Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
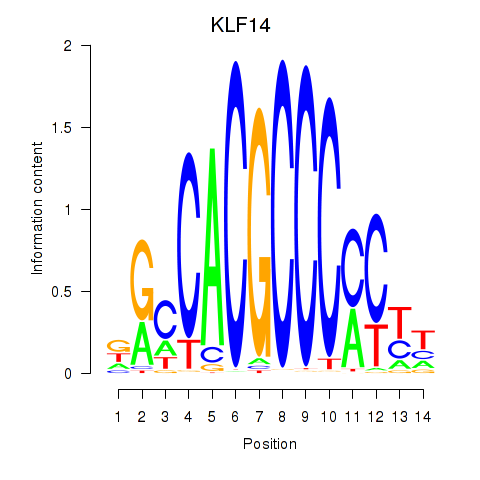
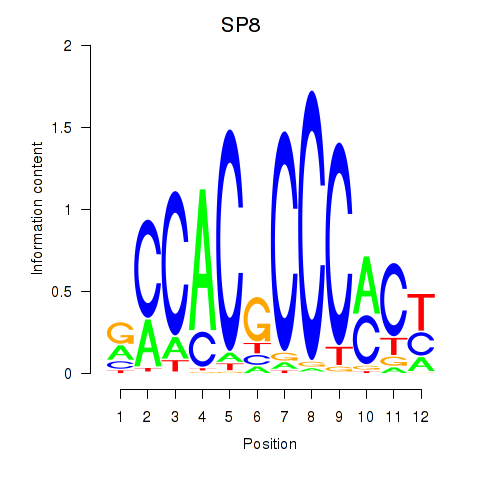
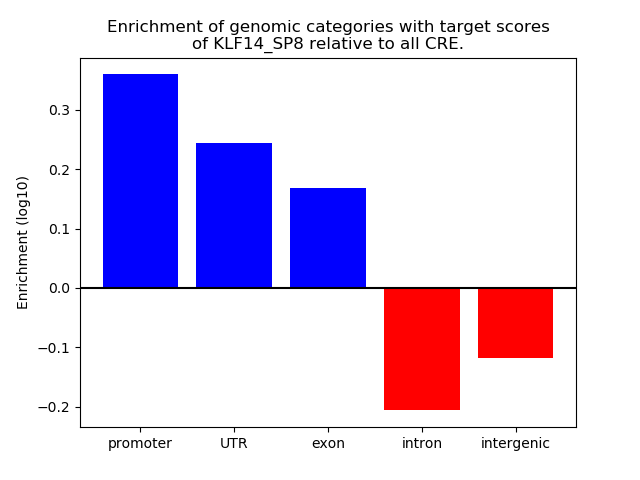
Results for KLF14_SP8
Z-value: 2.03
Transcription factors associated with KLF14_SP8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
KLF14
|
ENSG00000174595.4 | Kruppel like factor 14 |
|
SP8
|
ENSG00000164651.12 | Sp8 transcription factor |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr7_130419715_130419866 | KLF14 | 902 | 0.642700 | 0.66 | 5.3e-02 | Click! |
| chr7_130427792_130427943 | KLF14 | 8979 | 0.231836 | 0.45 | 2.2e-01 | Click! |
| chr7_130418289_130418568 | KLF14 | 460 | 0.849732 | 0.31 | 4.1e-01 | Click! |
| chr7_130419123_130419274 | KLF14 | 310 | 0.916207 | 0.31 | 4.2e-01 | Click! |
| chr7_130419356_130419681 | KLF14 | 630 | 0.767444 | 0.20 | 6.1e-01 | Click! |
| chr7_20876550_20876701 | SP8 | 50120 | 0.187855 | -0.65 | 5.9e-02 | Click! |
| chr7_20836375_20836526 | SP8 | 9945 | 0.313975 | 0.59 | 9.7e-02 | Click! |
| chr7_20950795_20950946 | SP8 | 124365 | 0.061043 | -0.56 | 1.1e-01 | Click! |
| chr7_20936732_20936883 | SP8 | 110302 | 0.073815 | 0.48 | 1.9e-01 | Click! |
| chr7_21003725_21003876 | SP8 | 177295 | 0.033174 | 0.44 | 2.4e-01 | Click! |
Activity of the KLF14_SP8 motif across conditions
Conditions sorted by the z-value of the KLF14_SP8 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
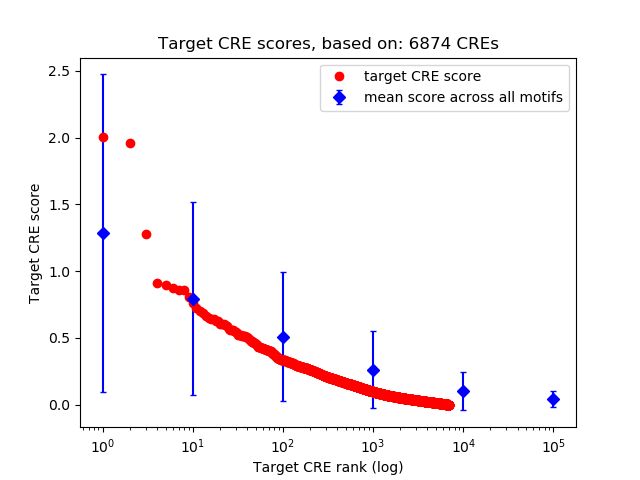
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr16_27879052_27879203 | 2.00 |
ENSG00000212382 |
. |
4555 |
0.25 |
| chr2_740601_740858 | 1.96 |
AC092159.3 |
|
47494 |
0.15 |
| chr4_1617240_1617391 | 1.28 |
FAM53A |
family with sequence similarity 53, member A |
39820 |
0.14 |
| chr22_39485961_39486281 | 0.91 |
RP4-742C19.12 |
|
1470 |
0.3 |
| chr20_58691613_58691835 | 0.90 |
C20orf197 |
chromosome 20 open reading frame 197 |
60744 |
0.14 |
| chr12_58165549_58165734 | 0.88 |
METTL1 |
methyltransferase like 1 |
261 |
0.49 |
| chr19_900208_900359 | 0.86 |
ENSG00000207507 |
. |
6799 |
0.08 |
| chr2_740331_740594 | 0.86 |
AC092159.3 |
|
47227 |
0.15 |
| chr7_100609089_100609240 | 0.81 |
ENSG00000225946 |
. |
1317 |
0.33 |
| chrX_154236957_154237108 | 0.76 |
F8 |
coagulation factor VIII, procoagulant component |
13996 |
0.15 |
| chr19_8657534_8657719 | 0.72 |
ADAMTS10 |
ADAM metallopeptidase with thrombospondin type 1 motif, 10 |
546 |
0.68 |
| chr20_3055475_3055737 | 0.70 |
OXT |
oxytocin/neurophysin I prepropeptide |
3340 |
0.15 |
| chrX_153169917_153170068 | 0.69 |
AVPR2 |
arginine vasopressin receptor 2 |
197 |
0.87 |
| chr2_740027_740237 | 0.67 |
AC092159.3 |
|
46897 |
0.15 |
| chr16_89259254_89259405 | 0.65 |
SLC22A31 |
solute carrier family 22, member 31 |
8743 |
0.12 |
| chr11_128493152_128493455 | 0.64 |
ETS1 |
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
35850 |
0.16 |
| chr17_8090983_8091134 | 0.64 |
ENSG00000266638 |
. |
565 |
0.51 |
| chr16_67719327_67719733 | 0.63 |
C16orf86 |
chromosome 16 open reading frame 86 |
18737 |
0.09 |
| chr19_8657183_8657409 | 0.63 |
ADAMTS10 |
ADAM metallopeptidase with thrombospondin type 1 motif, 10 |
216 |
0.9 |
| chr1_158768566_158768717 | 0.61 |
OR6N2 |
olfactory receptor, family 6, subfamily N, member 2 |
21216 |
0.14 |
| chr5_173145182_173145333 | 0.60 |
ENSG00000263401 |
. |
11824 |
0.27 |
| chr11_57406323_57406488 | 0.60 |
AP000662.4 |
|
556 |
0.59 |
| chr11_1864542_1864693 | 0.59 |
TNNI2 |
troponin I type 2 (skeletal, fast) |
3193 |
0.13 |
| chr15_82336812_82337149 | 0.59 |
ENSG00000222521 |
. |
345 |
0.84 |
| chr17_7184802_7184953 | 0.57 |
SLC2A4 |
solute carrier family 2 (facilitated glucose transporter), member 4 |
109 |
0.88 |
| chr16_58028490_58028941 | 0.56 |
USB1 |
U6 snRNA biogenesis 1 |
4735 |
0.14 |
| chr19_3573938_3574224 | 0.56 |
MFSD12 |
major facilitator superfamily domain containing 12 |
454 |
0.59 |
| chr14_91720187_91720656 | 0.56 |
GPR68 |
G protein-coupled receptor 68 |
152 |
0.95 |
| chr21_45574456_45574750 | 0.55 |
C21orf33 |
chromosome 21 open reading frame 33 |
18602 |
0.13 |
| chr19_14800378_14800529 | 0.55 |
ZNF333 |
zinc finger protein 333 |
258 |
0.61 |
| chr16_86600319_86600625 | 0.54 |
FOXC2 |
forkhead box C2 (MFH-1, mesenchyme forkhead 1) |
385 |
0.71 |
| chr16_84640133_84640303 | 0.52 |
COTL1 |
coactosin-like 1 (Dictyostelium) |
11447 |
0.16 |
| chr10_29971037_29971284 | 0.52 |
ENSG00000222092 |
. |
29684 |
0.19 |
| chr17_13984270_13984421 | 0.52 |
COX10 |
cytochrome c oxidase assembly homolog 10 (yeast) |
11424 |
0.22 |
| chr3_126422896_126423047 | 0.52 |
CHCHD6 |
coiled-coil-helix-coiled-coil-helix domain containing 6 |
92 |
0.98 |
| chr1_26873768_26874073 | 0.51 |
RPS6KA1 |
ribosomal protein S6 kinase, 90kDa, polypeptide 1 |
1577 |
0.3 |
| chr19_51228108_51228259 | 0.51 |
CLEC11A |
C-type lectin domain family 11, member A |
1403 |
0.27 |
| chr16_2028815_2028966 | 0.51 |
NOXO1 |
NADPH oxidase organizer 1 |
2294 |
0.08 |
| chr3_126184777_126184928 | 0.51 |
ZXDC |
ZXD family zinc finger C |
9856 |
0.17 |
| chr19_54402117_54402268 | 0.51 |
CACNG7 |
calcium channel, voltage-dependent, gamma subunit 7 |
10397 |
0.08 |
| chr12_54757135_54757286 | 0.50 |
GPR84 |
G protein-coupled receptor 84 |
1048 |
0.3 |
| chr17_62008717_62009009 | 0.49 |
CD79B |
CD79b molecule, immunoglobulin-associated beta |
758 |
0.5 |
| chr17_1617960_1618191 | 0.49 |
ENSG00000186594 |
. |
790 |
0.41 |
| chr17_1106444_1106595 | 0.48 |
ABR |
active BCR-related |
15903 |
0.17 |
| chr9_103190061_103190338 | 0.47 |
MSANTD3 |
Myb/SANT-like DNA-binding domain containing 3 |
480 |
0.78 |
| chr7_99752786_99752937 | 0.47 |
C7orf43 |
chromosome 7 open reading frame 43 |
1202 |
0.18 |
| chr19_4069404_4069640 | 0.47 |
ZBTB7A |
zinc finger and BTB domain containing 7A |
2579 |
0.17 |
| chr17_17109981_17110277 | 0.47 |
PLD6 |
phospholipase D family, member 6 |
500 |
0.69 |
| chr19_7771578_7772024 | 0.46 |
FCER2 |
Fc fragment of IgE, low affinity II, receptor for (CD23) |
4769 |
0.1 |
| chr19_36334847_36334998 | 0.46 |
NPHS1 |
nephrosis 1, congenital, Finnish type (nephrin) |
7817 |
0.09 |
| chr13_52496343_52496494 | 0.45 |
ATP7B |
ATPase, Cu++ transporting, beta polypeptide |
39633 |
0.15 |
| chr1_154297413_154297726 | 0.45 |
ATP8B2 |
ATPase, aminophospholipid transporter, class I, type 8B, member 2 |
460 |
0.66 |
| chr19_17873647_17873798 | 0.44 |
FCHO1 |
FCH domain only 1 |
210 |
0.91 |
| chr3_48411931_48412082 | 0.43 |
FBXW12 |
F-box and WD repeat domain containing 12 |
1703 |
0.23 |
| chr1_23853235_23853583 | 0.43 |
E2F2 |
E2F transcription factor 2 |
4303 |
0.21 |
| chr19_49931671_49931926 | 0.43 |
GFY |
golgi-associated, olfactory signaling regulator |
4792 |
0.07 |
| chr9_95892460_95892611 | 0.43 |
NINJ1 |
ninjurin 1 |
4035 |
0.2 |
| chr11_75946941_75947092 | 0.43 |
WNT11 |
wingless-type MMTV integration site family, member 11 |
25236 |
0.18 |
| chr20_61609083_61609372 | 0.43 |
SLC17A9 |
solute carrier family 17 (vesicular nucleotide transporter), member 9 |
24829 |
0.13 |
| chr11_64479576_64479837 | 0.42 |
NRXN2 |
neurexin 2 |
511 |
0.73 |
| chr19_46296144_46296295 | 0.42 |
DMWD |
dystrophia myotonica, WD repeat containing |
159 |
0.89 |
| chr12_122092000_122092151 | 0.42 |
MORN3 |
MORN repeat containing 3 |
