Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
Results for KLF15
Z-value: 1.00
Transcription factors associated with KLF15
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
KLF15
|
ENSG00000163884.3 | Kruppel like factor 15 |
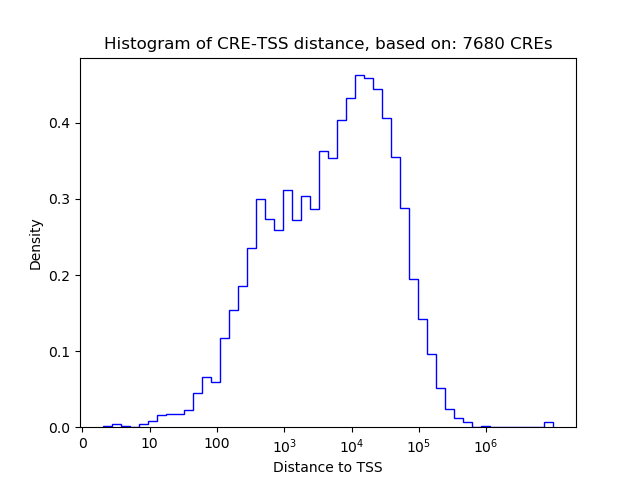
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr3_126072144_126072295 | KLF15 | 4066 | 0.228802 | 0.67 | 4.9e-02 | Click! |
| chr3_126005927_126006078 | KLF15 | 70283 | 0.082852 | 0.52 | 1.5e-01 | Click! |
| chr3_126076547_126076799 | KLF15 | 388 | 0.859998 | -0.33 | 3.9e-01 | Click! |
| chr3_126074913_126075289 | KLF15 | 1184 | 0.488770 | -0.26 | 4.9e-01 | Click! |
| chr3_126006706_126006857 | KLF15 | 69504 | 0.084084 | 0.18 | 6.4e-01 | Click! |
Activity of the KLF15 motif across conditions
Conditions sorted by the z-value of the KLF15 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
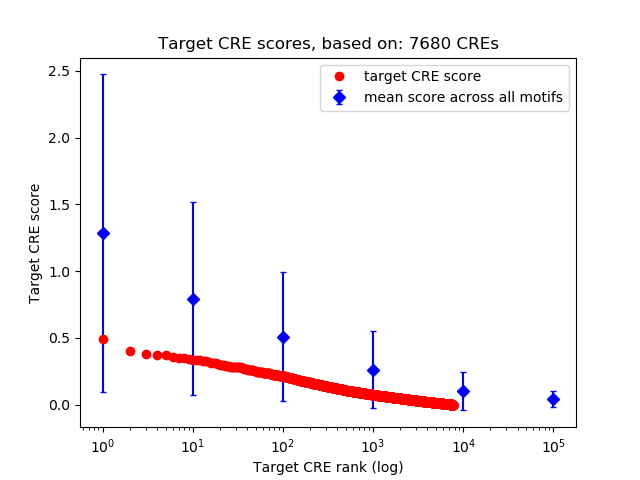
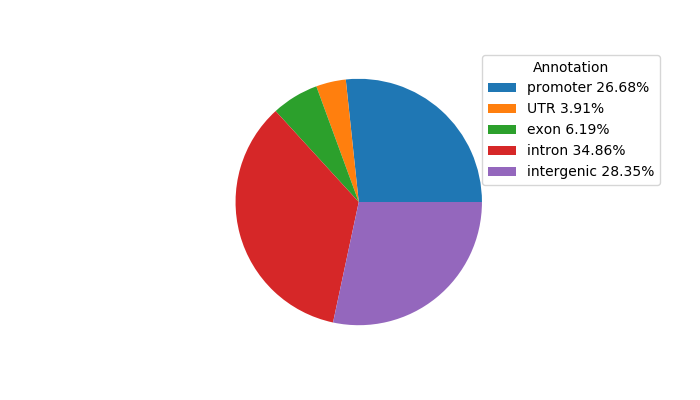
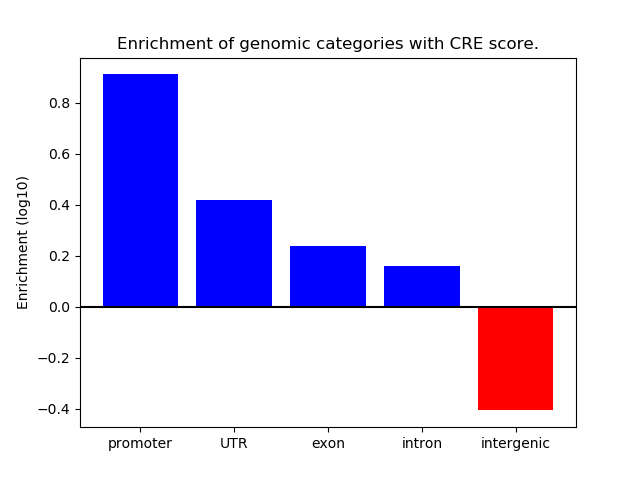
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr11_3159025_3159707 | 0.49 |
OSBPL5 |
oxysterol binding protein-like 5 |
8807 |
0.17 |
| chr21_46347768_46348484 | 0.40 |
ITGB2 |
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
507 |
0.62 |
| chr19_2982889_2983040 | 0.38 |
TLE6 |
transducin-like enhancer of split 6 (E(sp1) homolog, Drosophila) |
5386 |
0.14 |
| chr2_47089559_47089710 | 0.37 |
AC016722.3 |
|
8999 |
0.16 |
| chr13_114563717_114564199 | 0.37 |
GAS6 |
growth arrest-specific 6 |
3082 |
0.32 |
| chr1_115870179_115870464 | 0.36 |
NGF |
nerve growth factor (beta polypeptide) |
10536 |
0.26 |
| chr10_128770808_128771180 | 0.35 |
RP11-223P11.3 |
|
53692 |
0.14 |
| chr3_11253665_11253927 | 0.35 |
HRH1 |
histamine receptor H1 |
13921 |
0.26 |
| chr8_38429404_38429555 | 0.34 |
C8orf86 |
chromosome 8 open reading frame 86 |
43299 |
0.15 |
| chr11_65343918_65345038 | 0.34 |
EHBP1L1 |
EH domain binding protein 1-like 1 |
961 |
0.3 |
| chr15_74713092_74713309 | 0.34 |
SEMA7A |
semaphorin 7A, GPI membrane anchor (John Milton Hagen blood group) |
12801 |
0.14 |
| chr17_30411651_30412307 | 0.33 |
RP11-640N20.8 |
|
9972 |
0.15 |
| chr8_126557181_126557332 | 0.33 |
ENSG00000266452 |
. |
100449 |
0.08 |
| chr19_58907233_58907815 | 0.33 |
AC012313.1 |
uncharacterized protein LOC646862 |
67 |
0.92 |
| chr9_88150913_88151201 | 0.32 |
AGTPBP1 |
ATP/GTP binding protein 1 |
145144 |
0.05 |
| chr16_2084525_2084950 | 0.31 |
SLC9A3R2 |
solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3 regulator 2 |
1465 |
0.15 |
| chr17_17324288_17324439 | 0.31 |
ENSG00000201741 |
. |
39395 |
0.13 |
| chr12_52457538_52457815 | 0.31 |
C12orf44 |
chromosome 12 open reading frame 44 |
6079 |
0.11 |
| chr11_35562990_35563282 | 0.31 |
PAMR1 |
peptidase domain containing associated with muscle regeneration 1 |
11288 |
0.25 |
| chr19_5048583_5048873 | 0.30 |
KDM4B |
lysine (K)-specific demethylase 4B |
32392 |
0.18 |
| chr6_35393359_35394053 | 0.30 |
FANCE |
Fanconi anemia, complementation group E |
26432 |
0.15 |
| chr9_128507884_128508777 | 0.29 |
PBX3 |
pre-B-cell leukemia homeobox 3 |
1294 |
0.56 |
| chr11_8731101_8731252 | 0.29 |
ST5 |
suppression of tumorigenicity 5 |
1548 |
0.26 |
| chr8_126668581_126668732 | 0.29 |
ENSG00000266452 |
. |
211849 |
0.02 |
| chr17_28995530_28996050 | 0.29 |
ENSG00000241631 |
. |
34003 |
0.12 |
| chr9_140162312_140162640 | 0.28 |
TOR4A |
torsin family 4, member A |
9725 |
0.07 |
| chr15_79048825_79049161 | 0.28 |
RP11-160C18.4 |
|
3537 |
0.22 |
| chr7_73868587_73869056 | 0.28 |
GTF2IRD1 |
GTF2I repeat domain containing 1 |
382 |
0.88 |
| chr7_73686653_73687228 | 0.28 |
ENSG00000252538 |
. |
13994 |
0.15 |
| chr7_150048_150635 | 0.28 |
FAM20C |
family with sequence similarity 20, member C |
42628 |
0.16 |
| chr14_55243452_55244024 | 0.28 |
SAMD4A |
sterile alpha motif domain containing 4A |
22187 |
0.22 |
| chr17_80163150_80163535 | 0.28 |
CCDC57 |
coiled-coil domain containing 57 |
7347 |
0.11 |
| chr9_137037215_137037601 | 0.28 |
ENSG00000221676 |
. |
7722 |
0.2 |
| chr17_76339702_76340196 | 0.28 |
SOCS3 |
suppressor of cytokine signaling 3 |
16206 |
0.14 |
| chr15_46162513_46162674 | 0.28 |
SQRDL |
sulfide quinone reductase-like (yeast) |
187855 |
0.03 |
| chr1_94045560_94045711 | 0.28 |
BCAR3 |
breast cancer anti-estrogen resistance 3 |
5057 |
0.22 |
| chr15_60666127_60666304 | 0.27 |
ANXA2 |
annexin A2 |
489 |
0.87 |
| chr12_66297253_66297404 | 0.27 |
AC090673.2 |
Uncharacterized protein |
20639 |
0.16 |
| chr9_132096828_132097338 | 0.26 |
ENSG00000242281 |
. |
35657 |
0.16 |
| chr16_69951449_69951738 | 0.26 |
WWP2 |
WW domain containing E3 ubiquitin protein ligase 2 |
7300 |
0.16 |
| chr9_132077761_132077912 | 0.26 |
ENSG00000242281 |
. |
54904 |
0.11 |
| chr16_66521688_66521883 | 0.26 |
AC132186.1 |
CDNA FLJ27243 fis, clone SYN08134; Uncharacterized protein |
2038 |
0.21 |
| chr15_101741938_101742937 | 0.26 |
CHSY1 |
chondroitin sulfate synthase 1 |
49700 |
0.12 |
| chr11_124615638_124616224 | 0.26 |
RP11-677M14.2 |
|
302 |
0.81 |
| chr17_76388396_76388760 | 0.26 |
ENSG00000200063 |
. |
7438 |
0.14 |
| chr14_25518403_25519489 | 0.26 |
STXBP6 |
syntaxin binding protein 6 (amisyn) |
149 |
0.98 |
| chr3_72640698_72640849 | 0.26 |
ENSG00000199469 |
. |
14931 |
0.22 |
| chr10_133999367_134000256 | 0.25 |
RP11-140A10.3 |
|
345 |
0.67 |
| chr9_132244028_132244179 | 0.25 |
ENSG00000264298 |
. |
3268 |
0.28 |
| chr10_50362024_50362175 | 0.25 |
FAM170B-AS1 |
FAM170B antisense RNA 1 |
16559 |
0.15 |
| chr12_6809250_6809799 | 0.25 |
PIANP |
PILR alpha associated neural protein |
72 |
0.93 |
| chr9_132244264_132244728 | 0.25 |
ENSG00000264298 |
. |
3661 |
0.27 |
| chr19_18773709_18774267 | 0.25 |
CRTC1 |
CREB regulated transcription coactivator 1 |
20499 |
0.1 |
| chr17_40913377_40913932 | 0.25 |
RAMP2 |
receptor (G protein-coupled) activity modifying protein 2 |
378 |
0.71 |
| chr7_561603_561984 | 0.25 |
PDGFA |
platelet-derived growth factor alpha polypeptide |
1860 |
0.35 |
| chr3_183947384_183948074 | 0.24 |
VWA5B2 |
von Willebrand factor A domain containing 5B2 |
488 |
0.65 |
| chr11_126242895_126243350 | 0.24 |
ST3GAL4 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 4 |
17253 |
0.15 |
| chr10_120788305_120788993 | 0.24 |
NANOS1 |
nanos homolog 1 (Drosophila) |
579 |
0.67 |
| chr9_101812741_101812892 | 0.24 |
TGFBR1 |
transforming growth factor, beta receptor 1 |
53504 |
0.13 |
| chr9_126097759_126097969 | 0.24 |
CRB2 |
crumbs homolog 2 (Drosophila) |
20585 |
0.21 |
| chr22_44320967_44321118 | 0.24 |
PNPLA3 |
patatin-like phospholipase domain containing 3 |
1359 |
0.38 |
| chrX_193412_193843 | 0.24 |
PLCXD1 |
phosphatidylinositol-specific phospholipase C, X domain containing 1 |
607 |
0.8 |
| chr9_137494111_137494262 | 0.24 |
COL5A1 |
collagen, type V, alpha 1 |
39434 |
0.16 |
| chr8_124683990_124684169 | 0.24 |
KLHL38 |
kelch-like family member 38 |
18889 |
0.18 |
| chr1_228603985_228604375 | 0.24 |
TRIM17 |
tripartite motif containing 17 |
148 |
0.9 |
| chr2_85777153_85777517 | 0.24 |
MAT2A |
methionine adenosyltransferase II, alpha |
10570 |
0.09 |
| chr4_169577345_169577496 | 0.24 |
PALLD |
palladin, cytoskeletal associated protein |
1537 |
0.41 |
| chr18_60104933_60105472 | 0.24 |
RP11-640A1.3 |
|
59839 |
0.12 |
| chr11_68856171_68856322 | 0.24 |
ENSG00000265539 |
. |
5602 |
0.19 |
| chr20_10350533_10350684 | 0.23 |
SNAP25-AS1 |
SNAP25 antisense RNA 1 |
1184 |
0.51 |
| chr15_64200886_64201037 | 0.23 |
RP11-111E14.1 |
|
19512 |
0.19 |
| chr5_71929742_71929893 | 0.23 |
ZNF366 |
zinc finger protein 366 |
126263 |
0.05 |
| chr10_105452470_105452921 | 0.23 |
SH3PXD2A |
SH3 and PX domains 2A |
235 |
0.93 |
| chr1_153671480_153671631 | 0.23 |
ENSG00000242565 |
. |
5009 |
0.09 |
| chr8_145010136_145010660 | 0.23 |
PLEC |
plectin |
3360 |
0.14 |
| chr9_69786015_69786709 | 0.23 |
IGKV1OR-2 |
immunoglobulin kappa variable 1/OR-2 (pseudogene) |
8409 |
0.25 |
| chr22_29469276_29469693 | 0.23 |
KREMEN1 |
kringle containing transmembrane protein 1 |
365 |
0.83 |
| chr3_123600718_123600990 | 0.23 |
MYLK |
myosin light chain kinase |
2295 |
0.36 |
| chr22_39674017_39674168 | 0.23 |
RP3-333H23.8 |
|
14901 |
0.11 |
| chr7_73786713_73787112 | 0.22 |
CLIP2 |
CAP-GLY domain containing linker protein 2 |
4113 |
0.27 |
| chr9_136821628_136822276 | 0.22 |
VAV2 |
vav 2 guanine nucleotide exchange factor |
35451 |
0.17 |
| chr9_38024310_38024461 | 0.22 |
SHB |
Src homology 2 domain containing adaptor protein B |
44823 |
0.16 |
| chr3_45589006_45589723 | 0.22 |
LARS2-AS1 |
LARS2 antisense RNA 1 |
38327 |
0.14 |
| chr2_216001546_216001697 | 0.22 |
ABCA12 |
ATP-binding cassette, sub-family A (ABC1), member 12 |
1506 |
0.54 |
| chr10_103825823_103826651 | 0.22 |
HPS6 |
Hermansky-Pudlak syndrome 6 |
1090 |
0.45 |
| chr11_65257627_65258285 | 0.22 |
SCYL1 |
SCY1-like 1 (S. cerevisiae) |
34592 |
0.08 |
| chr13_31366120_31366271 | 0.22 |
ALOX5AP |
arachidonate 5-lipoxygenase-activating protein |
56550 |
0.14 |
| chr1_10961180_10961385 | 0.22 |
C1orf127 |
chromosome 1 open reading frame 127 |
46645 |
0.12 |
| chr9_130740072_130740785 | 0.22 |
FAM102A |
family with sequence similarity 102, member A |
2364 |
0.18 |
| chr10_101543672_101543823 | 0.22 |
ABCC2 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 2 |
1227 |
0.49 |
| chr9_139977440_139977591 | 0.22 |
AL807752.1 |
Uncharacterized protein |
3606 |
0.07 |
| chr5_135368758_135368909 | 0.22 |
TGFBI |
transforming growth factor, beta-induced, 68kDa |
4142 |
0.25 |
| chr7_1704828_1705261 | 0.22 |
ELFN1 |
extracellular leucine-rich repeat and fibronectin type III domain containing 1 |
22711 |
0.17 |
| chr2_42795325_42795726 | 0.22 |
MTA3 |
metastasis associated 1 family, member 3 |
132 |
0.97 |
| chr9_131398178_131398329 | 0.22 |
WDR34 |
WD repeat domain 34 |
20707 |
0.11 |
| chr22_41048488_41048852 | 0.22 |
MKL1 |
megakaryoblastic leukemia (translocation) 1 |
15975 |
0.19 |
| chr19_1168901_1169806 | 0.22 |
SBNO2 |
strawberry notch homolog 2 (Drosophila) |
411 |
0.76 |
| chr20_23735718_23735869 | 0.22 |
CST1 |
cystatin SN |
3888 |
0.24 |
| chr15_74232933_74233272 | 0.22 |
LOXL1-AS1 |
LOXL1 antisense RNA 1 |
12513 |
0.14 |
| chr15_78385903_78386242 | 0.21 |
SH2D7 |
SH2 domain containing 7 |
1145 |
0.36 |
| chr8_145751967_145752428 | 0.21 |
LRRC24 |
leucine rich repeat containing 24 |
205 |
0.81 |
| chr6_3389650_3389801 | 0.21 |
SLC22A23 |
solute carrier family 22, member 23 |
49927 |
0.15 |
| chr6_41673692_41674021 | 0.21 |
TFEB |
transcription factor EB |
178 |
0.92 |
| chr17_45867102_45867637 | 0.21 |
OSBPL7 |
oxysterol binding protein-like 7 |
31767 |
0.1 |
| chr17_77788487_77789261 | 0.21 |
ENSG00000238331 |
. |
4396 |
0.16 |
| chr2_164070691_164070842 | 0.21 |
ENSG00000200902 |
. |
74739 |
0.13 |
| chr14_100001157_100001308 | 0.21 |
CCDC85C |
coiled-coil domain containing 85C |
1193 |
0.44 |
| chr12_48146036_48146187 | 0.21 |
SLC48A1 |
solute carrier family 48 (heme transporter), member 1 |
1588 |
0.3 |
| chr3_185450728_185450956 | 0.21 |
C3orf65 |
chromosome 3 open reading frame 65 |
19762 |
0.2 |
| chr16_85034262_85034491 | 0.21 |
ZDHHC7 |
zinc finger, DHHC-type containing 7 |
4240 |
0.26 |
| chr15_48967599_48967750 | 0.21 |
FBN1 |
fibrillin 1 |
29628 |
0.21 |
| chr9_37964906_37965242 | 0.21 |
ENSG00000251745 |
. |
28309 |
0.2 |
| chr9_35960859_35961010 | 0.21 |
OR2S2 |
olfactory receptor, family 2, subfamily S, member 2 |
2783 |
0.23 |
| chr16_75290605_75290756 | 0.21 |
BCAR1 |
breast cancer anti-estrogen resistance 1 |
502 |
0.73 |
| chr17_55967739_55968110 | 0.20 |
CUEDC1 |
CUE domain containing 1 |
2125 |
0.29 |
| chr20_62576193_62576344 | 0.20 |
ENSG00000207554 |
. |
2189 |
0.12 |
| chr17_18124778_18124929 | 0.20 |
LLGL1 |
lethal giant larvae homolog 1 (Drosophila) |
4048 |
0.14 |
| chr7_101916989_101917140 | 0.20 |
CUX1 |
cut-like homeobox 1 |
381 |
0.8 |
| chr9_80066146_80066297 | 0.20 |
RP11-466A17.1 |
|
4990 |
0.3 |
| chr7_45921396_45921547 | 0.20 |
IGFBP1 |
insulin-like growth factor binding protein 1 |
6485 |
0.18 |
| chr15_78287228_78287379 | 0.20 |
RP11-114H24.6 |
|
1556 |
0.29 |
| chr2_20700806_20700957 | 0.20 |
ENSG00000200829 |
. |
20276 |
0.2 |
| chr17_36626163_36626314 | 0.20 |
ARHGAP23 |
Rho GTPase activating protein 23 |
1942 |
0.3 |
| chr6_81253336_81253487 | 0.20 |
BCKDHB |
branched chain keto acid dehydrogenase E1, beta polypeptide |
437028 |
0.01 |
| chr1_3459450_3459601 | 0.20 |
MEGF6 |
multiple EGF-like-domains 6 |
11513 |
0.17 |
| chr9_130099707_130099858 | 0.20 |
RP11-356B19.11 |
|
29267 |
0.15 |
| chr16_85429807_85430115 | 0.20 |
RP11-680G10.1 |
Uncharacterized protein |
38892 |
0.16 |
| chr6_33383327_33383874 | 0.20 |
PHF1 |
PHD finger protein 1 |
758 |
0.45 |
| chr6_44044467_44044818 | 0.20 |
RP5-1120P11.1 |
|
2253 |
0.28 |
| chr22_39417441_39417875 | 0.20 |
APOBEC3D |
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3D |
201 |
0.89 |
| chr17_60748078_60748229 | 0.19 |
ENSG00000207382 |
. |
9067 |
0.15 |
| chr16_67550113_67550264 | 0.19 |
FAM65A |
family with sequence similarity 65, member A |
2133 |
0.16 |
| chr8_119036254_119036405 | 0.19 |
EXT1 |
exostosin glycosyltransferase 1 |
86324 |
0.11 |
| chr4_76555269_76555772 | 0.19 |
CDKL2 |
cyclin-dependent kinase-like 2 (CDC2-related kinase) |
190 |
0.94 |
| chr10_3136297_3136448 | 0.19 |
PFKP |
phosphofructokinase, platelet |
10544 |
0.25 |
| chr3_50279084_50280004 | 0.19 |
GNAI2 |
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2 |
4000 |
0.12 |
| chr7_100751782_100751933 | 0.19 |
RP11-395B7.7 |
|
14529 |
0.1 |
| chr19_15559479_15560330 | 0.19 |
ENSG00000269782 |
. |
455 |
0.47 |
| chr12_88964078_88964229 | 0.19 |
KITLG |
KIT ligand |
10085 |
0.24 |
| chr20_48603753_48603932 | 0.19 |
SNAI1 |
snail family zinc finger 1 |
4306 |
0.18 |
| chr6_33576795_33576946 | 0.19 |
ITPR3 |
inositol 1,4,5-trisphosphate receptor, type 3 |
11272 |
0.13 |
| chr12_13326906_13327130 | 0.19 |
EMP1 |
epithelial membrane protein 1 |
22632 |
0.21 |
| chr16_69026070_69026221 | 0.19 |
ENSG00000202497 |
. |
14913 |
0.19 |
| chr19_41108139_41108638 | 0.19 |
LTBP4 |
latent transforming growth factor beta binding protein 4 |
1111 |
0.4 |
| chr20_56347824_56347975 | 0.19 |
PMEPA1 |
prostate transmembrane protein, androgen induced 1 |
61358 |
0.13 |
| chr16_85235840_85236043 | 0.19 |
CTC-786C10.1 |
|
31059 |
0.2 |
| chr6_150921076_150921438 | 0.19 |
PLEKHG1 |
pleckstrin homology domain containing, family G (with RhoGef domain) member 1 |
238 |
0.82 |
| chr16_84210184_84210335 | 0.19 |
DNAAF1 |
dynein, axonemal, assembly factor 1 |
512 |
0.67 |
| chr1_19253607_19253971 | 0.18 |
IFFO2 |
intermediate filament family orphan 2 |
15303 |
0.16 |
| chr2_236522839_236522990 | 0.18 |
ENSG00000221704 |
. |
42339 |
0.17 |
| chr19_872329_872946 | 0.18 |
CFD |
complement factor D (adipsin) |
12947 |
0.07 |
| chr1_227161852_227162199 | 0.18 |
ADCK3 |
aarF domain containing kinase 3 |
8643 |
0.21 |
| chr11_10139098_10139249 | 0.18 |
RP11-748C4.1 |
|
31566 |
0.21 |
| chr11_73032066_73032515 | 0.18 |
ARHGEF17 |
Rho guanine nucleotide exchange factor (GEF) 17 |
9680 |
0.14 |
| chr11_65293088_65294032 | 0.18 |
SCYL1 |
SCY1-like 1 (S. cerevisiae) |
676 |
0.53 |
| chr3_48731676_48731827 | 0.18 |
IP6K2 |
inositol hexakisphosphate kinase 2 |
1264 |
0.31 |
| chr22_39638721_39639222 | 0.18 |
PDGFB |
platelet-derived growth factor beta polypeptide |
50 |
0.97 |
| chr2_231279838_231280668 | 0.18 |
SP100 |
SP100 nuclear antigen |
404 |
0.89 |
| chr3_58012227_58012378 | 0.18 |
FLNB |
filamin B, beta |
18175 |
0.25 |
| chr14_103988319_103988805 | 0.18 |
CKB |
creatine kinase, brain |
471 |
0.64 |
| chr17_38174717_38174868 | 0.18 |
CSF3 |
colony stimulating factor 3 (granulocyte) |
3052 |
0.15 |
| chr1_156211867_156212112 | 0.18 |
BGLAP |
bone gamma-carboxyglutamate (gla) protein |
236 |
0.85 |
| chr7_75496335_75496486 | 0.18 |
RHBDD2 |
rhomboid domain containing 2 |
11856 |
0.16 |
| chr4_187767041_187767192 | 0.18 |
ENSG00000252382 |
. |
11494 |
0.3 |
| chr19_1513130_1513873 | 0.18 |
ADAMTSL5 |
ADAMTS-like 5 |
313 |
0.73 |
| chr15_62705556_62705707 | 0.18 |
RP11-299H22.5 |
|
142752 |
0.04 |
| chr11_119231679_119232072 | 0.18 |
USP2 |
ubiquitin specific peptidase 2 |
3042 |
0.12 |
| chr19_7989733_7990535 | 0.18 |
CTD-3193O13.8 |
|
752 |
0.29 |
| chr9_132949422_132949573 | 0.18 |
NCS1 |
neuronal calcium sensor 1 |
13375 |
0.21 |
| chr11_133989916_133990067 | 0.18 |
NCAPD3 |
non-SMC condensin II complex, subunit D3 |
48376 |
0.12 |
| chr6_152468161_152468312 | 0.18 |
SYNE1 |
spectrin repeat containing, nuclear envelope 1 |
21250 |
0.27 |
| chr15_79052113_79052552 | 0.17 |
RP11-160C18.4 |
|
6876 |
0.18 |
| chr6_35457763_35458156 | 0.17 |
TEAD3 |
TEA domain family member 3 |
6768 |
0.17 |
| chr18_9885538_9885689 | 0.17 |
TXNDC2 |
thioredoxin domain containing 2 (spermatozoa) |
150 |
0.95 |
| chr7_100762953_100763104 | 0.17 |
SERPINE1 |
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 |
7342 |
0.11 |
| chr17_70359556_70359707 | 0.17 |
SOX9 |
SRY (sex determining region Y)-box 9 |
242470 |
0.02 |
| chr11_1146453_1147024 | 0.17 |
MUC5AC |
mucin 5AC, oligomeric mucus/gel-forming |
4842 |
0.22 |
| chr9_124044170_124044436 | 0.17 |
RP11-477J21.6 |
|
705 |
0.59 |
| chr1_16500240_16500391 | 0.17 |
RP11-276H7.3 |
|
14554 |
0.1 |
| chr1_155223556_155224415 | 0.17 |
FAM189B |
family with sequence similarity 189, member B |
714 |
0.38 |
| chr17_79070617_79071314 | 0.17 |
BAIAP2 |
BAI1-associated protein 2 |
414 |
0.74 |
| chr5_139050769_139051130 | 0.17 |
CXXC5 |
CXXC finger protein 5 |
4072 |
0.25 |
| chr19_41318948_41319224 | 0.17 |
EGLN2 |
egl-9 family hypoxia-inducible factor 2 |
5938 |
0.13 |
| chr12_65918227_65918378 | 0.17 |
MSRB3 |
methionine sulfoxide reductase B3 |
197647 |
0.03 |
| chr9_140191332_140191483 | 0.17 |
RP13-122B23.8 |
|
1234 |
0.26 |
| chr1_856648_857113 | 0.17 |
SAMD11 |
sterile alpha motif domain containing 11 |
3380 |
0.14 |
| chr7_157072630_157072904 | 0.17 |
ENSG00000266453 |
. |
25720 |
0.21 |
| chr3_133282485_133282636 | 0.17 |
CDV3 |
CDV3 homolog (mouse) |
10014 |
0.2 |
| chr16_1086817_1086968 | 0.17 |
SSTR5 |
somatostatin receptor 5 |
35864 |
0.09 |
| chr17_18128026_18128293 | 0.17 |
LLGL1 |
lethal giant larvae homolog 1 (Drosophila) |
742 |
0.52 |
| chr5_150033063_150033214 | 0.17 |
MYOZ3 |
myozenin 3 |
7298 |
0.14 |
| chr11_65678995_65679525 | 0.17 |
C11orf68 |
chromosome 11 open reading frame 68 |
6571 |
0.08 |
| chr7_128550016_128550294 | 0.17 |
ENSG00000221401 |
. |
3330 |
0.19 |
| chr1_9375982_9376310 | 0.17 |
SPSB1 |
splA/ryanodine receptor domain and SOCS box containing 1 |
21559 |
0.19 |
| chr8_142143044_142143318 | 0.17 |
RP11-809O17.1 |
|
3121 |
0.25 |
| chr3_194293555_194293746 | 0.17 |
TMEM44-AS1 |
TMEM44 antisense RNA 1 |
11090 |
0.15 |
| chr3_66474645_66474796 | 0.17 |
LRIG1 |
leucine-rich repeats and immunoglobulin-like domains 1 |
76636 |
0.11 |
| chr18_10032471_10032622 | 0.17 |
ENSG00000263630 |
. |
27352 |
0.22 |
| chr7_1953149_1953300 | 0.17 |
MAD1L1 |
MAD1 mitotic arrest deficient-like 1 (yeast) |
26961 |
0.2 |
| chr2_216636638_216636908 | 0.17 |
ENSG00000212055 |
. |
106869 |
0.07 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0010511 | regulation of phosphatidylinositol biosynthetic process(GO:0010511) |
| 0.0 | 0.1 | GO:0030011 | maintenance of cell polarity(GO:0030011) |
| 0.0 | 0.2 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.2 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 0.1 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.0 | 0.2 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) chloride ion homeostasis(GO:0055064) |
| 0.0 | 0.1 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.0 | 0.1 | GO:0055022 | negative regulation of cardiac muscle tissue growth(GO:0055022) negative regulation of cardiac muscle tissue development(GO:0055026) negative regulation of cardiac muscle cell proliferation(GO:0060044) negative regulation of heart growth(GO:0061117) |
| 0.0 | 0.1 | GO:1903726 | negative regulation of phospholipid metabolic process(GO:1903726) |
| 0.0 | 0.3 | GO:0060581 | ventral spinal cord interneuron differentiation(GO:0021514) ventral spinal cord interneuron fate commitment(GO:0060579) cell fate commitment involved in pattern specification(GO:0060581) |
| 0.0 | 0.1 | GO:0016103 | diterpenoid catabolic process(GO:0016103) terpenoid catabolic process(GO:0016115) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.1 | GO:0071731 | response to nitric oxide(GO:0071731) |
| 0.0 | 0.1 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.1 | GO:0000470 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 0.1 | GO:0032455 | nerve growth factor processing(GO:0032455) |
| 0.0 | 0.3 | GO:0009629 | response to gravity(GO:0009629) |
| 0.0 | 0.1 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 0.2 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.1 | GO:0060737 | prostate gland morphogenetic growth(GO:0060737) |
| 0.0 | 0.1 | GO:0048670 | regulation of collateral sprouting(GO:0048670) |
| 0.0 | 0.1 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.0 | 0.2 | GO:0043954 | cellular component maintenance(GO:0043954) |
| 0.0 | 0.1 | GO:0042415 | norepinephrine metabolic process(GO:0042415) |
| 0.0 | 0.1 | GO:0030187 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.0 | 0.1 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 0.2 | GO:0002281 | macrophage activation involved in immune response(GO:0002281) |
| 0.0 | 0.1 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.4 | GO:0060840 | artery morphogenesis(GO:0048844) artery development(GO:0060840) |
| 0.0 | 0.1 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.0 | 0.1 | GO:0045112 | integrin biosynthetic process(GO:0045112) |
| 0.0 | 0.2 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.0 | 0.1 | GO:0061687 | detoxification of inorganic compound(GO:0061687) stress response to metal ion(GO:0097501) |
| 0.0 | 0.1 | GO:0007442 | hindgut morphogenesis(GO:0007442) hindgut development(GO:0061525) |
| 0.0 | 0.1 | GO:0008215 | spermine metabolic process(GO:0008215) |
| 0.0 | 0.0 | GO:0032328 | alanine transport(GO:0032328) |
| 0.0 | 0.1 | GO:0090292 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.1 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.1 | GO:0022601 | menstrual cycle phase(GO:0022601) |
| 0.0 | 0.1 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.0 | 0.0 | GO:0060014 | granulosa cell differentiation(GO:0060014) |
| 0.0 | 0.2 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.0 | 0.2 | GO:0051927 | obsolete negative regulation of calcium ion transport via voltage-gated calcium channel activity(GO:0051927) |
| 0.0 | 0.1 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 0.1 | GO:0007028 | cytoplasm organization(GO:0007028) |
| 0.0 | 0.0 | GO:0060068 | vagina development(GO:0060068) |
| 0.0 | 0.1 | GO:0045915 | positive regulation of catecholamine metabolic process(GO:0045915) positive regulation of dopamine metabolic process(GO:0045964) |
| 0.0 | 0.1 | GO:0009191 | nucleoside diphosphate catabolic process(GO:0009134) ribonucleoside diphosphate catabolic process(GO:0009191) |
| 0.0 | 0.1 | GO:0034371 | chylomicron remodeling(GO:0034371) |
| 0.0 | 0.2 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.1 | GO:0033600 | negative regulation of mammary gland epithelial cell proliferation(GO:0033600) |
| 0.0 | 0.1 | GO:0032897 | negative regulation of viral transcription(GO:0032897) |
| 0.0 | 0.2 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.0 | 0.1 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.1 | GO:0051492 | regulation of stress fiber assembly(GO:0051492) |
| 0.0 | 0.1 | GO:0032988 | ribonucleoprotein complex disassembly(GO:0032988) |
| 0.0 | 0.1 | GO:0006562 | proline catabolic process(GO:0006562) |
| 0.0 | 0.1 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.0 | 0.0 | GO:0009174 | UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.0 | 0.0 | GO:1903521 | apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 0.0 | 0.1 | GO:0002076 | osteoblast development(GO:0002076) |
| 0.0 | 0.0 | GO:0001774 | microglial cell activation(GO:0001774) |
| 0.0 | 0.0 | GO:0060687 | regulation of branching involved in prostate gland morphogenesis(GO:0060687) |
| 0.0 | 0.0 | GO:2000008 | regulation of protein localization to cell surface(GO:2000008) |
| 0.0 | 0.0 | GO:0030812 | negative regulation of nucleotide catabolic process(GO:0030812) |
| 0.0 | 0.0 | GO:0015864 | pyrimidine nucleoside transport(GO:0015864) |
| 0.0 | 0.2 | GO:0006893 | Golgi to plasma membrane transport(GO:0006893) vesicle-mediated transport to the plasma membrane(GO:0098876) |
| 0.0 | 0.1 | GO:0042940 | D-amino acid transport(GO:0042940) |
| 0.0 | 0.0 | GO:0007132 | meiotic metaphase I(GO:0007132) |
| 0.0 | 0.0 | GO:0042637 | catagen(GO:0042637) |
| 0.0 | 0.0 | GO:0010719 | negative regulation of epithelial to mesenchymal transition(GO:0010719) |
| 0.0 | 0.1 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.0 | GO:0048570 | notochord morphogenesis(GO:0048570) |
| 0.0 | 0.0 | GO:0035581 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.0 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.1 | GO:0061001 | dendritic spine morphogenesis(GO:0060997) regulation of dendritic spine morphogenesis(GO:0061001) dendritic spine organization(GO:0097061) |
| 0.0 | 0.1 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.0 | 0.2 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.0 | 0.0 | GO:0006927 | obsolete transformed cell apoptotic process(GO:0006927) |
| 0.0 | 0.0 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.0 | 0.1 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.0 | 0.1 | GO:0032055 | negative regulation of translation in response to stress(GO:0032055) |
| 0.0 | 0.0 | GO:0051665 | membrane raft localization(GO:0051665) |
| 0.0 | 0.0 | GO:0009732 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.0 | 0.1 | GO:0048525 | negative regulation of viral process(GO:0048525) negative regulation of viral life cycle(GO:1903901) |
| 0.0 | 0.0 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.0 | GO:0001547 | antral ovarian follicle growth(GO:0001547) |
| 0.0 | 0.0 | GO:0060083 | smooth muscle contraction involved in micturition(GO:0060083) |
| 0.0 | 0.0 | GO:0044320 | cellular response to leptin stimulus(GO:0044320) response to leptin(GO:0044321) |
| 0.0 | 0.0 | GO:0014832 | urinary bladder smooth muscle contraction(GO:0014832) |
| 0.0 | 0.1 | GO:0071450 | removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.0 | 0.0 | GO:0001921 | positive regulation of receptor recycling(GO:0001921) |
| 0.0 | 0.1 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.1 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.0 | 0.0 | GO:2000846 | corticosteroid hormone secretion(GO:0035930) regulation of steroid hormone secretion(GO:2000831) regulation of corticosteroid hormone secretion(GO:2000846) |
| 0.0 | 0.1 | GO:0018106 | peptidyl-histidine phosphorylation(GO:0018106) |
| 0.0 | 0.1 | GO:0018027 | peptidyl-lysine dimethylation(GO:0018027) |
| 0.0 | 0.0 | GO:2000344 | positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.1 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 0.0 | GO:1903053 | regulation of extracellular matrix disassembly(GO:0010715) negative regulation of extracellular matrix disassembly(GO:0010716) regulation of extracellular matrix organization(GO:1903053) negative regulation of extracellular matrix organization(GO:1903054) |
| 0.0 | 0.0 | GO:0045767 | obsolete regulation of anti-apoptosis(GO:0045767) |
| 0.0 | 0.0 | GO:0021631 | optic nerve morphogenesis(GO:0021631) |
| 0.0 | 0.0 | GO:0051176 | positive regulation of sulfur metabolic process(GO:0051176) |
| 0.0 | 0.1 | GO:0007619 | courtship behavior(GO:0007619) |
| 0.0 | 0.0 | GO:0006065 | UDP-glucuronate biosynthetic process(GO:0006065) |
| 0.0 | 0.1 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.0 | GO:2000300 | regulation of synaptic vesicle transport(GO:1902803) regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.0 | 0.0 | GO:0008054 | negative regulation of cyclin-dependent protein serine/threonine kinase by cyclin degradation(GO:0008054) |
| 0.0 | 0.0 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.1 | GO:0098810 | neurotransmitter reuptake(GO:0098810) |
| 0.0 | 0.1 | GO:0032288 | myelin assembly(GO:0032288) |
| 0.0 | 0.0 | GO:0010759 | positive regulation of macrophage chemotaxis(GO:0010759) |
| 0.0 | 0.0 | GO:0006296 | nucleotide-excision repair, DNA incision, 5'-to lesion(GO:0006296) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.2 | GO:0036126 | outer dense fiber(GO:0001520) sperm flagellum(GO:0036126) |
| 0.0 | 0.1 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.1 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.2 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.1 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 0.1 | GO:0060200 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.0 | 0.1 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.1 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.1 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.0 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.0 | 0.1 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.7 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.1 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.0 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.0 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.0 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.0 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.0 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.1 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 1.3 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.0 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.1 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.0 | 0.1 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.2 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.1 | GO:0098651 | basement membrane collagen trimer(GO:0098651) |
| 0.0 | 0.0 | GO:0042641 | actomyosin(GO:0042641) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.0 | 0.2 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.1 | GO:0004992 | platelet activating factor receptor activity(GO:0004992) |
| 0.0 | 0.1 | GO:0046980 | tapasin binding(GO:0046980) |
| 0.0 | 0.1 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.1 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.0 | 0.2 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.3 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.1 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.2 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.1 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.0 | 0.3 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.2 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.1 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.2 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.1 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.2 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 0.2 | GO:0016565 | obsolete general transcriptional repressor activity(GO:0016565) |
| 0.0 | 0.1 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.1 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.1 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) |
| 0.0 | 0.0 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.1 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.0 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.2 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.1 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.2 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.1 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.1 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.1 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.1 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.1 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.1 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.1 | GO:0008486 | diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) |
| 0.0 | 0.1 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.1 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.0 | 0.2 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.0 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.0 | 0.0 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.1 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 0.0 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.2 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.0 | 0.1 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.0 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.0 | 0.0 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.1 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.0 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.0 | 0.0 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.2 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.1 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.0 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.0 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.1 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.0 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.2 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.0 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.1 | GO:0003711 | obsolete transcription elongation regulator activity(GO:0003711) |
| 0.0 | 0.0 | GO:0003701 | obsolete RNA polymerase I transcription factor activity(GO:0003701) |
| 0.0 | 0.0 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.0 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.0 | 0.0 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.0 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.1 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.0 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.0 | 0.0 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.0 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.0 | 0.1 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 0.0 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.0 | 0.0 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.0 | 0.1 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.0 | GO:0015168 | glycerol transmembrane transporter activity(GO:0015168) |
| 0.0 | 0.0 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.0 | 0.1 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.1 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.1 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.0 | 0.0 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.0 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.1 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.0 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.0 | 0.0 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.0 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.4 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.2 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.2 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 0.3 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.1 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.3 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.2 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.3 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.0 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.2 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.1 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.1 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.3 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.0 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.1 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.1 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.1 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.1 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.1 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.1 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |