Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
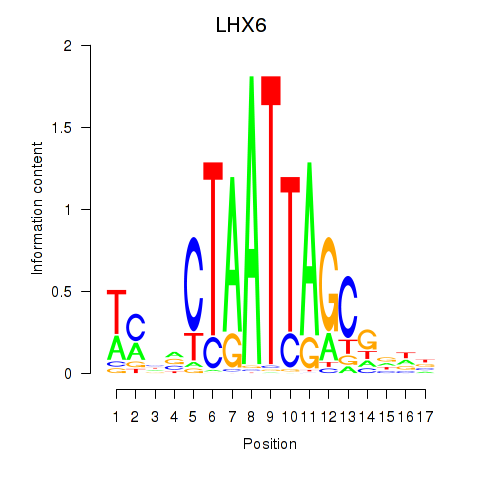
Results for LHX6
Z-value: 1.17
Transcription factors associated with LHX6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
LHX6
|
ENSG00000106852.11 | LIM homeobox 6 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr9_124980013_124980164 | LHX6 | 3887 | 0.177433 | 0.78 | 1.4e-02 | Click! |
| chr9_124976522_124976673 | LHX6 | 396 | 0.814498 | -0.62 | 7.4e-02 | Click! |
| chr9_124975632_124975793 | LHX6 | 460 | 0.773363 | -0.51 | 1.6e-01 | Click! |
| chr9_124976686_124976837 | LHX6 | 560 | 0.707628 | -0.47 | 2.0e-01 | Click! |
| chr9_124990834_124991046 | LHX6 | 184 | 0.927646 | -0.46 | 2.1e-01 | Click! |
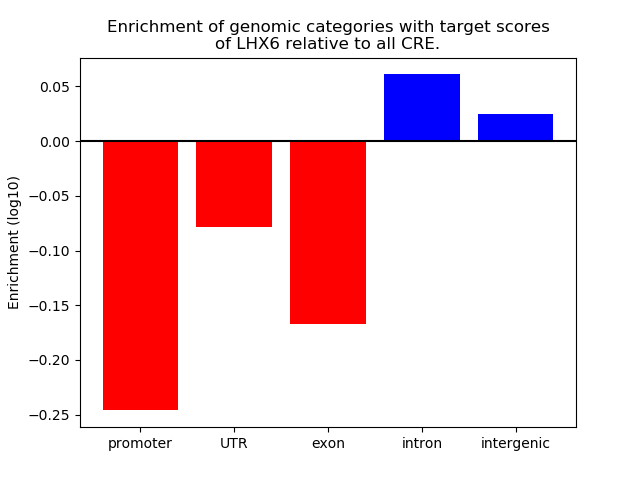
Activity of the LHX6 motif across conditions
Conditions sorted by the z-value of the LHX6 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
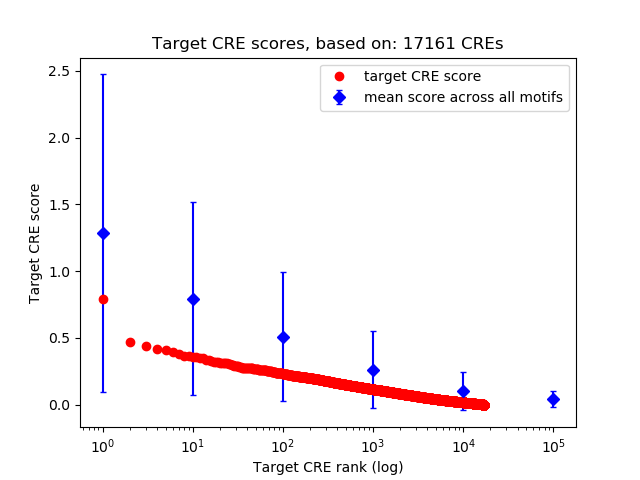
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr3_71030532_71030802 | 0.79 |
FOXP1 |
forkhead box P1 |
83410 |
0.1 |
| chr16_27257033_27257252 | 0.47 |
NSMCE1 |
non-SMC element 1 homolog (S. cerevisiae) |
12330 |
0.16 |
| chr4_187545719_187545870 | 0.44 |
FAT1 |
FAT atypical cadherin 1 |
26616 |
0.2 |
| chr17_13268235_13268519 | 0.42 |
ENSG00000266115 |
. |
135268 |
0.05 |
| chr17_26498050_26498459 | 0.41 |
PYY2 |
peptide YY, 2 (pseudogene) |
56078 |
0.09 |
| chr1_85904764_85904915 | 0.39 |
ENSG00000199459 |
. |
3872 |
0.26 |
| chr14_100240950_100241315 | 0.38 |
EML1 |
echinoderm microtubule associated protein like 1 |
1091 |
0.59 |
| chr11_14466972_14467123 | 0.37 |
ENSG00000251991 |
. |
33391 |
0.17 |
| chr11_1145070_1145742 | 0.36 |
MUC5AC |
mucin 5AC, oligomeric mucus/gel-forming |
6174 |
0.2 |
| chr17_66379311_66379462 | 0.36 |
ENSG00000207561 |
. |
41303 |
0.14 |
| chr5_158127271_158127455 | 0.36 |
CTD-2363C16.1 |
|
282651 |
0.01 |
| chr21_38799758_38799909 | 0.35 |
DYRK1A |
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A |
7156 |
0.25 |
| chr16_56227570_56227809 | 0.35 |
GNAO1 |
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O |
124 |
0.91 |
| chr2_47284202_47284907 | 0.34 |
AC073283.7 |
|
10407 |
0.19 |
| chr12_62931646_62931797 | 0.33 |
ENSG00000202034 |
. |
7738 |
0.19 |
| chr8_38790336_38790830 | 0.32 |
PLEKHA2 |
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 2 |
31721 |
0.12 |
| chr1_40887310_40887681 | 0.32 |
SMAP2 |
small ArfGAP2 |
24988 |
0.12 |
| chr10_17064484_17064884 | 0.32 |
CUBN |
cubilin (intrinsic factor-cobalamin receptor) |
38490 |
0.2 |
| chr8_121665009_121665299 | 0.32 |
RP11-713M15.1 |
|
108339 |
0.07 |
| chr10_75789745_75789896 | 0.32 |
RP11-417O11.5 |
|
24925 |
0.16 |
| chr5_154034007_154034158 | 0.31 |
ENSG00000221552 |
. |
31254 |
0.14 |
| chr15_56439129_56439280 | 0.31 |
RFX7 |
regulatory factor X, 7 |
42880 |
0.17 |
| chr9_81321983_81322311 | 0.31 |
PSAT1 |
phosphoserine aminotransferase 1 |
410088 |
0.01 |
| chr9_80524258_80524409 | 0.31 |
GNAQ |
guanine nucleotide binding protein (G protein), q polypeptide |
86418 |
0.1 |
| chr15_42720226_42720377 | 0.31 |
ZNF106 |
zinc finger protein 106 |
1 |
0.87 |
| chr7_20071186_20071471 | 0.31 |
AC005062.2 |
|
28783 |
0.23 |
| chr15_101815840_101816133 | 0.30 |
VIMP |
VCP-interacting membrane protein |
1506 |
0.33 |
| chr7_75952019_75952170 | 0.29 |
HSPB1 |
heat shock 27kDa protein 1 |
19200 |
0.14 |
| chr3_64326242_64326393 | 0.29 |
PRICKLE2 |
prickle homolog 2 (Drosophila) |
72662 |
0.11 |
| chr11_131479604_131479755 | 0.29 |
NTM |
neurotrimin |
27774 |
0.2 |
| chr11_69264697_69264848 | 0.29 |
CCND1 |
cyclin D1 |
191083 |
0.03 |
| chr10_29823814_29823965 | 0.29 |
ENSG00000207612 |
. |
10137 |
0.19 |
| chr3_45102056_45102207 | 0.28 |
ENSG00000252410 |
. |
26051 |
0.16 |
| chr6_148845405_148845556 | 0.28 |
ENSG00000223322 |
. |
104 |
0.98 |
| chr6_91072424_91072575 | 0.28 |
ENSG00000266760 |
. |
50038 |
0.15 |
| chr6_39787204_39787470 | 0.28 |
DAAM2 |
dishevelled associated activator of morphogenesis 2 |
26543 |
0.21 |
| chr9_89890769_89890954 | 0.28 |
ENSG00000212421 |
. |
15496 |
0.29 |
| chr5_68662452_68662603 | 0.28 |
TAF9 |
TAF9 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 32kDa |
121 |
0.95 |
| chr13_60664906_60665057 | 0.28 |
DIAPH3-AS2 |
DIAPH3 antisense RNA 2 |
53851 |
0.12 |
| chr3_120146387_120146538 | 0.28 |
FSTL1 |
follistatin-like 1 |
23376 |
0.21 |
| chr9_95007298_95007449 | 0.27 |
IARS |
isoleucyl-tRNA synthetase |
10 |
0.98 |
| chr20_19478003_19478154 | 0.27 |
ENSG00000216017 |
. |
92753 |
0.08 |
| chr4_86683324_86683672 | 0.27 |
ARHGAP24 |
Rho GTPase activating protein 24 |
16361 |
0.24 |
| chr1_53462333_53462484 | 0.27 |
SCP2 |
sterol carrier protein 2 |
18192 |
0.14 |
| chr1_67043331_67043482 | 0.27 |
SGIP1 |
SH3-domain GRB2-like (endophilin) interacting protein 1 |
43441 |
0.15 |
| chr3_190436866_190437153 | 0.27 |
ENSG00000223117 |
. |
77057 |
0.11 |
| chr9_80463474_80463724 | 0.27 |
GNAQ |
guanine nucleotide binding protein (G protein), q polypeptide |
25684 |
0.27 |
| chr3_71450784_71450957 | 0.27 |
FOXP1 |
forkhead box P1 |
96959 |
0.08 |
| chr17_59362891_59363042 | 0.27 |
RP11-332H18.3 |
|
97180 |
0.06 |
| chr10_17217462_17217735 | 0.27 |
TRDMT1 |
tRNA aspartic acid methyltransferase 1 |
939 |
0.58 |
| chr11_111848937_111849153 | 0.27 |
DIXDC1 |
DIX domain containing 1 |
1012 |
0.41 |
| chr8_8388322_8388473 | 0.26 |
ENSG00000263616 |
. |
51768 |
0.15 |
| chr1_193304758_193304909 | 0.26 |
B3GALT2 |
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2 |
149049 |
0.04 |
| chr20_39722951_39723102 | 0.26 |
PLCG1 |
phospholipase C, gamma 1 |
42574 |
0.14 |
| chr12_122412428_122412631 | 0.26 |
BCL7A |
B-cell CLL/lymphoma 7A |
44799 |
0.12 |
| chr7_141061897_141062048 | 0.26 |
RP5-842K16.2 |
|
69096 |
0.12 |
| chr16_85295971_85296292 | 0.26 |
ENSG00000266307 |
. |
43800 |
0.16 |
| chr15_50392799_50393114 | 0.26 |
ATP8B4 |
ATPase, class I, type 8B, member 4 |
13745 |
0.23 |
| chr10_111727206_111727357 | 0.26 |
RP11-451M19.3 |
Uncharacterized protein |
13632 |
0.18 |
| chr2_145270130_145270606 | 0.26 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
4747 |
0.27 |
| chr3_59761458_59761609 | 0.26 |
NPCDR1 |
nasopharyngeal carcinoma, down-regulated 1 |
196050 |
0.03 |
| chr11_75794475_75794626 | 0.26 |
UVRAG |
UV radiation resistance associated |
32407 |
0.17 |
| chr12_116596119_116596329 | 0.26 |
ENSG00000207967 |
. |
9765 |
0.21 |
| chr7_41910301_41910559 | 0.26 |
AC005027.3 |
|
165497 |
0.04 |
| chr3_16225459_16225610 | 0.26 |
GALNT15 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 15 |
9318 |
0.25 |
| chr1_92311778_92311983 | 0.26 |
TGFBR3 |
transforming growth factor, beta receptor III |
15270 |
0.21 |
| chr3_27208769_27208920 | 0.26 |
NEK10 |
NIMA-related kinase 10 |
49122 |
0.17 |
| chr15_50194108_50194259 | 0.25 |
ATP8B4 |
ATPase, class I, type 8B, member 4 |
6107 |
0.24 |
| chr1_244276859_244277010 | 0.25 |
ENSG00000244066 |
. |
9700 |
0.24 |
| chr14_73613505_73613656 | 0.25 |
PSEN1 |
presenilin 1 |
620 |
0.74 |
| chr17_29542627_29542778 | 0.25 |
NF1 |
neurofibromin 1 |
15148 |
0.18 |
| chr1_224564194_224564516 | 0.25 |
CNIH4 |
cornichon family AMPA receptor auxiliary protein 4 |
19760 |
0.13 |
| chr5_178703659_178703833 | 0.25 |
ADAMTS2 |
ADAM metallopeptidase with thrombospondin type 1 motif, 2 |
68685 |
0.12 |
| chr1_218891512_218891663 | 0.25 |
ENSG00000212610 |
. |
176346 |
0.03 |
| chr14_76326894_76327045 | 0.25 |
RP11-270M14.4 |
|
12928 |
0.25 |
| chr3_171039054_171039205 | 0.25 |
TNIK |
TRAF2 and NCK interacting kinase |
95629 |
0.08 |
| chr1_168329374_168329525 | 0.25 |
ENSG00000207974 |
. |
15313 |
0.23 |
| chr5_158448429_158448934 | 0.25 |
CTD-2363C16.2 |
|
35867 |
0.17 |
| chr11_16377737_16377888 | 0.25 |
SOX6 |
SRY (sex determining region Y)-box 6 |
14997 |
0.3 |
| chr16_68771111_68771428 | 0.24 |
CDH1 |
cadherin 1, type 1, E-cadherin (epithelial) |
14 |
0.97 |
| chr9_107630817_107631231 | 0.24 |
RP11-217B7.2 |
|
58810 |
0.12 |
| chr3_100601860_100602011 | 0.24 |
ABI3BP |
ABI family, member 3 (NESH) binding protein |
2487 |
0.39 |
| chr12_78797307_78797458 | 0.24 |
NAV3 |
neuron navigator 3 |
204526 |
0.03 |
| chr20_42213255_42213406 | 0.24 |
IFT52 |
intraflagellar transport 52 homolog (Chlamydomonas) |
6241 |
0.18 |
| chr4_57908197_57908348 | 0.24 |
ENSG00000251703 |
. |
39414 |
0.11 |
| chr10_44312151_44312815 | 0.24 |
ZNF32 |
zinc finger protein 32 |
168179 |
0.03 |
| chr8_130566904_130567142 | 0.24 |
ENSG00000266387 |
. |
57110 |
0.09 |
| chr19_47138317_47138577 | 0.24 |
GNG8 |
guanine nucleotide binding protein (G protein), gamma 8 |
505 |
0.62 |
| chr3_183886938_183887238 | 0.24 |
AP2M1 |
adaptor-related protein complex 2, mu 1 subunit |
5389 |
0.11 |
| chr4_16233802_16234018 | 0.24 |
TAPT1-AS1 |
TAPT1 antisense RNA 1 (head to head) |
5242 |
0.23 |
| chr4_157298853_157299004 | 0.24 |
ENSG00000221189 |
. |
81130 |
0.11 |
| chr9_79451688_79451839 | 0.24 |
PRUNE2 |
prune homolog 2 (Drosophila) |
69238 |
0.12 |
| chr6_106350521_106350672 | 0.24 |
ENSG00000200198 |
. |
1652 |
0.53 |
| chr4_119097025_119097176 | 0.24 |
ENSG00000269893 |
. |
103245 |
0.08 |
| chr17_66591627_66592081 | 0.23 |
FAM20A |
family with sequence similarity 20, member A |
5676 |
0.25 |
| chr9_4927697_4927848 | 0.23 |
ENSG00000238362 |
. |
40325 |
0.13 |
| chr9_88955484_88955635 | 0.23 |
ZCCHC6 |
zinc finger, CCHC domain containing 6 |
2479 |
0.34 |
| chr2_180911324_180911475 | 0.23 |
CWC22 |
CWC22 spliceosome-associated protein homolog (S. cerevisiae) |
39559 |
0.21 |
| chr2_36720605_36720756 | 0.23 |
CRIM1 |
cysteine rich transmembrane BMP regulator 1 (chordin-like) |
20153 |
0.19 |
| chrX_37702580_37702731 | 0.23 |
DYNLT3 |
dynein, light chain, Tctex-type 3 |
4006 |
0.26 |
| chr2_200604102_200604338 | 0.23 |
FTCDNL1 |
formiminotransferase cyclodeaminase N-terminal like |
111376 |
0.07 |
| chr8_121769766_121770271 | 0.23 |
RP11-713M15.1 |
|
3475 |
0.31 |
| chr2_46169357_46169508 | 0.23 |
PRKCE |
protein kinase C, epsilon |
58609 |
0.15 |
| chr10_81181755_81182122 | 0.23 |
ZCCHC24 |
zinc finger, CCHC domain containing 24 |
22037 |
0.18 |
| chr8_119923861_119924012 | 0.23 |
TNFRSF11B |
tumor necrosis factor receptor superfamily, member 11b |
40503 |
0.18 |
| chr4_89647157_89647308 | 0.23 |
FAM13A-AS1 |
FAM13A antisense RNA 1 |
4366 |
0.21 |
| chr12_4929077_4929347 | 0.23 |
KCNA6 |
potassium voltage-gated channel, shaker-related subfamily, member 6 |
10870 |
0.2 |
| chr16_25017302_25017551 | 0.23 |
ARHGAP17 |
Rho GTPase activating protein 17 |
9226 |
0.27 |
| chr6_44989881_44990300 | 0.23 |
SUPT3H |
suppressor of Ty 3 homolog (S. cerevisiae) |
66843 |
0.14 |
| chr5_78052251_78052402 | 0.23 |
LHFPL2 |
lipoma HMGIC fusion partner-like 2 |
107678 |
0.07 |
| chr10_49809064_49809215 | 0.23 |
ARHGAP22 |
Rho GTPase activating protein 22 |
3858 |
0.26 |
| chr6_34314260_34314411 | 0.23 |
NUDT3 |
nudix (nucleoside diphosphate linked moiety X)-type motif 3 |
46116 |
0.13 |
| chr1_178102184_178102335 | 0.22 |
RASAL2 |
RAS protein activator like 2 |
38983 |
0.22 |
| chr2_158326449_158327167 | 0.22 |
CYTIP |
cytohesin 1 interacting protein |
18533 |
0.22 |
| chr5_111085283_111085434 | 0.22 |
NREP |
neuronal regeneration related protein |
6590 |
0.22 |
| chr5_120632218_120632474 | 0.22 |
FTMT |
ferritin mitochondrial |
555304 |
0.0 |
| chr8_80226395_80226546 | 0.22 |
ENSG00000264969 |
. |
216872 |
0.02 |
| chr12_56917827_56917978 | 0.22 |
RBMS2 |
RNA binding motif, single stranded interacting protein 2 |
2119 |
0.23 |
| chr17_2120066_2120283 | 0.22 |
AC130689.5 |
|
865 |
0.42 |
| chr2_100625887_100626113 | 0.22 |
AFF3 |
AF4/FMR2 family, member 3 |
94992 |
0.08 |
| chr1_25097273_25097424 | 0.22 |
ENSG00000238482 |
. |
7122 |
0.22 |
| chr6_83664770_83664921 | 0.22 |
RP11-445L13__B.3 |
|
22525 |
0.24 |
| chr1_173822290_173822441 | 0.22 |
GAS5-AS1 |
GAS5 antisense RNA 1 |
10021 |
0.1 |
| chr3_105699405_105699556 | 0.22 |
CBLB |
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
111084 |
0.08 |
| chr15_38437640_38437791 | 0.22 |
SPRED1 |
sprouty-related, EVH1 domain containing 1 |
106812 |
0.08 |
| chr2_38054791_38055066 | 0.22 |
CDC42EP3 |
CDC42 effector protein (Rho GTPase binding) 3 |
89317 |
0.09 |
| chr6_37109268_37109585 | 0.22 |
PIM1 |
pim-1 oncogene |
28553 |
0.16 |
| chr18_12421274_12421492 | 0.22 |
SLMO1 |
slowmo homolog 1 (Drosophila) |
98 |
0.96 |
| chr6_155214377_155214528 | 0.22 |
ENSG00000238963 |
. |
11839 |
0.18 |
| chr6_113886507_113886658 | 0.22 |
ENSG00000266650 |
. |
37535 |
0.19 |
| chr16_72972238_72972389 | 0.22 |
ENSG00000221799 |
. |
46443 |
0.15 |
| chr8_108208326_108208625 | 0.22 |
ENSG00000211995 |
. |
58251 |
0.16 |
| chr17_80250217_80250392 | 0.22 |
CSNK1D |
casein kinase 1, delta |
18697 |
0.1 |
| chr1_2221391_2222142 | 0.22 |
RP4-713A8.1 |
|
36366 |
0.09 |
| chr2_98546607_98546758 | 0.22 |
ENSG00000238719 |
. |
17102 |
0.21 |
| chr11_33934050_33934291 | 0.22 |
LMO2 |
LIM domain only 2 (rhombotin-like 1) |
20334 |
0.2 |
| chr1_97604243_97604422 | 0.22 |
DPYD-AS1 |
DPYD antisense RNA 1 |
42853 |
0.21 |
| chr17_27887330_27887481 | 0.22 |
RP11-68I3.2 |
|
160 |
0.9 |
| chr9_79125250_79125401 | 0.22 |
GCNT1 |
glucosaminyl (N-acetyl) transferase 1, core 2 |
51179 |
0.15 |
| chr10_20884985_20885136 | 0.22 |
ENSG00000265372 |
. |
44161 |
0.21 |
| chr8_37640875_37641038 | 0.21 |
GPR124 |
G protein-coupled receptor 124 |
753 |
0.58 |
| chr2_44576037_44576404 | 0.21 |
PREPL |
prolyl endopeptidase-like |
10668 |
0.2 |
| chr9_125766060_125766211 | 0.21 |
RABGAP1 |
RAB GTPase activating protein 1 |
29653 |
0.12 |
| chr9_82676338_82676489 | 0.21 |
TLE4 |
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
354732 |
0.01 |
| chr2_235485735_235485886 | 0.21 |
ARL4C |
ADP-ribosylation factor-like 4C |
80113 |
0.12 |
| chr9_82227138_82227289 | 0.21 |
TLE4 |
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
38609 |
0.24 |
| chr12_66296581_66296811 | 0.21 |
RP11-366L20.2 |
Uncharacterized protein |
20749 |
0.16 |
| chr16_83934521_83934672 | 0.21 |
MLYCD |
malonyl-CoA decarboxylase |
1865 |
0.31 |
| chr13_33768544_33768695 | 0.21 |
STARD13 |
StAR-related lipid transfer (START) domain containing 13 |
8403 |
0.23 |
| chr3_45037433_45037728 | 0.21 |
EXOSC7 |
exosome component 7 |
19858 |
0.14 |
| chr9_114749197_114749348 | 0.21 |
ENSG00000222356 |
. |
9747 |
0.2 |
| chr13_28054385_28054536 | 0.21 |
ENSG00000252499 |
. |
7815 |
0.19 |
| chr2_65312285_65312436 | 0.21 |
RAB1A |
RAB1A, member RAS oncogene family |
21164 |
0.16 |
| chr16_72928505_72928656 | 0.21 |
ENSG00000251868 |
. |
72689 |
0.09 |
| chr19_54657228_54657379 | 0.21 |
CNOT3 |
CCR4-NOT transcription complex, subunit 3 |
158 |
0.88 |
| chr17_30586530_30586681 | 0.21 |
RHBDL3 |
rhomboid, veinlet-like 3 (Drosophila) |
6590 |
0.16 |
| chr1_170879044_170879195 | 0.21 |
MROH9 |
maestro heat-like repeat family member 9 |
25493 |
0.24 |
| chr13_31255786_31256279 | 0.21 |
ALOX5AP |
arachidonate 5-lipoxygenase-activating protein |
53613 |
0.14 |
| chr5_34497593_34497744 | 0.21 |
RAI14 |
retinoic acid induced 14 |
158674 |
0.04 |
| chr2_208116387_208116719 | 0.21 |
AC007879.5 |
|
2423 |
0.32 |
| chr11_71778137_71778288 | 0.21 |
NUMA1 |
nuclear mitotic apparatus protein 1 |
2921 |
0.12 |
| chr5_125339534_125339685 | 0.21 |
ENSG00000265637 |
. |
192919 |
0.03 |
| chr2_157313118_157313429 | 0.21 |
GPD2 |
glycerol-3-phosphate dehydrogenase 2 (mitochondrial) |
16811 |
0.19 |
| chr13_88323755_88324230 | 0.21 |
SLITRK5 |
SLIT and NTRK-like family, member 5 |
878 |
0.73 |
| chr17_69498538_69498689 | 0.21 |
ENSG00000222563 |
. |
191304 |
0.03 |
| chr10_115033205_115033385 | 0.21 |
ENSG00000238380 |
. |
79889 |
0.11 |
| chr9_79432035_79432186 | 0.21 |
PCA3 |
prostate cancer antigen 3 (non-protein coding) |
52758 |
0.16 |
| chr15_70620773_70621274 | 0.21 |
ENSG00000200216 |
. |
135448 |
0.05 |
| chr12_26811451_26811602 | 0.21 |
RP11-666F17.1 |
|
35224 |
0.22 |
| chr12_124483374_124483666 | 0.21 |
ZNF664 |
zinc finger protein 664 |
25678 |
0.08 |
| chr4_83281916_83282171 | 0.21 |
HNRNPD |
heterogeneous nuclear ribonucleoprotein D (AU-rich element RNA binding protein 1, 37kDa) |
1251 |
0.46 |
| chr19_19220612_19220870 | 0.21 |
TMEM161A |
transmembrane protein 161A |
22469 |
0.11 |
| chr4_15435517_15435668 | 0.21 |
RP11-665G4.1 |
|
6054 |
0.18 |
| chr14_103162755_103162994 | 0.21 |
ENSG00000202459 |
. |
1149 |
0.54 |
| chr16_50840051_50840280 | 0.21 |
RP11-327F22.4 |
|
2916 |
0.22 |
| chr4_71533612_71533824 | 0.21 |
IGJ |
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides |
1026 |
0.42 |
| chr7_94035985_94036136 | 0.21 |
COL1A2 |
collagen, type I, alpha 2 |
12187 |
0.27 |
| chr9_91752254_91752405 | 0.21 |
RP11-82L18.2 |
|
27480 |
0.2 |
| chr1_213499715_213500068 | 0.20 |
RPS6KC1 |
ribosomal protein S6 kinase, 52kDa, polypeptide 1 |
275287 |
0.01 |
| chr3_58030666_58030922 | 0.20 |
FLNB |
filamin B, beta |
33262 |
0.2 |
| chr1_109261913_109262064 | 0.20 |
FNDC7 |
fibronectin type III domain containing 7 |
3045 |
0.21 |
| chr17_8576822_8576973 | 0.20 |
MYH10 |
myosin, heavy chain 10, non-muscle |
42818 |
0.14 |
| chr12_19805873_19806024 | 0.20 |
ENSG00000200885 |
. |
71421 |
0.13 |
| chr15_50156516_50156667 | 0.20 |
ATP8B4 |
ATPase, class I, type 8B, member 4 |
12301 |
0.22 |
| chr13_98867933_98868084 | 0.20 |
ENSG00000263399 |
. |
7230 |
0.16 |
| chr5_9363080_9363231 | 0.20 |
CTD-2201E9.4 |
|
232 |
0.95 |
| chr9_77791417_77791568 | 0.20 |
ENSG00000200041 |
. |
5045 |
0.24 |
| chr8_27497354_27497505 | 0.20 |
SCARA3 |
scavenger receptor class A, member 3 |
5731 |
0.18 |
| chr17_40511255_40511412 | 0.20 |
STAT3 |
signal transducer and activator of transcription 3 (acute-phase response factor) |
29099 |
0.1 |
| chr10_97579295_97579446 | 0.20 |
RP11-429G19.3 |
|
13895 |
0.21 |
| chr3_193856714_193857102 | 0.20 |
HES1 |
hes family bHLH transcription factor 1 |
2974 |
0.25 |
| chr10_17151320_17151471 | 0.20 |
CUBN |
cubilin (intrinsic factor-cobalamin receptor) |
18441 |
0.23 |
| chr10_128259000_128259151 | 0.20 |
ENSG00000221717 |
. |
13442 |
0.26 |
| chr6_126142988_126143327 | 0.20 |
NCOA7-AS1 |
NCOA7 antisense RNA 1 |
3153 |
0.24 |
| chr5_77796656_77796807 | 0.20 |
LHFPL2 |
lipoma HMGIC fusion partner-like 2 |
48243 |
0.18 |
| chr7_2197941_2198092 | 0.20 |
MAD1L1 |
MAD1 mitotic arrest deficient-like 1 (yeast) |
48780 |
0.15 |
| chr11_8091043_8091194 | 0.20 |
RP11-236J17.5 |
|
9533 |
0.15 |
| chr1_198573317_198573468 | 0.20 |
PTPRC |
protein tyrosine phosphatase, receptor type, C |
34409 |
0.19 |
| chr10_80517839_80518027 | 0.20 |
ENSG00000223243 |
. |
7574 |
0.33 |
| chr18_56031230_56031381 | 0.20 |
RP11-845C23.2 |
|
7493 |
0.2 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.1 | 0.1 | GO:2000095 | regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.1 | 0.2 | GO:0090116 | DNA methylation on cytosine(GO:0032776) C-5 methylation of cytosine(GO:0090116) |
| 0.1 | 0.3 | GO:0071681 | response to indole-3-methanol(GO:0071680) cellular response to indole-3-methanol(GO:0071681) |
| 0.1 | 0.2 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.1 | 0.2 | GO:1900116 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.6 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.2 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 0.2 | GO:0002327 | immature B cell differentiation(GO:0002327) pre-B cell differentiation(GO:0002329) |
| 0.0 | 0.2 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.3 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 | 0.1 | GO:0035385 | Roundabout signaling pathway(GO:0035385) |
| 0.0 | 0.1 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.2 | GO:0002860 | natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002420) natural killer cell mediated immune response to tumor cell(GO:0002423) regulation of natural killer cell mediated immune response to tumor cell(GO:0002855) positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002858) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) |
| 0.0 | 0.2 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.2 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.0 | 0.3 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.1 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.1 | GO:0031061 | negative regulation of histone methylation(GO:0031061) |
| 0.0 | 0.1 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.3 | GO:0060760 | positive regulation of response to cytokine stimulus(GO:0060760) |
| 0.0 | 0.2 | GO:0050932 | regulation of melanocyte differentiation(GO:0045634) regulation of pigment cell differentiation(GO:0050932) |
| 0.0 | 0.1 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.1 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.1 | GO:0007132 | meiotic metaphase I(GO:0007132) |
| 0.0 | 0.2 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.1 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.1 | GO:0010837 | regulation of keratinocyte proliferation(GO:0010837) |
| 0.0 | 0.1 | GO:0007028 | cytoplasm organization(GO:0007028) |
| 0.0 | 0.1 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 | 0.1 | GO:0051124 | synaptic growth at neuromuscular junction(GO:0051124) |
| 0.0 | 0.1 | GO:0060710 | chorio-allantoic fusion(GO:0060710) |
| 0.0 | 0.2 | GO:0001547 | antral ovarian follicle growth(GO:0001547) |
| 0.0 | 0.2 | GO:1903670 | regulation of cell migration involved in sprouting angiogenesis(GO:0090049) regulation of sprouting angiogenesis(GO:1903670) |
| 0.0 | 0.1 | GO:0021535 | cell migration in hindbrain(GO:0021535) |
| 0.0 | 0.1 | GO:0071379 | cellular response to prostaglandin stimulus(GO:0071379) |
| 0.0 | 0.0 | GO:0009120 | deoxyribonucleoside metabolic process(GO:0009120) |
| 0.0 | 0.1 | GO:0018243 | protein O-linked glycosylation via serine(GO:0018242) protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.1 | GO:0006930 | substrate-dependent cell migration, cell extension(GO:0006930) |
| 0.0 | 0.1 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.1 | GO:0010700 | negative regulation of norepinephrine secretion(GO:0010700) |
| 0.0 | 0.1 | GO:0071221 | response to bacterial lipoprotein(GO:0032493) response to bacterial lipopeptide(GO:0070339) cellular response to bacterial lipoprotein(GO:0071220) cellular response to bacterial lipopeptide(GO:0071221) |
| 0.0 | 0.4 | GO:0014072 | response to isoquinoline alkaloid(GO:0014072) response to morphine(GO:0043278) |
| 0.0 | 0.0 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.0 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) |
| 0.0 | 0.0 | GO:0034392 | negative regulation of smooth muscle cell apoptotic process(GO:0034392) |
| 0.0 | 0.3 | GO:0045104 | intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 | 0.1 | GO:0019673 | GDP-mannose metabolic process(GO:0019673) |
| 0.0 | 0.0 | GO:0045631 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.0 | 0.0 | GO:0060083 | smooth muscle contraction involved in micturition(GO:0060083) |
| 0.0 | 0.1 | GO:0033137 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) |
| 0.0 | 0.0 | GO:0010766 | negative regulation of sodium ion transport(GO:0010766) |
| 0.0 | 0.0 | GO:0060197 | cloaca development(GO:0035844) cloacal septation(GO:0060197) |
| 0.0 | 0.1 | GO:0043206 | extracellular fibril organization(GO:0043206) fibril organization(GO:0097435) |
| 0.0 | 0.2 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.1 | GO:0071436 | sodium ion export(GO:0071436) |
| 0.0 | 0.1 | GO:0002040 | sprouting angiogenesis(GO:0002040) |
| 0.0 | 0.4 | GO:0001755 | neural crest cell migration(GO:0001755) |
| 0.0 | 0.1 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.0 | 0.0 | GO:0060789 | hair follicle placode formation(GO:0060789) |
| 0.0 | 0.1 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.0 | 0.1 | GO:0045630 | positive regulation of T-helper 2 cell differentiation(GO:0045630) |
| 0.0 | 0.1 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 0.5 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.1 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.1 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.1 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.3 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.1 | GO:0007210 | serotonin receptor signaling pathway(GO:0007210) |
| 0.0 | 0.1 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 | 0.1 | GO:0042983 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.0 | 0.0 | GO:0070254 | mucus secretion(GO:0070254) |
| 0.0 | 0.0 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.1 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.0 | 0.1 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.1 | GO:0060259 | regulation of feeding behavior(GO:0060259) |
| 0.0 | 0.0 | GO:0071364 | cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.0 | 0.1 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.0 | 0.1 | GO:0034695 | response to prostaglandin E(GO:0034695) |
| 0.0 | 0.2 | GO:0007263 | nitric oxide mediated signal transduction(GO:0007263) |
| 0.0 | 0.1 | GO:0008300 | isoprenoid catabolic process(GO:0008300) |
| 0.0 | 0.0 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.0 | GO:0032261 | purine nucleotide salvage(GO:0032261) |
| 0.0 | 0.1 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.0 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.0 | 0.0 | GO:0034694 | response to prostaglandin(GO:0034694) |
| 0.0 | 0.0 | GO:0016999 | antibiotic metabolic process(GO:0016999) antibiotic biosynthetic process(GO:0017000) |
| 0.0 | 0.1 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.0 | GO:0035511 | oxidative DNA demethylation(GO:0035511) |
| 0.0 | 0.0 | GO:0002072 | optic cup morphogenesis involved in camera-type eye development(GO:0002072) |
| 0.0 | 0.0 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.0 | 0.1 | GO:0006264 | mitochondrial DNA replication(GO:0006264) |
| 0.0 | 0.1 | GO:0002634 | regulation of germinal center formation(GO:0002634) |
| 0.0 | 0.1 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.0 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.0 | 0.1 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.1 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.1 | GO:0036465 | synaptic vesicle recycling(GO:0036465) synaptic vesicle endocytosis(GO:0048488) clathrin-mediated endocytosis(GO:0072583) |
| 0.0 | 0.1 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) negative regulation of neural precursor cell proliferation(GO:2000178) |
| 0.0 | 0.1 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 0.1 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.0 | 0.1 | GO:0033160 | positive regulation of protein import into nucleus, translocation(GO:0033160) |
| 0.0 | 0.0 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.0 | 0.1 | GO:0021801 | cerebral cortex radial glia guided migration(GO:0021801) telencephalon glial cell migration(GO:0022030) |
| 0.0 | 0.0 | GO:0002674 | negative regulation of acute inflammatory response(GO:0002674) |
| 0.0 | 0.0 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 0.0 | GO:0060638 | mesenchymal-epithelial cell signaling(GO:0060638) |
| 0.0 | 0.0 | GO:0009138 | pyrimidine nucleoside diphosphate metabolic process(GO:0009138) |
| 0.0 | 0.1 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.0 | GO:0072677 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.0 | 0.0 | GO:0010834 | obsolete telomere maintenance via telomere shortening(GO:0010834) |
| 0.0 | 0.0 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.0 | 0.0 | GO:0046469 | platelet activating factor biosynthetic process(GO:0006663) platelet activating factor metabolic process(GO:0046469) |
| 0.0 | 0.0 | GO:0010891 | negative regulation of sequestering of triglyceride(GO:0010891) |
| 0.0 | 0.0 | GO:0045989 | positive regulation of striated muscle contraction(GO:0045989) |
| 0.0 | 0.0 | GO:0033197 | response to vitamin E(GO:0033197) |
| 0.0 | 0.1 | GO:0060644 | mammary gland epithelial cell differentiation(GO:0060644) |
| 0.0 | 0.0 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.0 | 0.1 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) phosphatidylethanolamine metabolic process(GO:0046337) |
| 0.0 | 0.3 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.0 | GO:1903429 | regulation of neuron maturation(GO:0014041) regulation of cell maturation(GO:1903429) |
| 0.0 | 0.0 | GO:0051877 | pigment granule aggregation in cell center(GO:0051877) |
| 0.0 | 0.0 | GO:0071731 | response to nitric oxide(GO:0071731) |
| 0.0 | 0.0 | GO:0032641 | lymphotoxin A production(GO:0032641) lymphotoxin A biosynthetic process(GO:0042109) |
| 0.0 | 0.0 | GO:0002023 | reduction of food intake in response to dietary excess(GO:0002023) |
| 0.0 | 0.1 | GO:0015701 | bicarbonate transport(GO:0015701) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 0.3 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.3 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.0 | 0.1 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.3 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.0 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.1 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.2 | GO:0098642 | network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.1 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.2 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.2 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.3 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.1 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.1 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.1 | GO:0005954 | calcium- and calmodulin-dependent protein kinase complex(GO:0005954) |
| 0.0 | 0.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.2 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.2 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.1 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.1 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.3 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.6 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.1 | GO:0005606 | laminin-1 complex(GO:0005606) |
| 0.0 | 0.1 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.1 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.1 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.0 | GO:0036454 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.0 | 0.0 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.0 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.1 | GO:0098878 | ionotropic glutamate receptor complex(GO:0008328) neurotransmitter receptor complex(GO:0098878) |
| 0.0 | 0.1 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.1 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.0 | GO:0033268 | node of Ranvier(GO:0033268) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.1 | 0.2 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 0.2 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.2 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.3 | GO:0042153 | obsolete RPTP-like protein binding(GO:0042153) |
| 0.0 | 0.1 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.2 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.0 | GO:0043175 | RNA polymerase core enzyme binding(GO:0043175) |
| 0.0 | 0.1 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.2 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.0 | 0.4 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.1 | GO:0001098 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.0 | 0.1 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.3 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.2 | GO:0070696 | transmembrane receptor protein serine/threonine kinase binding(GO:0070696) |
| 0.0 | 0.1 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.1 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.0 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 0.1 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.1 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.2 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.1 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.1 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.1 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.3 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 0.1 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.1 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.1 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.2 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.1 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.0 | 0.1 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.0 | 0.1 | GO:0042910 | xenobiotic-transporting ATPase activity(GO:0008559) xenobiotic transporter activity(GO:0042910) |
| 0.0 | 0.2 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.3 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.3 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.1 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.1 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.2 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.0 | 0.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.2 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 0.1 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.1 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.0 | 0.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.1 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.0 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.0 | 0.0 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.1 | GO:0004075 | biotin carboxylase activity(GO:0004075) |
| 0.0 | 0.3 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.0 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.0 | 0.1 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.0 | GO:0043734 | DNA demethylase activity(GO:0035514) DNA-N1-methyladenine dioxygenase activity(GO:0043734) |
| 0.0 | 0.1 | GO:0042285 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.0 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.1 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.0 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.0 | GO:0033765 | steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) |
| 0.0 | 0.1 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.1 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.0 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.1 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.1 | GO:0008035 | high-density lipoprotein particle binding(GO:0008035) |
| 0.0 | 0.1 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.0 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.0 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.0 | 0.0 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.1 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.0 | 0.4 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.2 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.0 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.2 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.1 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.1 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.0 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.0 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.0 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.0 | 0.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.1 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.0 | 0.1 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.8 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.0 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.1 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.0 | GO:0050543 | icosatetraenoic acid binding(GO:0050543) |
| 0.0 | 0.0 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.1 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.0 | GO:0004556 | alpha-amylase activity(GO:0004556) amylase activity(GO:0016160) |
| 0.0 | 0.0 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.0 | 0.0 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.0 | 0.0 | GO:0019215 | intermediate filament binding(GO:0019215) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.0 | 0.1 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 0.9 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.4 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 0.0 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 0.3 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.1 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.0 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.2 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.2 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 0.0 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 0.3 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.2 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.2 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.5 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.4 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.2 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.2 | REACTOME NRIF SIGNALS CELL DEATH FROM THE NUCLEUS | Genes involved in NRIF signals cell death from the nucleus |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.3 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.0 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.0 | 0.0 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.0 | REACTOME SIGNALING BY NOTCH2 | Genes involved in Signaling by NOTCH2 |
| 0.0 | 0.7 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.1 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.0 | 0.2 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.0 | REACTOME PI 3K CASCADE | Genes involved in PI-3K cascade |
| 0.0 | 0.3 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 0.3 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.1 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.2 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 0.1 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 0.1 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.0 | REACTOME IL 3 5 AND GM CSF SIGNALING | Genes involved in Interleukin-3, 5 and GM-CSF signaling |