15485 |
0.16 |
| chr2_219258625_219258977 | 0.42 |
CTDSP1 |
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 1 |
4260 |
0.1 |
| chr15_91967474_91967944 | 0.42 |
SV2B |
synaptic vesicle glycoprotein 2B |
198609 |
0.03 |
| chr19_40871650_40871801 | 0.41 |
PLD3 |
phospholipase D family, member 3 |
176 |
0.9 |
| chr6_2651867_2652018 | 0.41 |
C6orf195 |
chromosome 6 open reading frame 195 |
17105 |
0.25 |
| chr15_99387637_99387788 | 0.41 |
IGF1R |
insulin-like growth factor 1 receptor |
45858 |
0.14 |
| chr11_117666974_117667125 | 0.41 |
DSCAML1 |
Down syndrome cell adhesion molecule like 1 |
757 |
0.61 |
| chr17_72735203_72735698 | 0.41 |
RAB37 |
RAB37, member RAS oncogene family |
2079 |
0.18 |
| chr1_228247522_228247860 | 0.41 |
ARF1 |
ADP-ribosylation factor 1 |
22670 |
0.12 |
| chr9_139237366_139237517 | 0.40 |
GPSM1 |
G-protein signaling modulator 1 |
8536 |
0.12 |
| chr15_62683922_62684073 | 0.40 |
RP11-299H22.5 |
|
121118 |
0.05 |
| chr17_43045837_43045988 | 0.40 |
C1QL1 |
complement component 1, q subcomponent-like 1 |
473 |
0.72 |
| chr16_727350_727692 | 0.39 |
RHBDL1 |
rhomboid, veinlet-like 1 (Drosophila) |
1446 |
0.13 |
| chr19_1064402_1064702 | 0.39 |
HMHA1 |
histocompatibility (minor) HA-1 |
1370 |
0.23 |
| chr19_50093322_50093473 | 0.39 |
NOSIP |
nitric oxide synthase interacting protein |
122 |
0.89 |
| chr15_74003742_74003893 | 0.38 |
CD276 |
CD276 molecule |
7567 |
0.23 |
| chr20_10530697_10531175 | 0.38 |
JAG1 |
jagged 1 |
112218 |
0.06 |
| chr22_39923418_39924173 | 0.38 |
RPS19BP1 |
ribosomal protein S19 binding protein 1 |
5065 |
0.16 |
| chr8_25153086_25153237 | 0.38 |
DOCK5 |
dedicator of cytokinesis 5 |
3297 |
0.35 |
| chr5_176862091_176862242 | 0.37 |
GRK6 |
G protein-coupled receptor kinase 6 |
8311 |
0.09 |
| chr3_57581712_57581923 | 0.37 |
ARF4 |
ADP-ribosylation factor 4 |
1235 |
0.28 |
| chr11_76341070_76341548 | 0.37 |
AP001189.4 |
|
26791 |
0.16 |
| chr17_78785304_78785677 | 0.37 |
RP11-28G8.1 |
|
6058 |
0.26 |
| chr12_56546362_56546609 | 0.36 |
MYL6B |
myosin, light chain 6B, alkali, smooth muscle and non-muscle |
94 |
0.9 |
| chr11_118309485_118309636 | 0.35 |
KMT2A |
lysine (K)-specific methyltransferase 2A |
2083 |
0.18 |
| chr13_21750174_21750509 | 0.35 |
SKA3 |
spindle and kinetochore associated complex subunit 3 |
345 |
0.55 |
| chr11_65545992_65546157 | 0.35 |
AP5B1 |
adaptor-related protein complex 5, beta 1 subunit |
2199 |
0.16 |
| chr19_18544394_18544545 | 0.35 |
SSBP4 |
single stranded DNA binding protein 4 |
384 |
0.71 |
| chr10_85954017_85954268 | 0.35 |
CDHR1 |
cadherin-related family member 1 |
268 |
0.89 |
| chr17_26205846_26205997 | 0.34 |
RP11-138P22.1 |
|
4250 |
0.18 |
| chr11_71955241_71955392 | 0.34 |
PHOX2A |
paired-like homeobox 2a |
96 |
0.94 |
| chr2_240196342_240196584 | 0.34 |
ENSG00000265215 |
. |
30694 |
0.15 |
| chr15_79603262_79603413 | 0.34 |
TMED3 |
transmembrane emp24 protein transport domain containing 3 |
67 |
0.97 |
| chr20_32856752_32856919 | 0.34 |
ASIP |
agouti signaling protein |
8664 |
0.15 |
| chr19_12936644_12936812 | 0.34 |
CTD-2265O21.3 |
|
203 |
0.82 |
| chr19_6161467_6161654 | 0.34 |
ACSBG2 |
acyl-CoA synthetase bubblegum family member 2 |
25850 |
0.13 |
| chr19_18700873_18701059 | 0.34 |
C19orf60 |
chromosome 19 open reading frame 60 |
554 |
0.55 |
| chr14_93081555_93081992 | 0.34 |
RIN3 |
Ras and Rab interactor 3 |
37073 |
0.2 |
| chr8_144655209_144655539 | 0.34 |
MROH6 |
maestro heat-like repeat family member 6 |
233 |
0.51 |
| chr19_47885013_47885403 | 0.33 |
MEIS3 |
Meis homeobox 3 |
27201 |
0.12 |
| chr22_20785961_20786186 | 0.33 |
SCARF2 |
scavenger receptor class F, member 2 |
6039 |
0.12 |
| chr19_41349423_41349599 | 0.33 |
CYP2A6 |
cytochrome P450, family 2, subfamily A, polypeptide 6 |
6841 |
0.14 |
| chr19_49713790_49713941 | 0.33 |
TRPM4 |
transient receptor potential cation channel, subfamily M, member 4 |
126 |
0.94 |
| chr6_7271411_7271677 | 0.33 |
RP11-69L16.4 |
|
4720 |
0.24 |
| chr17_39942887_39943167 | 0.33 |
JUP |
junction plakoglobin |
10 |
0.96 |
| chr2_121077549_121077700 | 0.33 |
INHBB |
inhibin, beta B |
26095 |
0.2 |
| chr9_139090141_139090292 | 0.33 |
LHX3 |
LIM homeobox 3 |
4788 |
0.21 |
| chr15_45353267_45353418 | 0.33 |
RP11-109D20.1 |
|
2437 |
0.16 |
| chr20_60635054_60635205 | 0.33 |
ENSG00000265306 |
. |
4729 |
0.19 |
| chr7_98467875_98468026 | 0.32 |
TMEM130 |
transmembrane protein 130 |
291 |
0.88 |
| chr2_131110050_131110232 | 0.32 |
PTPN18 |
protein tyrosine phosphatase, non-receptor type 18 (brain-derived) |
3439 |
0.19 |
| chrX_49033046_49033197 | 0.32 |
PLP2 |
proteolipid protein 2 (colonic epithelium-enriched) |
4848 |
0.09 |
| chr19_35466139_35466290 | 0.32 |
ZNF792 |
zinc finger protein 792 |
11261 |
0.12 |
| chr7_156800452_156800603 | 0.32 |
MNX1 |
motor neuron and pancreas homeobox 1 |
1242 |
0.34 |
| chr9_95788627_95788837 | 0.32 |
FGD3 |
FYVE, RhoGEF and PH domain containing 3 |
11390 |
0.19 |
| chr2_192110954_192111105 | 0.32 |
MYO1B |
myosin IB |
20 |
0.99 |
| chr5_171604379_171604532 | 0.32 |
STK10 |
serine/threonine kinase 10 |
10935 |
0.19 |
| chr22_24825444_24825875 | 0.32 |
ADORA2A |
adenosine A2a receptor |
2129 |
0.31 |
| chrX_49032595_49032858 | 0.32 |
PLP2 |
proteolipid protein 2 (colonic epithelium-enriched) |
4453 |
0.09 |
| chr6_36930970_36931726 | 0.32 |
PI16 |
peptidase inhibitor 16 |
9139 |
0.17 |
| chr10_121040558_121040709 | 0.31 |
RP11-79M19.2 |
|
55061 |
0.1 |
| chr17_42393670_42393821 | 0.31 |
AC003102.3 |
|
1062 |
0.28 |
| chr7_129420448_129420809 | 0.31 |
ENSG00000207691 |
. |
5774 |
0.16 |
| chrX_110253473_110253624 | 0.31 |
PAK3 |
p21 protein (Cdc42/Rac)-activated kinase 3 |
66035 |
0.14 |
| chr20_3062971_3063207 | 0.31 |
AVP |
arginine vasopressin |
2281 |
0.19 |
| chr16_75018526_75018758 | 0.31 |
WDR59 |
WD repeat domain 59 |
404 |
0.87 |
| chrX_134429664_134429815 | 0.31 |
ZNF75D |
zinc finger protein 75D |
220 |
0.95 |
| chr14_24033631_24033782 | 0.31 |
AP1G2 |
adaptor-related protein complex 1, gamma 2 subunit |
2091 |
0.17 |
| chr19_3606868_3607019 | 0.31 |
TBXA2R |
thromboxane A2 receptor |
105 |
0.73 |
| chr9_95715118_95715525 | 0.31 |
FGD3 |
FYVE, RhoGEF and PH domain containing 3 |
5588 |
0.23 |
| chr2_71814483_71814634 | 0.31 |
DYSF |
dysferlin |
120726 |
0.06 |
| chr11_124746668_124746852 | 0.31 |
ROBO3 |
roundabout, axon guidance receptor, homolog 3 (Drosophila) |
524 |
0.64 |
| chr10_6192977_6193257 | 0.30 |
ENSG00000263628 |
. |
1042 |
0.38 |
| chr9_92050627_92050944 | 0.30 |
SEMA4D |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D |
593 |
0.81 |
| chr1_15573599_15573750 | 0.30 |
FHAD1 |
forkhead-associated (FHA) phosphopeptide binding domain 1 |
94 |
0.98 |
| chr16_31093201_31093356 | 0.30 |
ZNF646 |
zinc finger protein 646 |
6055 |
0.07 |
| chr1_33565215_33565366 | 0.30 |
ADC |
arginine decarboxylase |
17439 |
0.18 |
| chr8_127570759_127570910 | 0.29 |
FAM84B |
family with sequence similarity 84, member B |
196 |
0.83 |
| chr16_66262127_66262278 | 0.29 |
ENSG00000201999 |
. |
73510 |
0.11 |
| chr22_38071514_38071665 | 0.29 |
LGALS1 |
lectin, galactoside-binding, soluble, 1 |
26 |
0.95 |
| chr19_46144336_46144530 | 0.29 |
C19orf83 |
chromosome 19 open reading frame 83 |
516 |
0.5 |
| chr10_134266352_134266533 | 0.29 |
C10orf91 |
chromosome 10 open reading frame 91 |
7749 |
0.19 |
| chr6_35703353_35703635 | 0.29 |
RP3-510O8.4 |
|
1230 |
0.28 |
| chr1_27463921_27464408 | 0.29 |
SLC9A1 |
solute carrier family 9, subfamily A (NHE1, cation proton antiporter 1), member 1 |
16809 |
0.19 |
| chr18_74811106_74811283 | 0.29 |
MBP |
myelin basic protein |
6023 |
0.31 |
| chr19_56879462_56879613 | 0.29 |
ZSCAN5A |
zinc finger and SCAN domain containing 5A |
215 |
0.85 |
| chr2_242823885_242824036 | 0.29 |
AC131097.3 |
|
139 |
0.93 |
| chr12_132111377_132111841 | 0.29 |
ENSG00000212154 |
. |
33999 |
0.2 |
| chr22_27066729_27066953 | 0.29 |
CRYBA4 |
crystallin, beta A4 |
48913 |
0.13 |
| chr9_130689855_130690006 | 0.29 |
PIP5KL1 |
phosphatidylinositol-4-phosphate 5-kinase-like 1 |
160 |
0.9 |
| chr7_5635756_5635907 | 0.29 |
FSCN1 |
fascin homolog 1, actin-bundling protein (Strongylocentrotus purpuratus) |
1514 |
0.35 |
| chr15_89173527_89174002 | 0.29 |
ISG20 |
interferon stimulated exonuclease gene 20kDa |
5620 |
0.16 |
| chr20_44640901_44641241 | 0.29 |
MMP9 |
matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase) |
3524 |
0.16 |
| chr7_135195139_135195313 | 0.29 |
CNOT4 |
CCR4-NOT transcription complex, subunit 4 |
351 |
0.91 |
| chr3_126209757_126209908 | 0.29 |
ZXDC |
ZXD family zinc finger C |
15070 |
0.15 |
| chr20_44650406_44650557 | 0.28 |
RP11-465L10.10 |
|
115 |
0.5 |
| chr17_40822405_40822817 | 0.28 |
PLEKHH3 |
pleckstrin homology domain containing, family H (with MyTH4 domain) member 3 |
6358 |
0.09 |
| chr6_80246853_80247004 | 0.28 |
LCA5 |
Leber congenital amaurosis 5 |
197 |
0.97 |
| chr3_100429243_100429647 | 0.28 |
TFG |
TRK-fused gene |
631 |
0.79 |
| chr19_14665471_14665622 | 0.28 |
TECR |
trans-2,3-enoyl-CoA reductase |
7233 |
0.1 |
| chr1_27852121_27852272 | 0.28 |
RP1-159A19.4 |
|
120 |
0.96 |
| chr7_75622709_75623133 | 0.28 |
POR |
P450 (cytochrome) oxidoreductase |
11692 |
0.16 |
| chr14_91859082_91859233 | 0.28 |
CCDC88C |
coiled-coil domain containing 88C |
24533 |
0.2 |
| chr3_42740299_42740539 | 0.28 |
HHATL |
hedgehog acyltransferase-like |
1828 |
0.2 |
| chrX_1653232_1653496 | 0.28 |
P2RY8 |
purinergic receptor P2Y, G-protein coupled, 8 |
2636 |
0.31 |
| chrY_1603232_1603496 | 0.28 |
NA |
NA |
> 106 |
NA |
| chr7_155331108_155331259 | 0.28 |
CNPY1 |
canopy FGF signaling regulator 1 |
4626 |
0.23 |
| chr17_49336576_49336727 | 0.28 |
MBTD1 |
mbt domain containing 1 |
747 |
0.52 |
| chr1_153619171_153619411 | 0.28 |
CHTOP |
chromatin target of PRMT1 |
8473 |
0.07 |
| chr5_171881171_171881322 | 0.28 |
SH3PXD2B |
SH3 and PX domains 2B |
116 |
0.97 |
| chr5_173229911_173230062 | 0.28 |
ENSG00000263401 |
. |
72905 |
0.11 |
| chr9_70489621_70490021 | 0.28 |
CBWD5 |
COBW domain containing 5 |
286 |
0.94 |
| chr16_147116_147267 | 0.28 |
Z69720.2 |
|
6562 |
0.1 |
| chr11_1890958_1891109 | 0.28 |
LSP1 |
lymphocyte-specific protein 1 |
161 |
0.9 |
| chr17_78194041_78194192 | 0.28 |
SGSH |
N-sulfoglucosamine sulfohydrolase |
16 |
0.59 |
| chr1_175152809_175152966 | 0.28 |
KIAA0040 |
KIAA0040 |
9003 |
0.27 |
| chr6_42337436_42337711 | 0.28 |
ENSG00000221252 |
. |
42561 |
0.15 |
| chr3_16365116_16365267 | 0.27 |
RP11-415F23.2 |
|
9245 |
0.17 |
| chr15_31125817_31125968 | 0.27 |
ENSG00000221379 |
. |
32383 |
0.14 |
| chr16_19443871_19444022 | 0.27 |
TMC5 |
transmembrane channel-like 5 |
14928 |
0.15 |
| chr7_4923362_4923513 | 0.27 |
RADIL |
Ras association and DIL domains |
87 |
0.97 |
| chr10_61665530_61665681 | 0.27 |
CCDC6 |
coiled-coil domain containing 6 |
809 |
0.73 |
| chrX_65242481_65242632 | 0.27 |
ENSG00000207939 |
. |
3844 |
0.24 |
| chr10_75504524_75504675 | 0.27 |
SEC24C |
SEC24 family member C |
441 |
0.65 |
| chr2_208364611_208364762 | 0.27 |
CREB1 |
cAMP responsive element binding protein 1 |
29775 |
0.18 |
| chr11_64067476_64067822 | 0.27 |
TEX40 |
testis expressed 40 |
214 |
0.8 |
| chr19_41381342_41381499 | 0.27 |
CYP2A7 |
cytochrome P450, family 2, subfamily A, polypeptide 7 |
7237 |
0.14 |
| chr15_77302641_77302975 | 0.27 |
PSTPIP1 |
proline-serine-threonine phosphatase interacting protein 1 |
5344 |
0.22 |
| chr21_47970216_47970537 | 0.27 |
ENSG00000272283 |
. |
14667 |
0.17 |
| chr15_90695555_90695706 | 0.27 |
SEMA4B |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B |
8206 |
0.13 |
| chr10_80814068_80814219 | 0.27 |
ZMIZ1-AS1 |
ZMIZ1 antisense RNA 1 |
13082 |
0.24 |
| chr3_194354391_194354771 | 0.27 |
TMEM44 |
transmembrane protein 44 |
163 |
0.88 |
| chr5_110848258_110848409 | 0.27 |
STARD4 |
StAR-related lipid transfer (START) domain containing 4 |
45 |
0.87 |
| chr8_145555628_145555779 | 0.26 |
SCRT1 |
scratch family zinc finger 1 |
4240 |
0.08 |
| chr16_88712969_88713120 | 0.26 |
CYBA |
cytochrome b-245, alpha polypeptide |
189 |
0.88 |
| chr19_2456239_2456463 | 0.26 |
LMNB2 |
lamin B2 |
607 |
0.65 |
| chr16_4303423_4303660 | 0.26 |
TFAP4 |
transcription factor AP-4 (activating enhancer binding protein 4) |
10197 |
0.16 |
| chr20_48837037_48837260 | 0.26 |
CEBPB |
CCAAT/enhancer binding protein (C/EBP), beta |
29772 |
0.18 |
| chr7_5609893_5610044 | 0.26 |
ACTB |
actin, beta |
6553 |
0.15 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0003077 | obsolete negative regulation of diuresis(GO:0003077) |
| 0.1 | 0.6 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) positive regulation of sprouting angiogenesis(GO:1903672) |
| 0.1 | 0.4 | GO:0003308 | negative regulation of Wnt signaling pathway involved in heart development(GO:0003308) |
| 0.1 | 0.3 | GO:0002407 | dendritic cell chemotaxis(GO:0002407) dendritic cell migration(GO:0036336) |
| 0.1 | 0.3 | GO:0002019 | regulation of renal output by angiotensin(GO:0002019) |
| 0.1 | 0.2 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.1 | 0.2 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.1 | 0.2 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.1 | 0.5 | GO:0014049 | positive regulation of glutamate secretion(GO:0014049) |
| 0.1 | 0.3 | GO:0045091 | regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045091) |
| 0.1 | 0.1 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.1 | 0.3 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) positive regulation of signal transduction by p53 class mediator(GO:1901798) |
| 0.1 | 0.2 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.1 | 0.2 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.0 | 0.1 | GO:0030187 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.0 | 0.2 | GO:0048541 | Peyer's patch development(GO:0048541) |
| 0.0 | 0.3 | GO:0051001 | negative regulation of nitric-oxide synthase activity(GO:0051001) |
| 0.0 | 0.2 | GO:0008216 | spermidine metabolic process(GO:0008216) |
| 0.0 | 0.3 | GO:0021548 | pons development(GO:0021548) |
| 0.0 | 0.1 | GO:0052257 | pathogen-associated molecular pattern dependent induction by symbiont of host innate immune response(GO:0052033) positive regulation by symbiont of host innate immune response(GO:0052166) modulation by symbiont of host innate immune response(GO:0052167) pathogen-associated molecular pattern dependent modulation by symbiont of host innate immune response(GO:0052169) pathogen-associated molecular pattern dependent induction by organism of innate immune response of other organism involved in symbiotic interaction(GO:0052257) positive regulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052305) modulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052306) pathogen-associated molecular pattern dependent modulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052308) |
| 0.0 | 0.1 | GO:0001774 | microglial cell activation(GO:0001774) |
| 0.0 | 0.2 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.2 | GO:0002281 | macrophage activation involved in immune response(GO:0002281) |
| 0.0 | 0.2 | GO:0010755 | regulation of plasminogen activation(GO:0010755) |
| 0.0 | 0.1 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.0 | 0.1 | GO:0001866 | NK T cell proliferation(GO:0001866) |
| 0.0 | 0.2 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.0 | 0.1 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) |
| 0.0 | 0.2 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.2 | GO:0051103 | DNA ligation involved in DNA repair(GO:0051103) |
| 0.0 | 0.1 | GO:0034723 | DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.1 | GO:0050665 | hydrogen peroxide biosynthetic process(GO:0050665) |
| 0.0 | 0.1 | GO:0071436 | sodium ion export(GO:0071436) |
| 0.0 | 0.1 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.0 | 0.1 | GO:0033197 | response to vitamin E(GO:0033197) |
| 0.0 | 0.2 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.0 | 0.1 | GO:0050765 | negative regulation of phagocytosis(GO:0050765) |
| 0.0 | 0.1 | GO:1903521 | apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 0.0 | 0.2 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.1 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.0 | 0.2 | GO:0010536 | regulation of activation of Janus kinase activity(GO:0010533) positive regulation of activation of Janus kinase activity(GO:0010536) |
| 0.0 | 0.1 | GO:0045341 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) |
| 0.0 | 0.0 | GO:0043380 | T-helper cell lineage commitment(GO:0002295) alpha-beta T cell lineage commitment(GO:0002363) CD4-positive, alpha-beta T cell lineage commitment(GO:0043373) memory T cell differentiation(GO:0043379) regulation of memory T cell differentiation(GO:0043380) positive regulation of memory T cell differentiation(GO:0043382) T-helper 17 type immune response(GO:0072538) T-helper 17 cell differentiation(GO:0072539) T-helper 17 cell lineage commitment(GO:0072540) regulation of T-helper 17 type immune response(GO:2000316) positive regulation of T-helper 17 type immune response(GO:2000318) regulation of T-helper 17 cell differentiation(GO:2000319) positive regulation of T-helper 17 cell differentiation(GO:2000321) regulation of T-helper 17 cell lineage commitment(GO:2000328) positive regulation of T-helper 17 cell lineage commitment(GO:2000330) |
| 0.0 | 0.1 | GO:0044320 | cellular response to leptin stimulus(GO:0044320) response to leptin(GO:0044321) |
| 0.0 | 0.1 | GO:0007028 | cytoplasm organization(GO:0007028) |
| 0.0 | 0.1 | GO:0046838 | phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) |
| 0.0 | 0.1 | GO:0015809 | arginine transport(GO:0015809) |
| 0.0 | 0.1 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.0 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.0 | 0.2 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.0 | 0.1 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.0 | 0.1 | GO:0016093 | polyprenol metabolic process(GO:0016093) dolichol metabolic process(GO:0019348) |
| 0.0 | 0.1 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.0 | GO:0072217 | negative regulation of metanephros development(GO:0072217) |
| 0.0 | 0.1 | GO:0051798 | positive regulation of hair cycle(GO:0042635) positive regulation of hair follicle development(GO:0051798) |
| 0.0 | 0.5 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.0 | 0.2 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.0 | 0.1 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.1 | GO:0015801 | aromatic amino acid transport(GO:0015801) |
| 0.0 | 0.5 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.0 | 0.1 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.1 | GO:0090273 | regulation of somatostatin secretion(GO:0090273) |
| 0.0 | 0.4 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.0 | GO:0045978 | negative regulation of nucleoside metabolic process(GO:0045978) negative regulation of ATP metabolic process(GO:1903579) |
| 0.0 | 0.1 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.0 | 0.6 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.0 | 0.1 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 | 0.0 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.0 | 0.0 | GO:0021938 | smoothened signaling pathway involved in regulation of cerebellar granule cell precursor cell proliferation(GO:0021938) |
| 0.0 | 0.1 | GO:0043374 | CD8-positive, alpha-beta T cell activation(GO:0036037) CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.2 | GO:0016446 | somatic diversification of immune receptors via somatic mutation(GO:0002566) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.0 | 0.0 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 0.0 | GO:0050703 | interleukin-1 alpha secretion(GO:0050703) |
| 0.0 | 0.1 | GO:0071636 | positive regulation of transforming growth factor beta production(GO:0071636) |
| 0.0 | 0.2 | GO:0015884 | folic acid transport(GO:0015884) |
| 0.0 | 0.1 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.0 | 0.1 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.2 | GO:0009812 | flavonoid metabolic process(GO:0009812) |
| 0.0 | 0.1 | GO:0051547 | regulation of keratinocyte migration(GO:0051547) positive regulation of keratinocyte migration(GO:0051549) |
| 0.0 | 0.1 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.0 | GO:0010803 | regulation of tumor necrosis factor-mediated signaling pathway(GO:0010803) |
| 0.0 | 0.1 | GO:0009698 | phenylpropanoid metabolic process(GO:0009698) coumarin metabolic process(GO:0009804) |
| 0.0 | 0.0 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 0.1 | GO:0017000 | antibiotic metabolic process(GO:0016999) antibiotic biosynthetic process(GO:0017000) |
| 0.0 | 0.0 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 0.1 | GO:0050872 | white fat cell differentiation(GO:0050872) |
| 0.0 | 0.2 | GO:0071333 | cellular response to monosaccharide stimulus(GO:0071326) cellular response to hexose stimulus(GO:0071331) cellular response to glucose stimulus(GO:0071333) |
| 0.0 | 0.1 | GO:0007008 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.1 | GO:0072081 | proximal/distal pattern formation involved in nephron development(GO:0072047) specification of nephron tubule identity(GO:0072081) |
| 0.0 | 0.1 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.0 | 0.1 | GO:0010642 | negative regulation of platelet-derived growth factor receptor signaling pathway(GO:0010642) |
| 0.0 | 0.1 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.0 | 0.2 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.0 | 0.0 | GO:0051709 | regulation of killing of cells of other organism(GO:0051709) |
| 0.0 | 0.1 | GO:0033683 | nucleotide-excision repair, DNA incision(GO:0033683) |
| 0.0 | 0.1 | GO:0032928 | regulation of superoxide anion generation(GO:0032928) |
| 0.0 | 0.2 | GO:1903078 | positive regulation of establishment of protein localization to plasma membrane(GO:0090004) positive regulation of protein localization to plasma membrane(GO:1903078) positive regulation of protein localization to cell periphery(GO:1904377) |
| 0.0 | 0.1 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.2 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.1 | GO:0090085 | regulation of protein deubiquitination(GO:0090085) |
| 0.0 | 0.0 | GO:0035058 | nonmotile primary cilium assembly(GO:0035058) |
| 0.0 | 0.0 | GO:0070339 | response to bacterial lipoprotein(GO:0032493) response to bacterial lipopeptide(GO:0070339) cellular response to bacterial lipoprotein(GO:0071220) cellular response to bacterial lipopeptide(GO:0071221) |
| 0.0 | 0.1 | GO:0034204 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.0 | 0.2 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.3 | GO:0071514 | genetic imprinting(GO:0071514) |
| 0.0 | 0.1 | GO:0035751 | lysosomal lumen acidification(GO:0007042) regulation of lysosomal lumen pH(GO:0035751) |
| 0.0 | 0.0 | GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0043619) |
| 0.0 | 0.1 | GO:0006772 | thiamine metabolic process(GO:0006772) |
| 0.0 | 0.1 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.0 | 0.1 | GO:0000089 | mitotic metaphase(GO:0000089) |
| 0.0 | 0.0 | GO:0060684 | epithelial-mesenchymal cell signaling(GO:0060684) |
| 0.0 | 0.1 | GO:0042178 | xenobiotic catabolic process(GO:0042178) |
| 0.0 | 0.0 | GO:0048302 | regulation of isotype switching to IgG isotypes(GO:0048302) |
| 0.0 | 0.1 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.0 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 0.1 | GO:0002085 | inhibition of neuroepithelial cell differentiation(GO:0002085) |
| 0.0 | 0.1 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.0 | GO:0090049 | regulation of cell migration involved in sprouting angiogenesis(GO:0090049) regulation of sprouting angiogenesis(GO:1903670) |
| 0.0 | 0.1 | GO:0006498 | N-terminal protein lipidation(GO:0006498) |
| 0.0 | 0.1 | GO:0009649 | entrainment of circadian clock(GO:0009649) |
| 0.0 | 0.2 | GO:0002029 | desensitization of G-protein coupled receptor protein signaling pathway(GO:0002029) negative adaptation of signaling pathway(GO:0022401) adaptation of signaling pathway(GO:0023058) |
| 0.0 | 0.1 | GO:0046968 | peptide antigen transport(GO:0046968) |
| 0.0 | 0.1 | GO:0021978 | telencephalon regionalization(GO:0021978) |
| 0.0 | 0.1 | GO:0046022 | regulation of transcription from RNA polymerase II promoter, mitotic(GO:0046021) positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 0.0 | 0.1 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 0.2 | GO:0021522 | spinal cord motor neuron differentiation(GO:0021522) |
| 0.0 | 0.1 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.0 | 0.0 | GO:0071681 | response to indole-3-methanol(GO:0071680) cellular response to indole-3-methanol(GO:0071681) |
| 0.0 | 0.1 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.0 | 0.3 | GO:0045646 | regulation of erythrocyte differentiation(GO:0045646) |
| 0.0 | 0.0 | GO:0006591 | ornithine metabolic process(GO:0006591) |
| 0.0 | 0.1 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.2 | GO:0042226 | interleukin-6 biosynthetic process(GO:0042226) |
| 0.0 | 0.0 | GO:2000542 | negative regulation of gastrulation(GO:2000542) |
| 0.0 | 0.3 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.0 | GO:0071699 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.0 | 0.0 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.0 | 0.2 | GO:0050798 | activated T cell proliferation(GO:0050798) |
| 0.0 | 0.3 | GO:0060079 | excitatory postsynaptic potential(GO:0060079) |
| 0.0 | 0.1 | GO:0032096 | negative regulation of response to food(GO:0032096) |
| 0.0 | 0.0 | GO:0042976 | activation of Janus kinase activity(GO:0042976) |
| 0.0 | 0.0 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.0 | 0.0 | GO:0042754 | negative regulation of circadian sleep/wake cycle, sleep(GO:0042321) negative regulation of circadian rhythm(GO:0042754) negative regulation of behavior(GO:0048521) |
| 0.0 | 0.1 | GO:0060914 | heart formation(GO:0060914) |
| 0.0 | 0.0 | GO:0046174 | polyol catabolic process(GO:0046174) |
| 0.0 | 0.1 | GO:0031579 | membrane raft organization(GO:0031579) |
| 0.0 | 0.1 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.1 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 0.0 | GO:0046831 | regulation of RNA export from nucleus(GO:0046831) |
| 0.0 | 0.0 | GO:0042661 | regulation of mesodermal cell fate specification(GO:0042661) regulation of mesoderm development(GO:2000380) |
| 0.0 | 0.0 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.0 | 0.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.0 | GO:0002369 | T cell cytokine production(GO:0002369) |
| 0.0 | 0.0 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.1 | GO:0006264 | mitochondrial DNA replication(GO:0006264) |
| 0.0 | 0.1 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 | 0.1 | GO:0033261 | obsolete regulation of S phase(GO:0033261) |
| 0.0 | 0.3 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.0 | 0.0 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.0 | 0.1 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.0 | GO:0022605 | oogenesis stage(GO:0022605) |
| 0.0 | 0.1 | GO:0010288 | response to lead ion(GO:0010288) |
| 0.0 | 0.0 | GO:0002693 | positive regulation of cellular extravasation(GO:0002693) |
| 0.0 | 0.0 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.0 | GO:0021913 | ventral spinal cord interneuron specification(GO:0021521) regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) cell fate specification involved in pattern specification(GO:0060573) |
| 0.0 | 0.0 | GO:0042538 | hyperosmotic salinity response(GO:0042538) |
| 0.0 | 0.0 | GO:0045007 | depurination(GO:0045007) |
| 0.0 | 0.0 | GO:0035246 | peptidyl-arginine N-methylation(GO:0035246) |
| 0.0 | 0.1 | GO:0000239 | pachytene(GO:0000239) |
| 0.0 | 0.1 | GO:0030889 | negative regulation of B cell proliferation(GO:0030889) |
| 0.0 | 0.1 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.0 | 0.1 | GO:0071168 | protein localization to chromatin(GO:0071168) |
| 0.0 | 0.0 | GO:0035269 | protein mannosylation(GO:0035268) protein O-linked mannosylation(GO:0035269) mannosylation(GO:0097502) |
| 0.0 | 0.1 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.1 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.1 | GO:0070498 | interleukin-1-mediated signaling pathway(GO:0070498) |
| 0.0 | 0.1 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.0 | 0.1 | GO:0030277 | epithelial structure maintenance(GO:0010669) maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.1 | GO:0006548 | histidine catabolic process(GO:0006548) imidazole-containing compound catabolic process(GO:0052805) |
| 0.0 | 0.2 | GO:0045777 | positive regulation of blood pressure(GO:0045777) |
| 0.0 | 0.0 | GO:0032075 | positive regulation of nuclease activity(GO:0032075) |
| 0.0 | 0.0 | GO:1901534 | positive regulation of megakaryocyte differentiation(GO:0045654) positive regulation of hematopoietic progenitor cell differentiation(GO:1901534) |
| 0.0 | 0.0 | GO:0071378 | growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.0 | 0.1 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.0 | GO:0033505 | floor plate morphogenesis(GO:0033505) |
| 0.0 | 0.0 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.0 | 0.0 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) telomere maintenance in response to DNA damage(GO:0043247) |
| 0.0 | 0.0 | GO:0015812 | gamma-aminobutyric acid secretion(GO:0014051) gamma-aminobutyric acid transport(GO:0015812) |
| 0.0 | 0.1 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.0 | GO:0032098 | regulation of appetite(GO:0032098) |
| 0.0 | 0.0 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.1 | GO:0006000 | fructose metabolic process(GO:0006000) |
| 0.0 | 0.0 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.0 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 0.0 | GO:0021898 | commitment of multipotent stem cells to neuronal lineage in forebrain(GO:0021898) |
| 0.0 | 0.2 | GO:0001709 | cell fate determination(GO:0001709) |
| 0.0 | 0.0 | GO:0060633 | negative regulation of transcription initiation from RNA polymerase II promoter(GO:0060633) negative regulation of DNA-templated transcription, initiation(GO:2000143) |
| 0.0 | 0.0 | GO:0006562 | proline catabolic process(GO:0006562) |
| 0.0 | 0.0 | GO:0008631 | intrinsic apoptotic signaling pathway in response to oxidative stress(GO:0008631) cell death in response to oxidative stress(GO:0036473) |
| 0.0 | 0.0 | GO:0061430 | bone trabecula formation(GO:0060346) bone trabecula morphogenesis(GO:0061430) |
| 0.0 | 0.1 | GO:1903963 | arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.0 | 0.0 | GO:0046469 | platelet activating factor metabolic process(GO:0046469) |
| 0.0 | 0.0 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) |
| 0.0 | 0.0 | GO:0033591 | response to L-ascorbic acid(GO:0033591) |
| 0.0 | 0.0 | GO:0051458 | corticotropin secretion(GO:0051458) regulation of corticotropin secretion(GO:0051459) positive regulation of corticotropin secretion(GO:0051461) |
| 0.0 | 0.1 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.0 | GO:0042535 | positive regulation of tumor necrosis factor biosynthetic process(GO:0042535) |
| 0.0 | 0.0 | GO:0009301 | snRNA transcription(GO:0009301) |
| 0.0 | 0.0 | GO:0060291 | long-term synaptic potentiation(GO:0060291) |
| 0.0 | 0.0 | GO:0032661 | interleukin-18 production(GO:0032621) regulation of interleukin-18 production(GO:0032661) |
| 0.0 | 0.0 | GO:0050857 | positive regulation of antigen receptor-mediated signaling pathway(GO:0050857) |
| 0.0 | 0.1 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 0.0 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.1 | GO:0048194 | COPI-coated vesicle budding(GO:0035964) Golgi vesicle budding(GO:0048194) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0070062 | extracellular exosome(GO:0070062) extracellular vesicle(GO:1903561) |
| 0.1 | 0.2 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.1 | 0.3 | GO:0043218 | compact myelin(GO:0043218) |
| 0.1 | 0.2 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.1 | 0.4 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.2 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.0 | 0.1 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.1 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.3 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.3 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.1 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.3 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.3 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.1 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.1 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.0 | 0.4 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.1 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.1 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.0 | 0.1 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.1 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.2 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.1 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 0.2 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.1 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 0.1 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.4 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.1 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.1 | GO:0042581 | specific granule(GO:0042581) |
| 0.0 | 0.1 | GO:0005678 | obsolete chromatin assembly complex(GO:0005678) |
| 0.0 | 0.1 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.0 | 0.1 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.1 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.0 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 0.1 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.0 | 0.0 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.1 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.1 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.1 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.1 | GO:0008274 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.0 | 0.1 | GO:0098984 | asymmetric synapse(GO:0032279) neuron to neuron synapse(GO:0098984) |
| 0.0 | 0.6 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.0 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.1 | GO:0071778 | obsolete WINAC complex(GO:0071778) |
| 0.0 | 0.1 | GO:0070688 | MLL5-L complex(GO:0070688) |
| 0.0 | 0.2 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.1 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.0 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.1 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.1 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.1 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 0.0 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.1 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.0 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.0 | GO:0070188 | obsolete Stn1-Ten1 complex(GO:0070188) |
| 0.0 | 0.0 | GO:0031082 | BLOC complex(GO:0031082) BLOC-1 complex(GO:0031083) |
| 0.0 | 0.0 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.1 | GO:0031105 | septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.1 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 0.1 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.0 | GO:0060201 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.0 | 0.0 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.1 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.1 | GO:0090533 | cation-transporting ATPase complex(GO:0090533) |
| 0.0 | 0.0 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.1 | 0.6 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.1 | 0.4 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.1 | 0.3 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.1 | 0.2 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.1 | 0.3 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 0.3 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.1 | 0.4 | GO:0042153 | obsolete RPTP-like protein binding(GO:0042153) |
| 0.1 | 0.3 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.1 | 0.2 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.1 | 0.2 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.1 | 0.2 | GO:0052834 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.1 | 0.4 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.1 | 0.2 | GO:0016972 | thiol oxidase activity(GO:0016972) |
| 0.1 | 0.2 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.0 | 0.1 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.1 | GO:0015137 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.0 | 0.1 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.0 | 0.2 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.2 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 0.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.2 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.5 | GO:0046970 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) histone deacetylase activity (H4-K16 specific)(GO:0034739) NAD-dependent histone deacetylase activity (H4-K16 specific)(GO:0046970) |
| 0.0 | 0.2 | GO:0097506 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.0 | 0.1 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.1 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.0 | 0.3 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.1 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.1 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.1 | GO:0070735 | protein-glycine ligase activity(GO:0070735) |
| 0.0 | 0.1 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.0 | 0.4 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.1 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.0 | 0.1 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.0 | 0.2 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.1 | GO:0042808 | obsolete neuronal Cdc2-like kinase binding(GO:0042808) |
| 0.0 | 0.1 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.0 | 0.1 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.0 | 0.0 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.0 | 0.2 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.1 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.1 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) |
| 0.0 | 0.1 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.0 | 0.1 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.0 | 0.1 | GO:0019863 | IgE binding(GO:0019863) |
| 0.0 | 0.0 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) |
| 0.0 | 0.1 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.4 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.1 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.1 | GO:0080031 | methyl indole-3-acetate esterase activity(GO:0080030) methyl salicylate esterase activity(GO:0080031) methyl jasmonate esterase activity(GO:0080032) |
| 0.0 | 0.3 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.0 | 0.0 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.4 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.0 | GO:0016717 | oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water(GO:0016717) |
| 0.0 | 0.2 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.0 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 0.0 | 0.1 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.2 | GO:0004954 | prostanoid receptor activity(GO:0004954) |
| 0.0 | 0.1 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.0 | 0.1 | GO:0030197 | extracellular matrix constituent, lubricant activity(GO:0030197) |
| 0.0 | 0.1 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.1 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.1 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.0 | 0.1 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 0.1 | GO:0004904 | interferon receptor activity(GO:0004904) |
| 0.0 | 0.2 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.1 | GO:0016273 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.0 | 0.1 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.1 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.0 | 0.3 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.1 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.0 | 0.2 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.1 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.1 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.4 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 0.1 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.0 | 0.1 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.1 | GO:0031779 | melanocortin receptor binding(GO:0031779) type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.0 | 0.1 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.0 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.1 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.2 | GO:0030553 | cGMP binding(GO:0030553) |
| 0.0 | 0.0 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.1 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.1 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.0 | 0.1 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.1 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.1 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.1 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.1 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.1 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) |
| 0.0 | 0.1 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.0 | 0.1 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.1 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 0.1 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.0 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.0 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.1 | GO:0019956 | chemokine binding(GO:0019956) |
| 0.0 | 0.3 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.0 | 0.1 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.0 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.2 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.0 | 0.1 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.1 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.1 | GO:0043295 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.0 | 0.0 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.0 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 0.0 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.0 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.0 | 0.0 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.1 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.0 | 0.1 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.3 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.1 | GO:0017089 | glycolipid transporter activity(GO:0017089) |
| 0.0 | 0.1 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.2 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.0 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.1 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.1 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.0 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.0 | 0.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.4 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 0.1 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.3 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.3 | GO:0008373 | sialyltransferase activity(GO:0008373) |
| 0.0 | 0.1 | GO:0004331 | fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.0 | 0.1 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.0 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.0 | 0.1 | GO:0010181 | FMN binding(GO:0010181) |
| 0.0 | 0.0 | GO:0042806 | fucose binding(GO:0042806) |
| 0.0 | 0.0 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.0 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.4 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.1 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 0.4 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 0.0 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 0.0 | 0.0 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.0 | 0.1 | GO:0003709 | obsolete RNA polymerase III transcription factor activity(GO:0003709) |
| 0.0 | 0.1 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.1 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.0 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 1.1 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.0 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.0 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 0.0 | 0.0 | GO:0016453 | C-acetyltransferase activity(GO:0016453) |
| 0.0 | 0.1 | GO:0016889 | endodeoxyribonuclease activity, producing 3'-phosphomonoesters(GO:0016889) |
| 0.0 | 0.2 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 0.0 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.0 | 0.0 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.0 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.0 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.1 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.4 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.7 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.4 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.3 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.0 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.3 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.6 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.0 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 0.2 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.2 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.0 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.0 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.2 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.1 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.0 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.4 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.2 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 0.1 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.2 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.5 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.2 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.3 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.3 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.2 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 0.1 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.1 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.0 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.4 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 0.4 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 0.1 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.0 | 0.2 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.2 | PID IL23 PATHWAY | IL23-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME GLUCAGON SIGNALING IN METABOLIC REGULATION | Genes involved in Glucagon signaling in metabolic regulation |
| 0.0 | 0.4 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.7 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.2 | REACTOME EARLY PHASE OF HIV LIFE CYCLE | Genes involved in Early Phase of HIV Life Cycle |
| 0.0 | 0.2 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.3 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.0 | 0.0 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.0 | 0.4 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.2 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.3 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 0.0 | REACTOME G ALPHA1213 SIGNALLING EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.0 | 0.5 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.1 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.1 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.1 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.0 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.4 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.2 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.2 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.6 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.0 | 0.2 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.0 | 0.0 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 0.2 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.1 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.3 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.0 | REACTOME RORA ACTIVATES CIRCADIAN EXPRESSION | Genes involved in RORA Activates Circadian Expression |
| 0.0 | 0.1 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.3 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.2 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.0 | 0.2 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.2 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.1 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.1 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.4 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.5 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.0 | 0.3 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.2 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.0 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.0 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.1 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.2 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.1 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.1 | REACTOME CA DEPENDENT EVENTS | Genes involved in Ca-dependent events |
| 0.0 | 0.2 | REACTOME APC C CDC20 MEDIATED DEGRADATION OF CYCLIN B | Genes involved in APC/C:Cdc20 mediated degradation of Cyclin B |
| 0.0 | 0.1 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.1 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.0 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.0 | 0.1 | REACTOME ENERGY DEPENDENT REGULATION OF MTOR BY LKB1 AMPK | Genes involved in Energy dependent regulation of mTOR by LKB1-AMPK |
| 0.0 | 0.3 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.1 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.0 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.2 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.1 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.0 | 0.2 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.2 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |