Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
Results for LHX3
Z-value: 1.16
Transcription factors associated with LHX3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
LHX3
|
ENSG00000107187.11 | LIM homeobox 3 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr9_139098395_139098546 | LHX3 | 1515 | 0.385643 | 0.43 | 2.5e-01 | Click! |
| chr9_139090141_139090292 | LHX3 | 4788 | 0.206786 | -0.38 | 3.2e-01 | Click! |
| chr9_139084589_139084740 | LHX3 | 10340 | 0.182530 | -0.37 | 3.3e-01 | Click! |
| chr9_139081692_139081843 | LHX3 | 13237 | 0.177542 | 0.33 | 3.8e-01 | Click! |
| chr9_139085334_139085485 | LHX3 | 9595 | 0.184074 | -0.29 | 4.5e-01 | Click! |
Activity of the LHX3 motif across conditions
Conditions sorted by the z-value of the LHX3 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
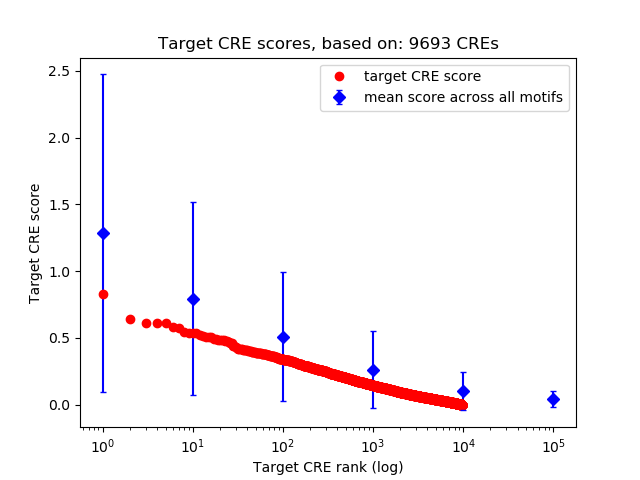
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr3_177651530_177651798 | 0.83 |
ENSG00000199858 |
. |
73903 |
0.13 |
| chr2_161232105_161232912 | 0.64 |
ENSG00000252465 |
. |
20971 |
0.19 |
| chr1_109912007_109912390 | 0.61 |
SORT1 |
sortilin 1 |
23781 |
0.14 |
| chr1_67397953_67399117 | 0.61 |
MIER1 |
mesoderm induction early response 1, transcriptional regulator |
2609 |
0.29 |
| chr3_182992678_182993398 | 0.61 |
B3GNT5 |
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5 |
9906 |
0.2 |
| chr7_40621099_40621486 | 0.58 |
AC004988.1 |
|
34765 |
0.24 |
| chr2_193011022_193011173 | 0.57 |
TMEFF2 |
transmembrane protein with EGF-like and two follistatin-like domains 2 |
48153 |
0.19 |
| chr9_93557837_93558119 | 0.54 |
SYK |
spleen tyrosine kinase |
6091 |
0.34 |
| chr6_144587139_144587431 | 0.54 |
UTRN |
utrophin |
19552 |
0.24 |
| chr4_53966535_53966686 | 0.54 |
RP11-752D24.2 |
|
154754 |
0.04 |
| chr9_90215657_90216062 | 0.53 |
DAPK1-IT1 |
DAPK1 intronic transcript 1 (non-protein coding) |
47490 |
0.16 |
| chr2_153277819_153278036 | 0.52 |
FMNL2 |
formin-like 2 |
86176 |
0.1 |
| chr8_108506998_108507182 | 0.51 |
ANGPT1 |
angiopoietin 1 |
133 |
0.98 |
| chr7_81303437_81303728 | 0.51 |
HGF |
hepatocyte growth factor (hepapoietin A; scatter factor) |
88730 |
0.1 |
| chr9_79065617_79065768 | 0.51 |
GCNT1 |
glucosaminyl (N-acetyl) transferase 1, core 2 |
8376 |
0.27 |
| chr22_33108393_33108637 | 0.51 |
LL22NC01-116C6.1 |
|
70648 |
0.1 |
| chr7_95239146_95239504 | 0.49 |
AC002451.3 |
|
217 |
0.94 |
| chr7_102532081_102532232 | 0.49 |
LRRC17 |
leucine rich repeat containing 17 |
21282 |
0.16 |
| chr1_222341096_222341247 | 0.49 |
ENSG00000212094 |
. |
154739 |
0.04 |
| chr11_111848937_111849153 | 0.49 |
DIXDC1 |
DIX domain containing 1 |
1012 |
0.41 |
| chr6_12266277_12266506 | 0.49 |
EDN1 |
endothelin 1 |
24205 |
0.26 |
| chr12_81256633_81256894 | 0.48 |
LIN7A |
lin-7 homolog A (C. elegans) |
26364 |
0.17 |
| chr10_31053149_31053377 | 0.48 |
SVILP1 |
supervillin pseudogene 1 |
68413 |
0.11 |
| chr5_158127271_158127455 | 0.47 |
CTD-2363C16.1 |
|
282651 |
0.01 |
| chr15_59641636_59642058 | 0.47 |
ENSG00000199512 |
. |
8231 |
0.13 |
| chr12_90127026_90127233 | 0.47 |
ENSG00000252207 |
. |
6416 |
0.21 |
| chr20_4046539_4046763 | 0.46 |
RP11-352D3.2 |
Uncharacterized protein |
9161 |
0.22 |
| chr6_114196551_114196945 | 0.44 |
MARCKS |
myristoylated alanine-rich protein kinase C substrate |
18207 |
0.17 |
| chr6_17933009_17933323 | 0.44 |
KIF13A |
kinesin family member 13A |
54528 |
0.15 |
| chr11_122002800_122002951 | 0.43 |
ENSG00000198975 |
. |
14426 |
0.14 |
| chr12_27430347_27430721 | 0.42 |
STK38L |
serine/threonine kinase 38 like |
16027 |
0.23 |
| chr3_55558379_55558651 | 0.42 |
WNT5A |
wingless-type MMTV integration site family, member 5A |
34542 |
0.2 |
| chr4_47215322_47215473 | 0.42 |
ENSG00000238301 |
. |
92657 |
0.09 |
| chr6_52425530_52425681 | 0.42 |
TRAM2 |
translocation associated membrane protein 2 |
16108 |
0.24 |
| chr3_149058191_149058568 | 0.42 |
TM4SF18 |
transmembrane 4 L six family member 18 |
6178 |
0.2 |
| chr10_90411662_90411938 | 0.41 |
LIPF |
lipase, gastric |
12398 |
0.19 |
| chr14_69555636_69555913 | 0.41 |
ENSG00000206768 |
. |
54896 |
0.12 |
| chr2_100218822_100218973 | 0.41 |
AFF3 |
AF4/FMR2 family, member 3 |
24065 |
0.23 |
| chr1_172248149_172248300 | 0.41 |
ENSG00000252354 |
. |
69059 |
0.1 |
| chr4_141866091_141866242 | 0.41 |
RNF150 |
ring finger protein 150 |
52024 |
0.17 |
| chr5_14399490_14399641 | 0.41 |
TRIO |
trio Rho guanine nucleotide exchange factor |
88992 |
0.1 |
| chr18_10142397_10142548 | 0.41 |
ENSG00000263630 |
. |
137278 |
0.05 |
| chr4_186196562_186196713 | 0.40 |
SNX25 |
sorting nexin 25 |
65404 |
0.09 |
| chr20_50543676_50543832 | 0.40 |
ENSG00000263659 |
. |
13618 |
0.19 |
| chr8_108325074_108325225 | 0.40 |
ANGPT1 |
angiopoietin 1 |
23601 |
0.25 |
| chr3_125019965_125020116 | 0.40 |
SLC12A8 |
solute carrier family 12, member 8 |
22019 |
0.2 |
| chr4_24553435_24554115 | 0.39 |
ENSG00000243005 |
. |
11309 |
0.21 |
| chr2_192121899_192122050 | 0.39 |
MYO1B |
myosin IB |
5751 |
0.29 |
| chr3_154604594_154604879 | 0.39 |
MME |
membrane metallo-endopeptidase |
137177 |
0.05 |
| chr3_136505593_136506225 | 0.39 |
SLC35G2 |
solute carrier family 35, member G2 |
31952 |
0.12 |
| chr2_27905704_27905855 | 0.39 |
SUPT7L |
suppressor of Ty 7 (S. cerevisiae)-like |
19103 |
0.11 |
| chr3_12235410_12235692 | 0.39 |
TIMP4 |
TIMP metallopeptidase inhibitor 4 |
34700 |
0.2 |
| chr4_38986506_38986933 | 0.39 |
TMEM156 |
transmembrane protein 156 |
47322 |
0.13 |
| chr12_65674928_65675079 | 0.39 |
RP11-305O6.3 |
|
434 |
0.84 |
| chr3_60921706_60921857 | 0.39 |
ENSG00000212211 |
. |
79504 |
0.12 |
| chr8_58980536_58980687 | 0.39 |
FAM110B |
family with sequence similarity 110, member B |
73498 |
0.13 |
| chr10_93221945_93222246 | 0.38 |
HECTD2 |
HECT domain containing E3 ubiquitin protein ligase 2 |
198 |
0.97 |
| chr1_44966238_44966535 | 0.38 |
ENSG00000263381 |
. |
44779 |
0.13 |
| chr7_137276747_137277035 | 0.38 |
PTN |
pleiotrophin |
248280 |
0.02 |
| chr7_42233492_42233928 | 0.38 |
GLI3 |
GLI family zinc finger 3 |
33610 |
0.25 |
| chr6_3140070_3140221 | 0.38 |
BPHL |
biphenyl hydrolase-like (serine hydrolase) |
514 |
0.72 |
| chr5_142253906_142254057 | 0.38 |
ARHGAP26-AS1 |
ARHGAP26 antisense RNA 1 |
5494 |
0.28 |
| chr11_3980211_3980997 | 0.38 |
STIM1 |
stromal interaction molecule 1 |
12031 |
0.18 |
| chrX_134122511_134122662 | 0.38 |
SMIM10 |
small integral membrane protein 10 |
2382 |
0.26 |
| chr1_87801300_87801913 | 0.38 |
LMO4 |
LIM domain only 4 |
4255 |
0.35 |
| chr6_140892392_140892543 | 0.38 |
ENSG00000221336 |
. |
89383 |
0.1 |
| chr1_95009721_95009872 | 0.37 |
F3 |
coagulation factor III (thromboplastin, tissue factor) |
2440 |
0.41 |
| chr9_130302299_130302610 | 0.37 |
FAM129B |
family with sequence similarity 129, member B |
28913 |
0.13 |
| chr2_46325558_46325709 | 0.37 |
AC017006.2 |
|
19666 |
0.24 |
| chr9_14251921_14252072 | 0.37 |
NFIB |
nuclear factor I/B |
56016 |
0.15 |
| chr4_86713976_86714229 | 0.37 |
ARHGAP24 |
Rho GTPase activating protein 24 |
14243 |
0.26 |
| chr13_33766329_33766480 | 0.37 |
STARD13 |
StAR-related lipid transfer (START) domain containing 13 |
6188 |
0.24 |
| chr6_39772853_39773004 | 0.37 |
DAAM2 |
dishevelled associated activator of morphogenesis 2 |
12134 |
0.26 |
| chr16_53809610_53810340 | 0.37 |
FTO |
fat mass and obesity associated |
71881 |
0.11 |
| chr6_56591218_56591369 | 0.37 |
DST |
dystonin |
59384 |
0.15 |
| chr6_36403738_36403889 | 0.36 |
KCTD20 |
potassium channel tetramerization domain containing 20 |
6731 |
0.14 |
| chr4_125335710_125335861 | 0.36 |
ANKRD50 |
ankyrin repeat domain 50 |
298102 |
0.01 |
| chr1_55699646_55699797 | 0.36 |
ENSG00000265822 |
. |
8407 |
0.23 |
| chr4_2869241_2869562 | 0.36 |
ADD1 |
adducin 1 (alpha) |
8222 |
0.19 |
| chr1_152022964_152023546 | 0.36 |
S100A11 |
S100 calcium binding protein A11 |
13744 |
0.14 |
| chr3_99500056_99500207 | 0.36 |
CMSS1 |
cms1 ribosomal small subunit homolog (yeast) |
36547 |
0.18 |
| chr2_153441109_153441260 | 0.36 |
FMNL2 |
formin-like 2 |
34908 |
0.22 |
| chr8_80274236_80274387 | 0.35 |
STMN2 |
stathmin-like 2 |
248738 |
0.02 |
| chr7_82224652_82224988 | 0.35 |
CACNA2D1 |
calcium channel, voltage-dependent, alpha 2/delta subunit 1 |
151706 |
0.05 |
| chr3_152745972_152746123 | 0.35 |
RP11-529G21.2 |
|
133517 |
0.05 |
| chr8_128937773_128937924 | 0.35 |
TMEM75 |
transmembrane protein 75 |
22743 |
0.19 |
| chr16_69255951_69256102 | 0.35 |
RP11-70O5.2 |
|
18426 |
0.12 |
| chr4_111029289_111029440 | 0.35 |
ENSG00000263940 |
. |
9528 |
0.26 |
| chr10_59804431_59804582 | 0.35 |
ENSG00000238970 |
. |
194015 |
0.03 |
| chr3_188995375_188995558 | 0.35 |
TPRG1-AS2 |
TPRG1 antisense RNA 2 |
37083 |
0.21 |
| chr5_167097874_167098025 | 0.35 |
CTB-78F1.1 |
|
10463 |
0.25 |
| chr13_41627976_41628416 | 0.35 |
WBP4 |
WW domain binding protein 4 |
7214 |
0.17 |
| chr5_52627031_52627182 | 0.34 |
FST |
follistatin |
149133 |
0.04 |
| chr17_35567851_35568148 | 0.34 |
ACACA |
acetyl-CoA carboxylase alpha |
5273 |
0.28 |
| chr9_91346743_91346894 | 0.34 |
ENSG00000265873 |
. |
14002 |
0.29 |
| chr12_27779794_27779945 | 0.34 |
PPFIBP1 |
PTPRF interacting protein, binding protein 1 (liprin beta 1) |
27588 |
0.17 |
| chr10_16952291_16952577 | 0.34 |
CUBN |
cubilin (intrinsic factor-cobalamin receptor) |
73760 |
0.11 |
| chr17_61351351_61351502 | 0.34 |
RP11-269G24.2 |
|
39456 |
0.15 |
| chr10_95235571_95235788 | 0.34 |
MYOF |
myoferlin |
6272 |
0.2 |
| chr3_24407013_24407164 | 0.34 |
THRB-IT1 |
THRB intronic transcript 1 (non-protein coding) |
93837 |
0.08 |
| chr3_188560859_188561010 | 0.34 |
TPRG1 |
tumor protein p63 regulated 1 |
104069 |
0.08 |
| chr7_36487948_36488099 | 0.34 |
AC006960.7 |
|
14609 |
0.19 |
| chr4_178156273_178156424 | 0.34 |
RP11-487E13.1 |
Uncharacterized protein |
13579 |
0.23 |
| chr12_8309941_8310303 | 0.34 |
ZNF705A |
zinc finger protein 705A |
492 |
0.74 |
| chr13_52928121_52928272 | 0.34 |
ENSG00000199605 |
. |
45943 |
0.13 |
| chr10_54079967_54080118 | 0.34 |
DKK1 |
dickkopf WNT signaling pathway inhibitor 1 |
5986 |
0.26 |
| chr2_216529775_216530114 | 0.34 |
ENSG00000212055 |
. |
213698 |
0.02 |
| chr2_145135578_145135895 | 0.34 |
GTDC1 |
glycosyltransferase-like domain containing 1 |
45653 |
0.17 |
| chr14_52122560_52122711 | 0.34 |
FRMD6 |
FERM domain containing 6 |
3937 |
0.24 |
| chr2_118804547_118804712 | 0.34 |
ENSG00000243510 |
. |
30671 |
0.13 |
| chr1_183776609_183776992 | 0.34 |
RGL1 |
ral guanine nucleotide dissociation stimulator-like 1 |
2510 |
0.36 |
| chr1_178143791_178143942 | 0.34 |
RASAL2 |
RAS protein activator like 2 |
80590 |
0.11 |
| chr8_32432955_32433282 | 0.33 |
NRG1 |
neuregulin 1 |
26873 |
0.26 |
| chr2_55328902_55329053 | 0.33 |
ENSG00000266376 |
. |
6869 |
0.2 |
| chr13_27159400_27159551 | 0.33 |
WASF3 |
WAS protein family, member 3 |
27588 |
0.22 |
| chr3_110132390_110132541 | 0.33 |
ENSG00000221206 |
. |
138347 |
0.05 |
| chr12_124515572_124515723 | 0.33 |
ZNF664 |
zinc finger protein 664 |
57805 |
0.07 |
| chr3_98701653_98701804 | 0.33 |
ENSG00000207331 |
. |
74425 |
0.1 |
| chr2_200907594_200907745 | 0.33 |
C2orf47 |
chromosome 2 open reading frame 47 |
87125 |
0.08 |
| chr6_147234613_147234825 | 0.33 |
STXBP5-AS1 |
STXBP5 antisense RNA 1 |
1977 |
0.49 |
| chr6_126268678_126268829 | 0.33 |
HINT3 |
histidine triad nucleotide binding protein 3 |
9174 |
0.18 |
| chr13_24462234_24462908 | 0.33 |
C1QTNF9B-AS1 |
C1QTNF9B antisense RNA 1 |
457 |
0.68 |
| chr8_59745127_59745278 | 0.33 |
ENSG00000201231 |
. |
18091 |
0.27 |
| chr2_208123731_208123882 | 0.33 |
AC007879.5 |
|
4830 |
0.24 |
| chr8_57806614_57806867 | 0.33 |
ENSG00000206975 |
. |
22903 |
0.26 |
| chr5_124748352_124748503 | 0.33 |
ENSG00000222107 |
. |
61603 |
0.16 |
| chr11_113981742_113981893 | 0.32 |
ENSG00000221112 |
. |
24166 |
0.2 |
| chr3_159542327_159542478 | 0.32 |
ENSG00000264146 |
. |
1918 |
0.3 |
| chr2_114040706_114040961 | 0.32 |
PAX8 |
paired box 8 |
4306 |
0.2 |
| chr15_49336349_49337349 | 0.32 |
SECISBP2L |
SECIS binding protein 2-like |
1781 |
0.28 |
| chr6_158460883_158461210 | 0.32 |
SYNJ2 |
synaptojanin 2 |
22549 |
0.18 |
| chr12_18498312_18498592 | 0.32 |
PIK3C2G |
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 gamma |
63474 |
0.15 |
| chr8_93978348_93978807 | 0.32 |
TRIQK |
triple QxxK/R motif containing |
204 |
0.96 |
| chr5_9888608_9888759 | 0.32 |
ENSG00000222054 |
. |
127664 |
0.05 |
| chr8_96574856_96575007 | 0.32 |
ENSG00000202112 |
. |
36411 |
0.22 |
| chr4_38771269_38771420 | 0.32 |
TLR10 |
toll-like receptor 10 |
6473 |
0.17 |
| chr17_20092898_20093141 | 0.32 |
SPECC1 |
sperm antigen with calponin homology and coiled-coil domains 1 |
15830 |
0.22 |
| chr19_8605989_8606140 | 0.31 |
ENSG00000265528 |
. |
5679 |
0.12 |
| chr10_78157189_78157340 | 0.31 |
RP11-369F10.2 |
|
41269 |
0.18 |
| chr9_133720557_133720776 | 0.31 |
ABL1 |
c-abl oncogene 1, non-receptor tyrosine kinase |
10213 |
0.21 |
| chr8_25066500_25066825 | 0.31 |
DOCK5 |
dedicator of cytokinesis 5 |
24282 |
0.21 |
| chr6_56588753_56588904 | 0.31 |
DST |
dystonin |
61849 |
0.14 |
| chrX_12229658_12229809 | 0.31 |
ENSG00000206795 |
. |
62488 |
0.12 |
| chr5_16890159_16890310 | 0.31 |
MYO10 |
myosin X |
26402 |
0.18 |
| chr6_114290051_114291212 | 0.31 |
RP3-399L15.3 |
|
234 |
0.87 |
| chr9_3912136_3912287 | 0.31 |
GLIS3-AS1 |
GLIS3 antisense RNA 1 |
13569 |
0.25 |
| chr3_69813958_69814283 | 0.31 |
MITF |
microphthalmia-associated transcription factor |
1158 |
0.56 |
| chr12_52348007_52348213 | 0.31 |
ACVR1B |
activin A receptor, type IB |
946 |
0.5 |
| chr6_140859760_140859911 | 0.31 |
ENSG00000221336 |
. |
122015 |
0.06 |
| chr3_111521650_111521801 | 0.31 |
PHLDB2 |
pleckstrin homology-like domain, family B, member 2 |
56302 |
0.14 |
| chr1_17275864_17276015 | 0.31 |
CROCC |
ciliary rootlet coiled-coil, rootletin |
26689 |
0.11 |
| chr2_40510156_40510307 | 0.31 |
AC007377.1 |
|
22084 |
0.23 |
| chr17_35851980_35852131 | 0.30 |
DUSP14 |
dual specificity phosphatase 14 |
485 |
0.81 |
| chr15_57513605_57513971 | 0.30 |
TCF12 |
transcription factor 12 |
2124 |
0.42 |
| chr7_38209227_38209494 | 0.30 |
STARD3NL |
STARD3 N-terminal like |
8464 |
0.31 |
| chr14_61041455_61041606 | 0.30 |
ENSG00000253014 |
. |
32255 |
0.15 |
| chr2_145779005_145779156 | 0.30 |
ENSG00000253036 |
. |
313558 |
0.01 |
| chr4_57909352_57909761 | 0.30 |
ENSG00000251703 |
. |
40698 |
0.11 |
| chr2_225681896_225682047 | 0.30 |
DOCK10 |
dedicator of cytokinesis 10 |
602 |
0.85 |
| chr2_75819618_75819769 | 0.30 |
EVA1A |
eva-1 homolog A (C. elegans) |
22845 |
0.13 |
| chr4_7881276_7881455 | 0.30 |
AFAP1 |
actin filament associated protein 1 |
7325 |
0.25 |
| chr12_120131477_120131628 | 0.30 |
RP1-127H14.3 |
Uncharacterized protein |
6021 |
0.18 |
| chr3_72338599_72338998 | 0.30 |
ENSG00000212070 |
. |
27219 |
0.26 |
| chr15_39636633_39636784 | 0.30 |
RP11-624L4.1 |
|
70624 |
0.12 |
| chr7_92295493_92295644 | 0.30 |
ENSG00000206763 |
. |
35560 |
0.18 |
| chr4_182984434_182984585 | 0.30 |
AC108142.1 |
|
21728 |
0.23 |
| chr6_44414403_44414554 | 0.30 |
ENSG00000266619 |
. |
11100 |
0.18 |
| chr5_52079021_52079172 | 0.29 |
ITGA1 |
integrin, alpha 1 |
4634 |
0.21 |
| chr1_101108683_101108834 | 0.29 |
VCAM1 |
vascular cell adhesion molecule 1 |
76540 |
0.1 |
| chr1_23610078_23610229 | 0.29 |
HNRNPR |
heterogeneous nuclear ribonucleoprotein R |
60602 |
0.09 |
| chr4_48942200_48942351 | 0.29 |
ENSG00000252913 |
. |
7404 |
0.21 |
| chr15_70556363_70556608 | 0.29 |
ENSG00000200216 |
. |
70910 |
0.13 |
| chr7_152010807_152011572 | 0.29 |
KMT2C |
lysine (K)-specific methyltransferase 2C |
66085 |
0.11 |
| chr2_69417973_69418124 | 0.29 |
ENSG00000199460 |
. |
8039 |
0.21 |
| chr2_138722580_138722731 | 0.29 |
HNMT |
histamine N-methyltransferase |
612 |
0.86 |
| chr1_186426484_186426635 | 0.29 |
PDC |
phosducin |
3695 |
0.21 |
| chr3_167577259_167577557 | 0.29 |
SERPINI1 |
serpin peptidase inhibitor, clade I (neuroserpin), member 1 |
52382 |
0.17 |
| chr8_32573551_32573702 | 0.29 |
NRG1 |
neuregulin 1 |
5645 |
0.34 |
| chr9_110573374_110573525 | 0.29 |
ENSG00000244104 |
. |
746 |
0.68 |
| chr7_28002187_28002338 | 0.29 |
JAZF1 |
JAZF zinc finger 1 |
29413 |
0.22 |
| chr10_104956679_104957044 | 0.29 |
NT5C2 |
5'-nucleotidase, cytosolic II |
3805 |
0.25 |
| chr4_138229530_138229827 | 0.29 |
ENSG00000222526 |
. |
80396 |
0.12 |
| chrX_39832066_39832217 | 0.29 |
BCOR |
BCL6 corepressor |
90049 |
0.09 |
| chr12_24383335_24383486 | 0.29 |
ENSG00000216192 |
. |
18055 |
0.3 |
| chr6_128390722_128390873 | 0.29 |
PTPRK |
protein tyrosine phosphatase, receptor type, K |
4889 |
0.29 |
| chr17_57075608_57075759 | 0.29 |
ENSG00000216168 |
. |
9368 |
0.15 |
| chr9_138861135_138861286 | 0.29 |
UBAC1 |
UBA domain containing 1 |
7984 |
0.21 |
| chr9_91389382_91389533 | 0.29 |
ENSG00000265873 |
. |
28637 |
0.25 |
| chr5_163012032_163012183 | 0.29 |
MAT2B |
methionine adenosyltransferase II, beta |
79487 |
0.09 |
| chr2_69469610_69469761 | 0.29 |
ENSG00000199460 |
. |
59676 |
0.11 |
| chr6_15066355_15066506 | 0.29 |
ENSG00000242989 |
. |
46769 |
0.18 |
| chr15_56617819_56617970 | 0.28 |
TEX9 |
testis expressed 9 |
39731 |
0.17 |
| chr12_89698996_89699278 | 0.28 |
ENSG00000238302 |
. |
23075 |
0.22 |
| chr11_75657287_75657506 | 0.28 |
ENSG00000223013 |
. |
11416 |
0.16 |
| chr15_34913517_34913668 | 0.28 |
ENSG00000252425 |
. |
23833 |
0.16 |
| chr2_150601107_150601258 | 0.28 |
ENSG00000207270 |
. |
133894 |
0.05 |
| chr1_88305877_88306028 | 0.28 |
ENSG00000199318 |
. |
386896 |
0.01 |
| chr8_99571908_99572059 | 0.28 |
KB-1205A7.2 |
|
43498 |
0.17 |
| chr1_178313576_178313822 | 0.28 |
RASAL2 |
RAS protein activator like 2 |
3093 |
0.37 |
| chr16_66604769_66604920 | 0.28 |
CMTM1 |
CKLF-like MARVEL transmembrane domain containing 1 |
4427 |
0.11 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.1 | 0.3 | GO:0035385 | Roundabout signaling pathway(GO:0035385) |
| 0.1 | 0.3 | GO:0051305 | chromosome movement towards spindle pole(GO:0051305) |
| 0.1 | 0.2 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 0.2 | GO:0030091 | protein repair(GO:0030091) |
| 0.1 | 0.2 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) |
| 0.1 | 0.1 | GO:0010837 | regulation of keratinocyte proliferation(GO:0010837) |
| 0.1 | 0.2 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.1 | 0.3 | GO:0002138 | retinoic acid biosynthetic process(GO:0002138) diterpenoid biosynthetic process(GO:0016102) |
| 0.1 | 0.2 | GO:0045425 | positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) |
| 0.1 | 0.2 | GO:0061318 | renal filtration cell differentiation(GO:0061318) glomerular epithelium development(GO:0072010) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.1 | 0.3 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.1 | 0.3 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.1 | 0.2 | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.1 | 0.2 | GO:0072201 | negative regulation of mesenchymal cell proliferation(GO:0072201) |
| 0.1 | 0.1 | GO:1903224 | endodermal cell fate specification(GO:0001714) regulation of endodermal cell fate specification(GO:0042663) regulation of endodermal cell differentiation(GO:1903224) |
| 0.0 | 0.0 | GO:0021834 | chemorepulsion involved in embryonic olfactory bulb interneuron precursor migration(GO:0021834) |
| 0.0 | 0.1 | GO:0060594 | mammary gland specification(GO:0060594) |
| 0.0 | 0.1 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.0 | 0.1 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.2 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 | 0.1 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.0 | 0.2 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.2 | GO:0009649 | entrainment of circadian clock(GO:0009649) |
| 0.0 | 0.1 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.0 | 0.1 | GO:0048669 | collateral sprouting in absence of injury(GO:0048669) |
| 0.0 | 0.1 | GO:0042726 | flavin-containing compound metabolic process(GO:0042726) |
| 0.0 | 0.1 | GO:0060638 | mesenchymal-epithelial cell signaling(GO:0060638) |
| 0.0 | 0.1 | GO:0090081 | regulation of heart induction by regulation of canonical Wnt signaling pathway(GO:0090081) |
| 0.0 | 0.1 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.0 | 0.1 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.2 | GO:0010886 | positive regulation of cholesterol storage(GO:0010886) |
| 0.0 | 0.2 | GO:0097061 | dendritic spine morphogenesis(GO:0060997) regulation of dendritic spine morphogenesis(GO:0061001) dendritic spine organization(GO:0097061) |
| 0.0 | 0.0 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.0 | 0.1 | GO:0048631 | regulation of skeletal muscle tissue growth(GO:0048631) |
| 0.0 | 0.2 | GO:0001547 | antral ovarian follicle growth(GO:0001547) |
| 0.0 | 0.2 | GO:0002634 | regulation of germinal center formation(GO:0002634) |
| 0.0 | 0.1 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.0 | GO:0060197 | cloaca development(GO:0035844) cloacal septation(GO:0060197) |
| 0.0 | 0.1 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.1 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.1 | GO:0009128 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.0 | 0.1 | GO:0070340 | detection of bacterial lipoprotein(GO:0042494) detection of bacterial lipopeptide(GO:0070340) |
| 0.0 | 0.1 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.1 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.0 | 0.1 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 0.0 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 0.0 | 0.2 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 0.4 | GO:0021846 | cell proliferation in forebrain(GO:0021846) |
| 0.0 | 0.1 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.1 | GO:0060129 | thyroid-stimulating hormone-secreting cell differentiation(GO:0060129) |
| 0.0 | 0.1 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.0 | 0.1 | GO:0071694 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.1 | GO:0001957 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 | 0.1 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.1 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
| 0.0 | 0.1 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.0 | GO:0061525 | hindgut morphogenesis(GO:0007442) hindgut development(GO:0061525) |
| 0.0 | 0.1 | GO:0060754 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 0.1 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.0 | 0.2 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.1 | GO:0015864 | pyrimidine nucleoside transport(GO:0015864) |
| 0.0 | 0.1 | GO:0071379 | cellular response to prostaglandin stimulus(GO:0071379) |
| 0.0 | 0.1 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.0 | 0.1 | GO:1903170 | negative regulation of release of sequestered calcium ion into cytosol(GO:0051280) positive regulation of sequestering of calcium ion(GO:0051284) negative regulation of calcium ion transmembrane transport(GO:1903170) |
| 0.0 | 0.1 | GO:0033024 | mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) |
| 0.0 | 0.1 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.1 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.0 | GO:2000095 | regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.0 | 0.0 | GO:0031392 | regulation of prostaglandin biosynthetic process(GO:0031392) positive regulation of prostaglandin biosynthetic process(GO:0031394) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.0 | 0.1 | GO:0051877 | pigment granule aggregation in cell center(GO:0051877) |
| 0.0 | 0.1 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.0 | 0.0 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.1 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.1 | GO:0008215 | spermine metabolic process(GO:0008215) |
| 0.0 | 0.1 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 0.0 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.0 | 0.0 | GO:0070672 | response to interleukin-15(GO:0070672) |
| 0.0 | 0.1 | GO:1901722 | regulation of cell proliferation involved in kidney development(GO:1901722) |
| 0.0 | 0.1 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.0 | 0.1 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.1 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.0 | 0.1 | GO:0031558 | obsolete induction of apoptosis in response to chemical stimulus(GO:0031558) |
| 0.0 | 0.1 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) positive regulation of signal transduction by p53 class mediator(GO:1901798) |
| 0.0 | 0.0 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.0 | 0.1 | GO:0042483 | negative regulation of odontogenesis(GO:0042483) |
| 0.0 | 0.0 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
| 0.0 | 0.1 | GO:0033860 | regulation of NAD(P)H oxidase activity(GO:0033860) |
| 0.0 | 0.0 | GO:0072393 | microtubule anchoring at microtubule organizing center(GO:0072393) |
| 0.0 | 0.1 | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.0 | 0.3 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 0.1 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.1 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.0 | 0.1 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.0 | 0.1 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.0 | 0.2 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.0 | 0.1 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.1 | GO:0002063 | chondrocyte development(GO:0002063) |
| 0.0 | 0.1 | GO:0071436 | sodium ion export(GO:0071436) |
| 0.0 | 0.3 | GO:0032330 | regulation of chondrocyte differentiation(GO:0032330) |
| 0.0 | 0.1 | GO:0010566 | regulation of ketone biosynthetic process(GO:0010566) regulation of aldosterone metabolic process(GO:0032344) regulation of aldosterone biosynthetic process(GO:0032347) |
| 0.0 | 0.4 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.2 | GO:0006972 | hyperosmotic response(GO:0006972) |
| 0.0 | 0.1 | GO:0060710 | chorio-allantoic fusion(GO:0060710) |
| 0.0 | 0.0 | GO:0046882 | regulation of follicle-stimulating hormone secretion(GO:0046880) negative regulation of follicle-stimulating hormone secretion(GO:0046882) |
| 0.0 | 0.1 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.0 | GO:0072193 | ureter smooth muscle development(GO:0072191) ureter smooth muscle cell differentiation(GO:0072193) |
| 0.0 | 0.1 | GO:0045197 | establishment or maintenance of epithelial cell apical/basal polarity(GO:0045197) |
| 0.0 | 0.1 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.1 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.0 | 0.1 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.1 | GO:2000757 | negative regulation of histone acetylation(GO:0035067) negative regulation of protein acetylation(GO:1901984) negative regulation of peptidyl-lysine acetylation(GO:2000757) |
| 0.0 | 0.0 | GO:0016999 | antibiotic metabolic process(GO:0016999) antibiotic biosynthetic process(GO:0017000) |
| 0.0 | 0.1 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.0 | 0.3 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.0 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.0 | 0.0 | GO:0070932 | histone H3 deacetylation(GO:0070932) histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.0 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 0.1 | GO:0009200 | deoxyribonucleoside triphosphate metabolic process(GO:0009200) |
| 0.0 | 0.1 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.0 | GO:0060789 | hair follicle placode formation(GO:0060789) |
| 0.0 | 0.1 | GO:0007320 | insemination(GO:0007320) |
| 0.0 | 0.1 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.0 | 0.1 | GO:0008655 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine nucleoside salvage(GO:0043097) |
| 0.0 | 0.0 | GO:0060083 | smooth muscle contraction involved in micturition(GO:0060083) |
| 0.0 | 0.0 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) |
| 0.0 | 0.0 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.0 | GO:0021873 | forebrain neuroblast division(GO:0021873) |
| 0.0 | 0.0 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.0 | 0.0 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.0 | 0.0 | GO:0045992 | negative regulation of embryonic development(GO:0045992) |
| 0.0 | 0.2 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 0.1 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.0 | 0.0 | GO:0018342 | protein prenylation(GO:0018342) prenylation(GO:0097354) |
| 0.0 | 0.0 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.0 | 0.1 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 0.0 | GO:0070254 | mucus secretion(GO:0070254) |
| 0.0 | 0.0 | GO:0060535 | lung growth(GO:0060437) trachea cartilage development(GO:0060534) trachea cartilage morphogenesis(GO:0060535) cartilage morphogenesis(GO:0060536) |
| 0.0 | 0.1 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 | 0.1 | GO:1902603 | carnitine shuttle(GO:0006853) carnitine transmembrane transport(GO:1902603) |
| 0.0 | 0.1 | GO:0071360 | cellular response to exogenous dsRNA(GO:0071360) |
| 0.0 | 0.1 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.1 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.0 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.0 | 0.0 | GO:0051549 | regulation of keratinocyte migration(GO:0051547) positive regulation of keratinocyte migration(GO:0051549) |
| 0.0 | 0.1 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.0 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.0 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.0 | 0.0 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.0 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.2 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.0 | GO:0000966 | RNA 5'-end processing(GO:0000966) |
| 0.0 | 0.1 | GO:0046825 | regulation of protein export from nucleus(GO:0046825) |
| 0.0 | 0.0 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.0 | GO:1903332 | regulation of protein folding(GO:1903332) |
| 0.0 | 0.0 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 | 0.0 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.1 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.0 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.0 | GO:0071680 | response to indole-3-methanol(GO:0071680) cellular response to indole-3-methanol(GO:0071681) |
| 0.0 | 0.1 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.0 | 0.1 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.0 | 0.1 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.0 | GO:0060839 | endothelial cell fate commitment(GO:0060839) |
| 0.0 | 0.0 | GO:0032959 | inositol trisphosphate biosynthetic process(GO:0032959) |
| 0.0 | 0.1 | GO:0007603 | phototransduction, visible light(GO:0007603) |
| 0.0 | 0.0 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.0 | GO:0002689 | negative regulation of leukocyte chemotaxis(GO:0002689) |
| 0.0 | 0.0 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.0 | 0.2 | GO:0045104 | intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 | 0.2 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 0.1 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.1 | GO:0060644 | mammary gland epithelial cell differentiation(GO:0060644) |
| 0.0 | 0.1 | GO:0001556 | oocyte maturation(GO:0001556) |
| 0.0 | 0.0 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.0 | 0.1 | GO:0048844 | artery morphogenesis(GO:0048844) |
| 0.0 | 0.0 | GO:0014041 | regulation of neuron maturation(GO:0014041) regulation of cell maturation(GO:1903429) |
| 0.0 | 0.0 | GO:0022601 | menstrual cycle phase(GO:0022601) |
| 0.0 | 0.2 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.0 | 0.1 | GO:0046498 | S-adenosylhomocysteine metabolic process(GO:0046498) |
| 0.0 | 0.0 | GO:0007571 | age-dependent response to oxidative stress(GO:0001306) age-dependent general metabolic decline(GO:0007571) |
| 0.0 | 0.1 | GO:0051593 | response to folic acid(GO:0051593) |
| 0.0 | 0.0 | GO:0060174 | limb bud formation(GO:0060174) |
| 0.0 | 0.1 | GO:0006551 | leucine metabolic process(GO:0006551) |
| 0.0 | 0.0 | GO:0045627 | positive regulation of T-helper 1 cell differentiation(GO:0045627) |
| 0.0 | 0.1 | GO:0071675 | mononuclear cell migration(GO:0071674) regulation of mononuclear cell migration(GO:0071675) |
| 0.0 | 0.1 | GO:0003156 | regulation of organ formation(GO:0003156) |
| 0.0 | 0.1 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.1 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.0 | GO:0033600 | negative regulation of mammary gland epithelial cell proliferation(GO:0033600) |
| 0.0 | 0.0 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 0.2 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.2 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.2 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.2 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.1 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.1 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.1 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.4 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.2 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.8 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.1 | GO:0031312 | extrinsic component of organelle membrane(GO:0031312) |
| 0.0 | 0.1 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.1 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.0 | GO:0044215 | host(GO:0018995) host cell part(GO:0033643) host intracellular part(GO:0033646) intracellular region of host(GO:0043656) host cell(GO:0043657) other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) |
| 0.0 | 0.2 | GO:0014704 | intercalated disc(GO:0014704) cell-cell contact zone(GO:0044291) |
| 0.0 | 0.1 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.1 | GO:0005954 | calcium- and calmodulin-dependent protein kinase complex(GO:0005954) |
| 0.0 | 0.1 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.1 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.1 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.1 | GO:0098645 | network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.0 | 0.0 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.1 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.0 | 0.4 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.1 | GO:0001527 | microfibril(GO:0001527) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.1 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.1 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.1 | GO:0001520 | outer dense fiber(GO:0001520) sperm flagellum(GO:0036126) |
| 0.0 | 0.1 | GO:0005749 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.1 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.0 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.0 | GO:0005827 | polar microtubule(GO:0005827) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 0.3 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 0.2 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.1 | 0.3 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.1 | 0.2 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.1 | 0.2 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 0.2 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.1 | 0.3 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 0.2 | GO:0031708 | bombesin receptor binding(GO:0031705) endothelin B receptor binding(GO:0031708) |
| 0.1 | 0.2 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.1 | 0.3 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.1 | 0.3 | GO:0009374 | biotin binding(GO:0009374) |
| 0.1 | 0.2 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.1 | 0.3 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 0.1 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.2 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.2 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.1 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.0 | 0.0 | GO:0070696 | transmembrane receptor protein serine/threonine kinase binding(GO:0070696) |
| 0.0 | 0.1 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.1 | GO:0008252 | nucleotidase activity(GO:0008252) |
| 0.0 | 0.7 | GO:1990782 | receptor tyrosine kinase binding(GO:0030971) protein tyrosine kinase binding(GO:1990782) |
| 0.0 | 0.2 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.1 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.1 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.1 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.3 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.3 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 0.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.1 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.1 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.1 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.2 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.0 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.1 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.0 | 0.1 | GO:0004515 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.0 | 0.2 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 0.2 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.1 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.0 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.0 | 0.0 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.0 | 0.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.0 | 0.1 | GO:0030343 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.0 | 0.1 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.1 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.1 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.0 | 0.1 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.0 | 0.1 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.1 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.1 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.3 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.2 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.1 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.1 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.0 | 0.1 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.1 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 0.2 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.1 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.1 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.1 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.2 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.1 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.1 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.4 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.1 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 0.0 | 0.4 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.1 | GO:0016615 | malate dehydrogenase activity(GO:0016615) |
| 0.0 | 0.1 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.0 | GO:0004075 | biotin carboxylase activity(GO:0004075) |
| 0.0 | 0.0 | GO:0032558 | purine deoxyribonucleotide binding(GO:0032554) adenyl deoxyribonucleotide binding(GO:0032558) |
| 0.0 | 0.0 | GO:0001098 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.0 | 0.0 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.0 | GO:0005415 | nucleoside:sodium symporter activity(GO:0005415) |
| 0.0 | 0.2 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.0 | GO:0004803 | transposase activity(GO:0004803) |
| 0.0 | 0.0 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 0.0 | GO:0034648 | histone demethylase activity (H3-K4 specific)(GO:0032453) histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.0 | 0.1 | GO:0051636 | obsolete Gram-negative bacterial cell surface binding(GO:0051636) |
| 0.0 | 0.1 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.0 | 0.1 | GO:0034483 | heparan sulfate sulfotransferase activity(GO:0034483) |
| 0.0 | 0.0 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.0 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.2 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.0 | 0.0 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.0 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.0 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.0 | 0.1 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.2 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.1 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) |
| 0.0 | 0.1 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.0 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.0 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.0 | 0.0 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.0 | 0.0 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.0 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.0 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.0 | 0.0 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.0 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.0 | 0.0 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.1 | GO:0005536 | glucose binding(GO:0005536) |
| 0.0 | 0.1 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.0 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.1 | GO:0004945 | angiotensin receptor activity(GO:0001595) angiotensin type II receptor activity(GO:0004945) |
| 0.0 | 0.0 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.0 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.0 | 0.2 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.1 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.0 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.1 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.1 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.1 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.1 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.0 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 0.0 | 0.1 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.1 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.1 | GO:0016406 | carnitine O-acyltransferase activity(GO:0016406) |
| 0.0 | 0.1 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.0 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.0 | 0.1 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.1 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.0 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.4 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 0.0 | GO:0050693 | LBD domain binding(GO:0050693) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.5 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.1 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.3 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.2 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.8 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.1 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.4 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.2 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.0 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.0 | 0.0 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.3 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.0 | REACTOME PLC BETA MEDIATED EVENTS | Genes involved in PLC beta mediated events |
| 0.0 | 0.0 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.7 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.0 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS | Genes involved in Synthesis of bile acids and bile salts |
| 0.0 | 0.2 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.1 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.4 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.0 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.2 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.2 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.1 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.2 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.3 | REACTOME INTEGRIN ALPHAIIB BETA3 SIGNALING | Genes involved in Integrin alphaIIb beta3 signaling |
| 0.0 | 0.1 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.2 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.2 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.3 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.0 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.0 | 0.1 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.0 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.0 | 0.4 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.0 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 0.1 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.1 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.6 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |