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr17_7184802_7184953 | 0.73 |
SLC2A4 |
solute carrier family 2 (facilitated glucose transporter), member 4 |
109 |
0.88 |
| chr1_161146927_161147078 | 0.51 |
B4GALT3 |
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 3 |
273 |
0.76 |
| chr19_46103286_46103456 | 0.51 |
GPR4 |
G protein-coupled receptor 4 |
2095 |
0.16 |
| chr10_134265737_134265934 | 0.48 |
C10orf91 |
chromosome 10 open reading frame 91 |
7142 |
0.19 |
| chr22_18269517_18269668 | 0.46 |
BID |
BH3 interacting domain death agonist |
12161 |
0.18 |
| chr19_55952161_55952312 | 0.46 |
SHISA7 |
shisa family member 7 |
1468 |
0.21 |
| chr9_131971017_131971168 | 0.44 |
RP11-247A12.2 |
|
7498 |
0.15 |
| chr2_219258625_219258977 | 0.42 |
CTDSP1 |
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 1 |
4260 |
0.1 |
| chr6_6886561_6886823 | 0.41 |
ENSG00000240936 |
. |
52438 |
0.17 |
| chr2_71295991_71296142 | 0.40 |
NAGK |
N-acetylglucosamine kinase |
274 |
0.84 |
| chr6_158993431_158993582 | 0.40 |
TMEM181 |
transmembrane protein 181 |
36038 |
0.15 |
| chr15_99601132_99601283 | 0.40 |
SYNM |
synemin, intermediate filament protein |
37213 |
0.12 |
| chr1_7130417_7130568 | 0.39 |
RP11-334N17.1 |
|
56153 |
0.15 |
| chr5_171604379_171604532 | 0.37 |
STK10 |
serine/threonine kinase 10 |
10935 |
0.19 |
| chr16_57174332_57174517 | 0.37 |
CPNE2 |
copine II |
21313 |
0.13 |
| chr18_5057426_5057577 | 0.36 |
ENSG00000222463 |
. |
69333 |
0.13 |
| chr19_18898466_18898690 | 0.36 |
COMP |
cartilage oligomeric matrix protein |
3536 |
0.19 |
| chr2_740601_740858 | 0.35 |
AC092159.3 |
|
47494 |
0.15 |
| chr19_49931671_49931926 | 0.35 |
GFY |
golgi-associated, olfactory signaling regulator |
4792 |
0.07 |
| chr5_139487909_139488137 | 0.35 |
LINC01024 |
long intergenic non-protein coding RNA 1024 |
127 |
0.95 |
| chr19_15580914_15581065 | 0.35 |
RASAL3 |
RAS protein activator like 3 |
5607 |
0.13 |
| chr19_55972386_55972537 | 0.35 |
ISOC2 |
isochorismatase domain containing 2 |
475 |
0.62 |
| chr1_68151591_68151742 | 0.35 |
GADD45A |
growth arrest and DNA-damage-inducible, alpha |
782 |
0.69 |
| chr11_336653_336830 | 0.34 |
IFITM3 |
interferon induced transmembrane protein 3 |
9204 |
0.07 |
| chr17_2696074_2696428 | 0.34 |
RAP1GAP2 |
RAP1 GTPase activating protein 2 |
3481 |
0.22 |
| chr19_46144336_46144530 | 0.33 |
C19orf83 |
chromosome 19 open reading frame 83 |
516 |
0.5 |
| chr20_3102729_3102880 | 0.33 |
UBOX5-AS1 |
UBOX5 antisense RNA 1 |
11521 |
0.11 |
| chr2_85059019_85059256 | 0.33 |
TRABD2A |
TraB domain containing 2A |
49069 |
0.13 |
| chr17_74493523_74493698 | 0.33 |
RHBDF2 |
rhomboid 5 homolog 2 (Drosophila) |
3843 |
0.12 |
| chr15_64126438_64126618 | 0.32 |
HERC1 |
HECT and RLD domain containing E3 ubiquitin protein ligase family member 1 |
387 |
0.89 |
| chr20_61609083_61609372 | 0.32 |
SLC17A9 |
solute carrier family 17 (vesicular nucleotide transporter), member 9 |
24829 |
0.13 |
| chr2_85153301_85153532 | 0.32 |
TMSB10 |
thymosin beta 10 |
20667 |
0.17 |
| chr14_74416480_74416631 | 0.32 |
COQ6 |
coenzyme Q6 monooxygenase |
74 |
0.75 |
| chr6_37467874_37468104 | 0.32 |
CCDC167 |
coiled-coil domain containing 167 |
291 |
0.92 |
| chr17_64962073_64962262 | 0.31 |
CACNG4 |
calcium channel, voltage-dependent, gamma subunit 4 |
1141 |
0.49 |
| chr8_134508796_134508947 | 0.31 |
ST3GAL1 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
2733 |
0.41 |
| chr6_15017519_15017967 | 0.31 |
ENSG00000242989 |
. |
95456 |
0.09 |
| chr14_75802560_75802738 | 0.30 |
FOS |
FBJ murine osteosarcoma viral oncogene homolog |
55753 |
0.1 |
| chr10_6192977_6193257 | 0.30 |
ENSG00000263628 |
. |
1042 |
0.38 |
| chr2_16080025_16080176 | 0.30 |
MYCN |
v-myc avian myelocytomatosis viral oncogene neuroblastoma derived homolog |
586 |
0.63 |
| chr19_862652_862803 | 0.29 |
CFD |
complement factor D (adipsin) |
3037 |
0.11 |
| chr13_50918820_50919100 | 0.29 |
ENSG00000221198 |
. |
18413 |
0.28 |
| chr3_37285712_37286013 | 0.29 |
RP11-259K5.2 |
|
194 |
0.89 |
| chr14_93313883_93314055 | 0.29 |
GOLGA5 |
golgin A5 |
14498 |
0.25 |
| chr3_158519751_158519902 | 0.29 |
MFSD1 |
major facilitator superfamily domain containing 1 |
54 |
0.56 |
| chr1_1118104_1118264 | 0.28 |
TTLL10 |
tubulin tyrosine ligase-like family, member 10 |
3107 |
0.1 |
| chr1_160684089_160684457 | 0.28 |
CD48 |
CD48 molecule |
2632 |
0.23 |
| chr17_72735203_72735698 | 0.28 |
RAB37 |
RAB37, member RAS oncogene family |
2079 |
0.18 |
| chr2_99364356_99364519 | 0.28 |
MGAT4A |
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A |
16848 |
0.2 |
| chr8_11324526_11324725 | 0.28 |
FAM167A |
family with sequence similarity 167, member A |
349 |
0.85 |
| chr10_62727947_62728098 | 0.28 |
RHOBTB1 |
Rho-related BTB domain containing 1 |
24017 |
0.24 |
| chr1_90396399_90396550 | 0.28 |
ENSG00000252797 |
. |
57029 |
0.13 |
| chr8_142130442_142130593 | 0.27 |
DENND3 |
DENN/MADD domain containing 3 |
3140 |
0.26 |
| chr3_196372798_196372949 | 0.27 |
PIGX |
phosphatidylinositol glycan anchor biosynthesis, class X |
6227 |
0.14 |
| chr17_25870024_25870175 | 0.27 |
KSR1 |
kinase suppressor of ras 1 |
39848 |
0.15 |
| chr17_47817453_47817604 | 0.27 |
FAM117A |
family with sequence similarity 117, member A |
15639 |
0.14 |
| chr8_143385130_143385421 | 0.27 |
TSNARE1 |
t-SNARE domain containing 1 |
50849 |
0.13 |
| chr8_82543299_82543455 | 0.26 |
IMPA1P |
inositol(myo)-1(or 4)-monophosphatase 1 pseudogene |
4193 |
0.23 |
| chrX_129243058_129243209 | 0.26 |
ELF4 |
E74-like factor 4 (ets domain transcription factor) |
1203 |
0.51 |
| chr15_99258837_99259137 | 0.26 |
IGF1R |
insulin-like growth factor 1 receptor |
7927 |
0.26 |
| chr15_75055139_75055425 | 0.26 |
CYP1A2 |
cytochrome P450, family 1, subfamily A, polypeptide 2 |
14097 |
0.11 |
| chr19_39971056_39971294 | 0.26 |
TIMM50 |
translocase of inner mitochondrial membrane 50 homolog (S. cerevisiae) |
123 |
0.92 |
| chr1_26100922_26101094 | 0.26 |
SEPN1 |
selenoprotein N, 1 |
25659 |
0.11 |
| chr11_118309485_118309636 | 0.26 |
KMT2A |
lysine (K)-specific methyltransferase 2A |
2083 |
0.18 |
| chr14_104338342_104338493 | 0.25 |
PPP1R13B |
protein phosphatase 1, regulatory subunit 13B |
24490 |
0.13 |
| chr9_131267547_131267753 | 0.25 |
GLE1 |
GLE1 RNA export mediator |
565 |
0.62 |
| chr19_10528216_10528367 | 0.25 |
PDE4A |
phosphodiesterase 4A, cAMP-specific |
672 |
0.52 |
| chr11_61658320_61658531 | 0.25 |
FADS3 |
fatty acid desaturase 3 |
428 |
0.76 |
| chr10_71078022_71078173 | 0.25 |
HK1 |
hexokinase 1 |
503 |
0.8 |
| chr16_57177310_57177461 | 0.25 |
CPNE2 |
copine II |
24274 |
0.12 |
| chr3_69134778_69134936 | 0.24 |
ARL6IP5 |
ADP-ribosylation-like factor 6 interacting protein 5 |
685 |
0.62 |
| chr17_46660068_46660219 | 0.24 |
HOXB3 |
homeobox B3 |
159 |
0.76 |
| chr20_10530697_10531175 | 0.24 |
JAG1 |
jagged 1 |
112218 |
0.06 |
| chr19_6261889_6262159 | 0.24 |
CTC-503J8.4 |
|
2856 |
0.2 |
| chr19_18775251_18775402 | 0.24 |
CRTC1 |
CREB regulated transcription coactivator 1 |
19161 |
0.11 |
| chr9_92063731_92064015 | 0.24 |
SEMA4D |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D |
6995 |
0.26 |
| chrX_153169917_153170068 | 0.23 |
AVPR2 |
arginine vasopressin receptor 2 |
197 |
0.87 |
| chr16_58018074_58018383 | 0.23 |
TEPP |
testis, prostate and placenta expressed |
7889 |
0.13 |
| chr14_105641458_105641667 | 0.23 |
NUDT14 |
nudix (nucleoside diphosphate linked moiety X)-type motif 14 |
6078 |
0.17 |
| chrX_109543624_109543775 | 0.23 |
AMMECR1-IT1 |
AMMECR1 intronic transcript 1 (non-protein coding) |
6750 |
0.2 |
| chr18_43913133_43913284 | 0.23 |
RNF165 |
ring finger protein 165 |
723 |
0.78 |
| chr2_60618463_60618614 | 0.23 |
ENSG00000266078 |
. |
3958 |
0.27 |
| chr10_94582178_94582340 | 0.23 |
EXOC6 |
exocyst complex component 6 |
8676 |
0.28 |
| chr18_77450365_77450516 | 0.23 |
CTDP1 |
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) phosphatase, subunit 1 |
9591 |
0.23 |
| chr10_74384901_74385052 | 0.22 |
MICU1 |
mitochondrial calcium uptake 1 |
860 |
0.57 |
| chr19_900208_900359 | 0.22 |
ENSG00000207507 |
. |
6799 |
0.08 |
| chr20_58691613_58691835 | 0.22 |
C20orf197 |
chromosome 20 open reading frame 197 |
60744 |
0.14 |
| chr1_221052508_221052659 | 0.22 |
HLX |
H2.0-like homeobox |
884 |
0.41 |
| chr3_194354391_194354771 | 0.22 |
TMEM44 |
transmembrane protein 44 |
163 |
0.88 |
| chr2_208364611_208364762 | 0.22 |
CREB1 |
cAMP responsive element binding protein 1 |
29775 |
0.18 |
| chr4_141077881_141078032 | 0.22 |
MAML3 |
mastermind-like 3 (Drosophila) |
2618 |
0.33 |
| chr2_178937506_178937762 | 0.21 |
PDE11A |
phosphodiesterase 11A |
152 |
0.96 |
| chr12_48357535_48357686 | 0.21 |
TMEM106C |
transmembrane protein 106C |
31 |
0.97 |
| chr1_39665289_39665440 | 0.21 |
MACF1 |
microtubule-actin crosslinking factor 1 |
4554 |
0.2 |
| chr11_124746668_124746852 | 0.21 |
ROBO3 |
roundabout, axon guidance receptor, homolog 3 (Drosophila) |
524 |
0.64 |
| chr9_77567865_77568016 | 0.21 |
RP11-197P3.4 |
|
56 |
0.61 |
| chr10_77168496_77168860 | 0.21 |
ENSG00000237149 |
. |
5866 |
0.2 |
| chr6_144607266_144607540 | 0.21 |
UTRN |
utrophin |
566 |
0.83 |
| chr4_71599845_71600265 | 0.21 |
RUFY3 |
RUN and FYVE domain containing 3 |
8 |
0.97 |
| chr1_167585648_167585802 | 0.21 |
RCSD1 |
RCSD domain containing 1 |
13605 |
0.19 |
| chr8_144416616_144416856 | 0.21 |
TOP1MT |
topoisomerase (DNA) I, mitochondrial |
229 |
0.86 |
| chr2_47175075_47175226 | 0.21 |
MCFD2 |
multiple coagulation factor deficiency 2 |
6156 |
0.16 |
| chr15_86036305_86036456 | 0.21 |
AKAP13 |
A kinase (PRKA) anchor protein 13 |
50891 |
0.13 |
| chr9_249028_249455 | 0.21 |
RP11-59O6.3 |
|
23761 |
0.16 |
| chr20_60606558_60606856 | 0.21 |
TAF4 |
TAF4 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 135kDa |
24090 |
0.16 |
| chr1_181059536_181059906 | 0.21 |
IER5 |
immediate early response 5 |
2083 |
0.37 |
| chr12_109125460_109125611 | 0.20 |
CORO1C |
coronin, actin binding protein, 1C |
163 |
0.94 |
| chr9_99775939_99776257 | 0.20 |
HIATL2 |
hippocampus abundant transcript-like 2 |
236 |
0.94 |
| chr1_12148511_12148915 | 0.20 |
ENSG00000201135 |
. |
10672 |
0.15 |
| chr19_8213404_8213659 | 0.20 |
FBN3 |
fibrillin 3 |
222 |
0.93 |
| chr11_74855211_74855408 | 0.20 |
CTD-2562J17.2 |
|
4116 |
0.2 |
| chr4_42637963_42638114 | 0.20 |
ATP8A1 |
ATPase, aminophospholipid transporter (APLT), class I, type 8A, member 1 |
21072 |
0.22 |
| chr19_52148720_52148871 | 0.19 |
SIGLEC5 |
sialic acid binding Ig-like lectin 5 |
3 |
0.95 |
| chr20_32255270_32255449 | 0.19 |
NECAB3 |
N-terminal EF-hand calcium binding protein 3 |
894 |
0.32 |
| chr11_4107967_4108118 | 0.19 |
RRM1 |
ribonucleotide reductase M1 |
7997 |
0.2 |
| chr19_55667901_55668169 | 0.19 |
TNNI3 |
troponin I type 3 (cardiac) |
58 |
0.93 |
| chr1_27945410_27945570 | 0.19 |
FGR |
feline Gardner-Rasheed sarcoma viral oncogene homolog |
5083 |
0.16 |
| chr9_123659112_123659288 | 0.19 |
TRAF1 |
TNF receptor-associated factor 1 |
17650 |
0.17 |
| chr11_59435711_59435871 | 0.19 |
PATL1 |
protein associated with topoisomerase II homolog 1 (yeast) |
662 |
0.47 |
| chr19_36236721_36236922 | 0.19 |
PSENEN |
presenilin enhancer gamma secretase subunit |
301 |
0.53 |
| chr15_62683922_62684073 | 0.19 |
RP11-299H22.5 |
|
121118 |
0.05 |
| chr19_19843996_19844147 | 0.19 |
ZNF14 |
zinc finger protein 14 |
165 |
0.94 |
| chr3_42633530_42633694 | 0.19 |
SS18L2 |
synovial sarcoma translocation gene on chromosome 18-like 2 |
1314 |
0.33 |
| chr16_75284233_75284384 | 0.19 |
BCAR1 |
breast cancer anti-estrogen resistance 1 |
444 |
0.77 |
| chr22_20000686_20000837 | 0.18 |
ARVCF |
armadillo repeat gene deleted in velocardiofacial syndrome |
3570 |
0.13 |
| chr3_57581712_57581923 | 0.18 |
ARF4 |
ADP-ribosylation factor 4 |
1235 |
0.28 |
| chr19_8657534_8657719 | 0.18 |
ADAMTS10 |
ADAM metallopeptidase with thrombospondin type 1 motif, 10 |
546 |
0.68 |
| chr19_39616574_39617021 | 0.18 |
PAK4 |
p21 protein (Cdc42/Rac)-activated kinase 4 |
323 |
0.84 |
| chr16_27879052_27879203 | 0.18 |
ENSG00000212382 |
. |
4555 |
0.25 |
| chr6_7161361_7161773 | 0.18 |
RP11-405O10.2 |
|
21749 |
0.17 |
| chr6_7053651_7053882 | 0.18 |
ENSG00000251762 |
. |
12987 |
0.24 |
| chr10_73105302_73105453 | 0.18 |
ENSG00000238918 |
. |
20292 |
0.18 |
| chr11_117587082_117587362 | 0.18 |
DSCAML1 |
Down syndrome cell adhesion molecule like 1 |
80584 |
0.08 |
| chr5_72744482_72744675 | 0.18 |
FOXD1 |
forkhead box D1 |
226 |
0.94 |
| chr9_136924579_136924782 | 0.18 |
BRD3 |
bromodomain containing 3 |
3546 |
0.2 |
| chr4_174113548_174113766 | 0.18 |
RP11-10K16.1 |
|
22854 |
0.17 |
| chr2_7084781_7084993 | 0.18 |
RNF144A |
ring finger protein 144A |
11713 |
0.2 |
| chr17_41173335_41173521 | 0.18 |
VAT1 |
vesicle amine transport 1 |
945 |
0.34 |
| chr5_1315108_1315672 | 0.17 |
ENSG00000263670 |
. |
5898 |
0.18 |
| chr5_173145182_173145333 | 0.17 |
ENSG00000263401 |
. |
11824 |
0.27 |
| chr22_42304434_42304780 | 0.17 |
SHISA8 |
shisa family member 8 |
5963 |
0.1 |
| chr19_19043290_19043516 | 0.17 |
HOMER3 |
homer homolog 3 (Drosophila) |
6388 |
0.1 |
| chr2_740027_740237 | 0.17 |
AC092159.3 |
|
46897 |
0.15 |
| chrX_119150159_119150382 | 0.17 |
RP4-755D9.1 |
|
19931 |
0.17 |
| chr5_139145540_139145730 | 0.17 |
PSD2 |
pleckstrin and Sec7 domain containing 2 |
29771 |
0.16 |
| chrX_83443667_83443818 | 0.17 |
RPS6KA6 |
ribosomal protein S6 kinase, 90kDa, polypeptide 6 |
809 |
0.74 |
| chr14_100771176_100771436 | 0.17 |
SLC25A29 |
solute carrier family 25 (mitochondrial carnitine/acylcarnitine carrier), member 29 |
1472 |
0.2 |
| chr16_84121193_84121344 | 0.17 |
MBTPS1 |
membrane-bound transcription factor peptidase, site 1 |
29132 |
0.11 |
| chr10_50009128_50009364 | 0.17 |
WDFY4 |
WDFY family member 4 |
1537 |
0.47 |
| chr1_16276231_16276382 | 0.17 |
ZBTB17 |
zinc finger and BTB domain containing 17 |
5383 |
0.16 |
| chr1_232055123_232055279 | 0.17 |
DISC1-IT1 |
DISC1 intronic transcript 1 (non-protein coding) |
6379 |
0.33 |
| chr1_9136576_9136727 | 0.17 |
SLC2A5 |
solute carrier family 2 (facilitated glucose/fructose transporter), member 5 |
3470 |
0.2 |
| chr11_121315919_121316119 | 0.17 |
SORL1 |
sortilin-related receptor, L(DLR class) A repeats containing |
6893 |
0.25 |
| chr6_14225312_14225463 | 0.17 |
ENSG00000238987 |
. |
68694 |
0.13 |
| chrX_153619109_153619401 | 0.17 |
RPL10 |
ribosomal protein L10 |
6240 |
0.08 |
| chr4_186049872_186050054 | 0.17 |
ENSG00000265619 |
. |
6487 |
0.2 |
| chr2_9934112_9934263 | 0.17 |
TAF1B |
TATA box binding protein (TBP)-associated factor, RNA polymerase I, B, 63kDa |
49296 |
0.14 |
| chrX_103175016_103175167 | 0.17 |
LL0XNC01-116E7.2 |
|
960 |
0.43 |
| chr2_160143487_160143638 | 0.17 |
WDSUB1 |
WD repeat, sterile alpha motif and U-box domain containing 1 |
252 |
0.95 |
| chr11_76860343_76860494 | 0.17 |
MYO7A |
myosin VIIA |
21070 |
0.19 |
| chr2_114195746_114195897 | 0.16 |
CBWD2 |
COBW domain containing 2 |
481 |
0.76 |
| chr16_88844539_88844690 | 0.16 |
PIEZO1 |
piezo-type mechanosensitive ion channel component 1 |
7005 |
0.09 |
| chr10_85954017_85954268 | 0.16 |
CDHR1 |
cadherin-related family member 1 |
268 |
0.89 |
| chr16_56622840_56623034 | 0.16 |
MT3 |
metallothionein 3 |
49 |
0.95 |
| chr15_47958076_47958227 | 0.16 |
SEMA6D |
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D |
51044 |
0.14 |
| chr21_37693189_37693340 | 0.16 |
AP000692.10 |
|
296 |
0.76 |
| chr6_116937723_116937948 | 0.16 |
RSPH4A |
radial spoke head 4 homolog A (Chlamydomonas) |
48 |
0.97 |
| chr19_41324095_41324416 | 0.16 |
EGLN2 |
egl-9 family hypoxia-inducible factor 2 |
11107 |
0.12 |
| chr1_21655784_21655935 | 0.16 |
ECE1 |
endothelin converting enzyme 1 |
16138 |
0.2 |
| chr11_115580446_115580732 | 0.16 |
ENSG00000239153 |
. |
82686 |
0.11 |
| chr6_150326385_150326536 | 0.16 |
RAET1K |
retinoic acid early transcript 1K pseudogene |
167 |
0.94 |
| chr22_37614574_37614752 | 0.16 |
SSTR3 |
somatostatin receptor 3 |
6301 |
0.14 |
| chr18_25755759_25755910 | 0.16 |
CDH2 |
cadherin 2, type 1, N-cadherin (neuronal) |
1576 |
0.58 |
| chr1_154946234_154946385 | 0.16 |
SHC1 |
SHC (Src homology 2 domain containing) transforming protein 1 |
503 |
0.44 |
| chr8_9414176_9414915 | 0.16 |
RP11-375N15.2 |
|
515 |
0.7 |
| chr14_92997995_92998245 | 0.16 |
RIN3 |
Ras and Rab interactor 3 |
17972 |
0.26 |
| chr1_155024637_155024788 | 0.16 |
ADAM15 |
ADAM metallopeptidase domain 15 |
884 |
0.33 |
| chr1_156814971_156815214 | 0.16 |
INSRR |
insulin receptor-related receptor |
13718 |
0.12 |
| chr3_32911345_32911496 | 0.16 |
TRIM71 |
tripartite motif containing 71, E3 ubiquitin protein ligase |
51910 |
0.14 |
| chr3_23792642_23792891 | 0.15 |
ENSG00000238672 |
. |
16336 |
0.21 |
| chr19_532690_532841 | 0.15 |
CDC34 |
cell division cycle 34 |
705 |
0.48 |
| chr19_55951851_55952002 | 0.15 |
SHISA7 |
shisa family member 7 |
1778 |
0.17 |
| chr1_226257523_226257818 | 0.15 |
H3F3A |
H3 histone, family 3A |
5992 |
0.18 |
| chr22_39485961_39486281 | 0.15 |
RP4-742C19.12 |
|
1470 |
0.3 |
| chr8_143139430_143139581 | 0.15 |
ENSG00000221123 |
. |
112645 |
0.06 |
| chr19_18700873_18701059 | 0.15 |
C19orf60 |
chromosome 19 open reading frame 60 |
554 |
0.55 |
| chr7_138917063_138917328 | 0.15 |
UBN2 |
ubinuclein 2 |
964 |
0.61 |
| chr12_4550645_4550796 | 0.15 |
FGF6 |
fibroblast growth factor 6 |
2665 |
0.31 |
| chr15_29561283_29561435 | 0.15 |
NDNL2 |
necdin-like 2 |
674 |
0.77 |
| chr1_2226282_2226433 | 0.15 |
RP4-713A8.1 |
|
31775 |
0.1 |
| chr12_110799070_110799221 | 0.15 |
ENSG00000264626 |
. |
594 |
0.71 |
| chr17_16697326_16697519 | 0.15 |
USP32P1 |
ubiquitin specific peptidase 32 pseudogene 1 |
1935 |
0.24 |
| chr2_101991834_101991985 | 0.15 |
CREG2 |
cellular repressor of E1A-stimulated genes 2 |
12148 |
0.22 |
| chr16_31404523_31404674 | 0.15 |
ITGAD |
integrin, alpha D |
35 |
0.96 |
| chr12_282069_282229 | 0.15 |
RP11-598F7.5 |
|
6662 |
0.14 |
| chr19_42723833_42723998 | 0.15 |
DEDD2 |
death effector domain containing 2 |
360 |
0.53 |
| chr17_46521887_46522400 | 0.15 |
SKAP1 |
src kinase associated phosphoprotein 1 |
14562 |
0.13 |
| chr12_131706127_131706426 | 0.15 |
AC092850.1 |
Protein LOC100996701 |
73791 |
0.11 |
| chr4_109572628_109573042 | 0.15 |
ENSG00000206601 |
. |
471 |
0.69 |
| chr11_18742754_18742995 | 0.14 |
IGSF22 |
immunoglobulin superfamily, member 22 |
4853 |
0.16 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0003077 | obsolete negative regulation of diuresis(GO:0003077) |
| 0.1 | 0.2 | GO:0021563 | glossopharyngeal nerve development(GO:0021563) glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.1 | 0.2 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.0 | 0.2 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.1 | GO:0036336 | dendritic cell chemotaxis(GO:0002407) dendritic cell migration(GO:0036336) |
| 0.0 | 0.2 | GO:0072268 | pattern specification involved in metanephros development(GO:0072268) |
| 0.0 | 0.3 | GO:0006744 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 0.2 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.0 | 0.1 | GO:0001957 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 | 0.1 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.0 | 0.2 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.7 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.0 | 0.1 | GO:0060340 | positive regulation of type I interferon-mediated signaling pathway(GO:0060340) |
| 0.0 | 0.1 | GO:0003078 | obsolete regulation of natriuresis(GO:0003078) |
| 0.0 | 0.1 | GO:0060831 | smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:0060831) |
| 0.0 | 0.2 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.0 | 0.1 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.3 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 0.1 | GO:0060596 | mammary placode formation(GO:0060596) |
| 0.0 | 0.1 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.1 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.0 | 0.1 | GO:0006591 | ornithine metabolic process(GO:0006591) |
| 0.0 | 0.0 | GO:0072143 | mesangial cell differentiation(GO:0072007) kidney interstitial fibroblast differentiation(GO:0072071) renal interstitial fibroblast development(GO:0072141) mesangial cell development(GO:0072143) pericyte cell differentiation(GO:1904238) |
| 0.0 | 0.1 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.1 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.0 | 0.1 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.1 | GO:0002693 | positive regulation of cellular extravasation(GO:0002693) |
| 0.0 | 0.1 | GO:0046838 | phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) |
| 0.0 | 0.1 | GO:0016999 | antibiotic metabolic process(GO:0016999) antibiotic biosynthetic process(GO:0017000) |
| 0.0 | 0.1 | GO:0033962 | cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.0 | 0.1 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.3 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.0 | 0.0 | GO:0015809 | arginine transport(GO:0015809) |
| 0.0 | 0.3 | GO:0071479 | cellular response to ionizing radiation(GO:0071479) |
| 0.0 | 0.1 | GO:0048563 | post-embryonic organ morphogenesis(GO:0048563) |
| 0.0 | 0.1 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 0.1 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.0 | 0.0 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.0 | 0.0 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 0.1 | GO:0031641 | regulation of myelination(GO:0031641) |
| 0.0 | 0.1 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.0 | GO:0032212 | positive regulation of telomere maintenance via telomerase(GO:0032212) positive regulation of telomere maintenance via telomere lengthening(GO:1904358) |
| 0.0 | 0.1 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 | 0.1 | GO:0001866 | NK T cell proliferation(GO:0001866) |
| 0.0 | 0.1 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.1 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 0.0 | GO:0015917 | aminophospholipid transport(GO:0015917) |
| 0.0 | 0.0 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.0 | 0.1 | GO:0042987 | amyloid precursor protein catabolic process(GO:0042987) |
| 0.0 | 0.1 | GO:0021903 | rostrocaudal neural tube patterning(GO:0021903) |
| 0.0 | 0.1 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.0 | GO:0060120 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.0 | 0.0 | GO:0072201 | negative regulation of mesenchymal cell proliferation(GO:0072201) |
| 0.0 | 0.1 | GO:0071364 | cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.0 | 0.0 | GO:0055078 | sodium ion homeostasis(GO:0055078) |
| 0.0 | 0.0 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.0 | 0.1 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.0 | 0.0 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 0.0 | GO:0032203 | telomere formation via telomerase(GO:0032203) |
| 0.0 | 0.0 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.0 | 0.2 | GO:0001836 | release of cytochrome c from mitochondria(GO:0001836) |
| 0.0 | 0.0 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 0.0 | 0.0 | GO:0016246 | RNA interference(GO:0016246) |
| 0.0 | 0.3 | GO:0050771 | negative regulation of axonogenesis(GO:0050771) |
| 0.0 | 0.0 | GO:0009698 | phenylpropanoid metabolic process(GO:0009698) coumarin metabolic process(GO:0009804) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0070062 | extracellular exosome(GO:0070062) extracellular vesicle(GO:1903561) |
| 0.0 | 0.2 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.1 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.0 | 0.2 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.2 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 0.1 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.1 | GO:0042584 | chromaffin granule(GO:0042583) chromaffin granule membrane(GO:0042584) |
| 0.0 | 0.2 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.1 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.0 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.1 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.2 | GO:0044447 | axoneme part(GO:0044447) |
| 0.0 | 0.0 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.1 | 0.2 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.1 | 0.6 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.1 | 0.2 | GO:0004340 | glucokinase activity(GO:0004340) |
| 0.1 | 0.2 | GO:0070735 | protein-glycine ligase activity(GO:0070735) |
| 0.1 | 0.2 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.2 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.2 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.1 | GO:0008934 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.1 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.1 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.4 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.0 | 0.1 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.1 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.1 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.4 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.3 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.1 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.1 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.1 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.1 | GO:0016717 | oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water(GO:0016717) |
| 0.0 | 0.0 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.0 | 0.0 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.0 | GO:0052659 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) inositol-1,3,4,5-tetrakisphosphate 5-phosphatase activity(GO:0052659) |
| 0.0 | 0.1 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.0 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.0 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.0 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0008187 | poly-pyrimidine tract binding(GO:0008187) |
| 0.0 | 0.1 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.0 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.0 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) |
| 0.0 | 0.6 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.0 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 0.0 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.2 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.3 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.0 | 0.2 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 0.2 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.4 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.2 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.3 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.2 | REACTOME SIGNALING BY NOTCH3 | Genes involved in Signaling by NOTCH3 |
| 0.0 | 0.2 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.1 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |