Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
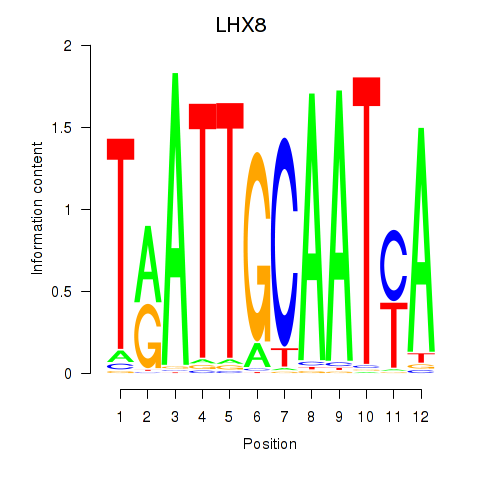
Results for LHX8
Z-value: 1.48
Transcription factors associated with LHX8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
LHX8
|
ENSG00000162624.10 | LIM homeobox 8 |
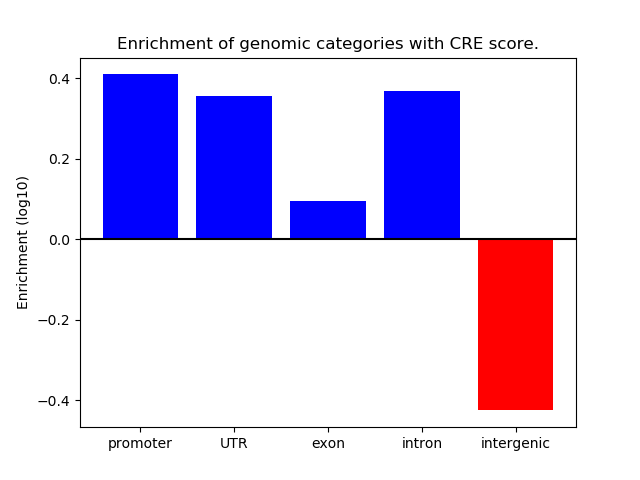
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr1_75600824_75600996 | LHX8 | 343 | 0.839693 | -0.85 | 3.6e-03 | Click! |
| chr1_75600579_75600730 | LHX8 | 87 | 0.957213 | -0.78 | 1.3e-02 | Click! |
| chr1_75601220_75601458 | LHX8 | 772 | 0.585136 | -0.75 | 2.0e-02 | Click! |
| chr1_75601640_75601791 | LHX8 | 1148 | 0.437031 | -0.42 | 2.6e-01 | Click! |
| chr1_75430278_75430429 | LHX8 | 163766 | 0.033665 | -0.28 | 4.7e-01 | Click! |
Activity of the LHX8 motif across conditions
Conditions sorted by the z-value of the LHX8 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
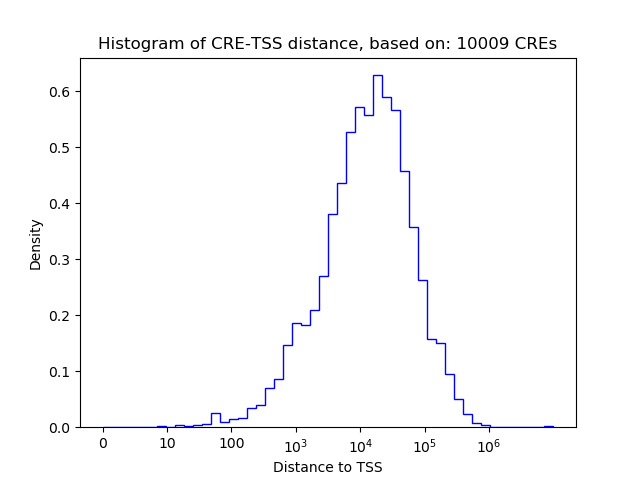
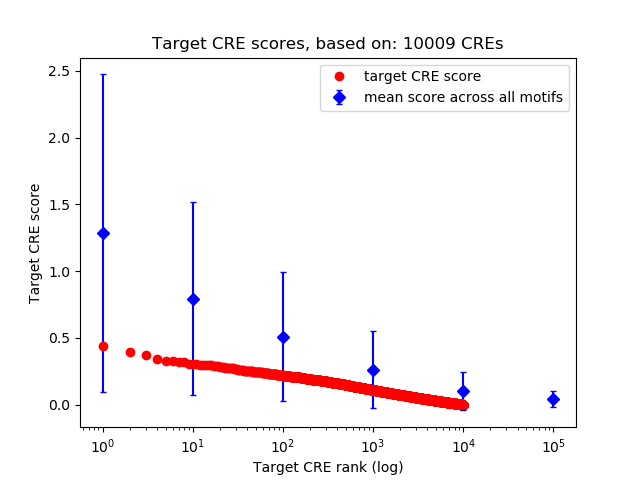
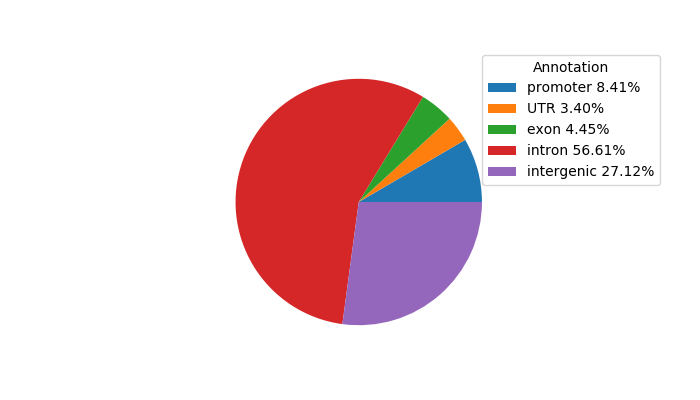
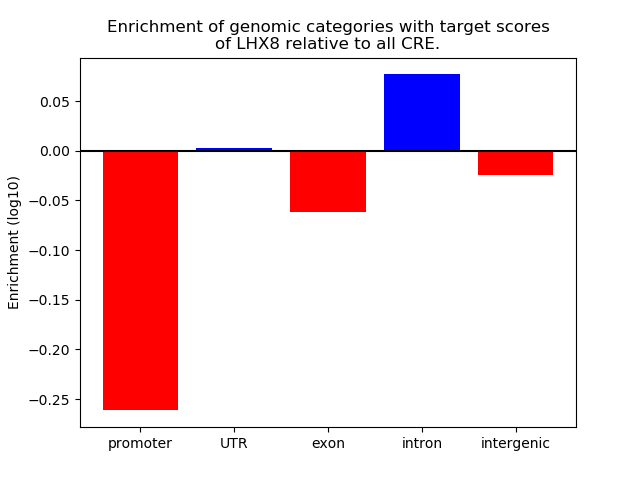
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr3_171547228_171547513 | 0.44 |
TMEM212 |
transmembrane protein 212 |
13769 |
0.18 |
| chr6_35588984_35589141 | 0.39 |
ENSG00000212579 |
. |
30533 |
0.14 |
| chr7_132524694_132524845 | 0.37 |
AC009518.8 |
|
9316 |
0.27 |
| chr5_118718463_118718642 | 0.34 |
TNFAIP8 |
tumor necrosis factor, alpha-induced protein 8 |
26822 |
0.17 |
| chr2_224800348_224800499 | 0.33 |
WDFY1 |
WD repeat and FYVE domain containing 1 |
9651 |
0.2 |
| chr12_110816955_110817106 | 0.33 |
ANAPC7 |
anaphase promoting complex subunit 7 |
2633 |
0.23 |
| chr10_95734237_95734388 | 0.32 |
PLCE1 |
phospholipase C, epsilon 1 |
19434 |
0.21 |
| chr4_100863849_100864145 | 0.32 |
DNAJB14 |
DnaJ (Hsp40) homolog, subfamily B, member 14 |
3886 |
0.2 |
| chr5_74798216_74798367 | 0.31 |
COL4A3BP |
collagen, type IV, alpha 3 (Goodpasture antigen) binding protein |
9163 |
0.19 |
| chr20_43646840_43647043 | 0.31 |
ENSG00000252021 |
. |
13952 |
0.16 |
| chr1_202125548_202126022 | 0.30 |
PTPN7 |
protein tyrosine phosphatase, non-receptor type 7 |
3334 |
0.18 |
| chr3_137935329_137935480 | 0.30 |
ARMC8 |
armadillo repeat containing 8 |
7074 |
0.23 |
| chr22_17597863_17598182 | 0.30 |
CECR6 |
cat eye syndrome chromosome region, candidate 6 |
4121 |
0.17 |
| chr3_5029460_5029611 | 0.30 |
BHLHE40-AS1 |
BHLHE40 antisense RNA 1 |
7889 |
0.21 |
| chr2_42331828_42332250 | 0.30 |
PKDCC |
protein kinase domain containing, cytoplasmic |
56879 |
0.12 |
| chr12_46018307_46018458 | 0.30 |
ENSG00000239178 |
. |
69940 |
0.13 |
| chrX_11784442_11784722 | 0.29 |
MSL3 |
male-specific lethal 3 homolog (Drosophila) |
6835 |
0.32 |
| chr9_88953538_88953925 | 0.29 |
ZCCHC6 |
zinc finger, CCHC domain containing 6 |
4307 |
0.27 |
| chr6_130509586_130509737 | 0.29 |
SAMD3 |
sterile alpha motif domain containing 3 |
26774 |
0.22 |
| chr19_17505749_17506030 | 0.28 |
BST2 |
bone marrow stromal cell antigen 2 |
10568 |
0.08 |
| chr4_101961643_101961893 | 0.28 |
EMCN-IT1 |
EMCN intronic transcript 1 (non-protein coding) |
226331 |
0.02 |
| chr5_132393541_132393692 | 0.28 |
HSPA4 |
heat shock 70kDa protein 4 |
5962 |
0.2 |
| chr1_198158796_198158947 | 0.28 |
NEK7 |
NIMA-related kinase 7 |
17133 |
0.3 |
| chr4_106087217_106087368 | 0.28 |
TET2-AS1 |
TET2 antisense RNA 1 |
11928 |
0.21 |
| chr9_101882526_101882813 | 0.28 |
TGFBR1 |
transforming growth factor, beta receptor 1 |
7388 |
0.21 |
| chr5_100043082_100043233 | 0.27 |
ENSG00000207269 |
. |
25605 |
0.25 |
| chr2_114664564_114664715 | 0.27 |
ACTR3 |
ARP3 actin-related protein 3 homolog (yeast) |
16464 |
0.22 |
| chr8_104117881_104118032 | 0.27 |
ENSG00000265667 |
. |
13007 |
0.13 |
| chr1_159045897_159046203 | 0.27 |
AIM2 |
absent in melanoma 2 |
641 |
0.71 |
| chr3_2951081_2951232 | 0.27 |
CNTN4 |
contactin 4 |
8761 |
0.3 |
| chr12_69041617_69041845 | 0.26 |
ENSG00000206650 |
. |
20576 |
0.18 |
| chr14_59812487_59812638 | 0.26 |
ENSG00000252869 |
. |
54179 |
0.14 |
| chr18_9793644_9793795 | 0.26 |
ENSG00000242651 |
. |
36905 |
0.15 |
| chr11_86661115_86661399 | 0.26 |
FZD4 |
frizzled family receptor 4 |
5176 |
0.26 |
| chr21_23086135_23086286 | 0.26 |
ENSG00000221210 |
. |
316064 |
0.01 |
| chr11_109945056_109945207 | 0.26 |
ZC3H12C |
zinc finger CCCH-type containing 12C |
18956 |
0.29 |
| chr1_98368422_98368711 | 0.26 |
DPYD |
dihydropyrimidine dehydrogenase |
17987 |
0.28 |
| chr2_144031553_144031826 | 0.25 |
RP11-190J23.1 |
|
101948 |
0.08 |
| chr6_11365715_11365866 | 0.25 |
NEDD9 |
neural precursor cell expressed, developmentally down-regulated 9 |
16742 |
0.26 |
| chr22_37677600_37677751 | 0.25 |
CYTH4 |
cytohesin 4 |
393 |
0.83 |
| chr11_77050584_77050735 | 0.25 |
PAK1 |
p21 protein (Cdc42/Rac)-activated kinase 1 |
9673 |
0.24 |
| chr13_74288455_74288993 | 0.25 |
KLF12 |
Kruppel-like factor 12 |
280462 |
0.01 |
| chr2_118854222_118854373 | 0.25 |
INSIG2 |
insulin induced gene 2 |
8247 |
0.24 |
| chr10_22906078_22906308 | 0.25 |
PIP4K2A |
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha |
25551 |
0.25 |
| chr18_2687837_2688110 | 0.25 |
SMCHD1 |
structural maintenance of chromosomes flexible hinge domain containing 1 |
32087 |
0.13 |
| chr4_15644909_15645060 | 0.25 |
FBXL5 |
F-box and leucine-rich repeat protein 5 |
1288 |
0.49 |
| chr1_114393658_114393809 | 0.25 |
AP4B1-AS1 |
AP4B1 antisense RNA 1 |
5524 |
0.14 |
| chr6_16687204_16687395 | 0.25 |
RP1-151F17.1 |
|
74070 |
0.11 |
| chr3_107802086_107802279 | 0.25 |
CD47 |
CD47 molecule |
7679 |
0.31 |
| chr5_114877836_114878017 | 0.24 |
FEM1C |
fem-1 homolog c (C. elegans) |
2665 |
0.3 |
| chr10_90709918_90710069 | 0.24 |
ACTA2 |
actin, alpha 2, smooth muscle, aorta |
2537 |
0.23 |
| chr9_86050414_86050565 | 0.24 |
ENSG00000238608 |
. |
16699 |
0.21 |
| chr2_46665689_46665843 | 0.24 |
ENSG00000241791 |
. |
9884 |
0.2 |
| chr3_43515210_43515361 | 0.24 |
ENSG00000251811 |
. |
1316 |
0.55 |
| chr10_112720074_112720225 | 0.24 |
SHOC2 |
soc-2 suppressor of clear homolog (C. elegans) |
4460 |
0.19 |
| chr3_81801729_81801880 | 0.24 |
GBE1 |
glucan (1,4-alpha-), branching enzyme 1 |
9024 |
0.33 |
| chr6_22366815_22366966 | 0.24 |
PRL |
prolactin |
69160 |
0.13 |
| chr3_39250957_39251108 | 0.24 |
XIRP1 |
xin actin-binding repeat containing 1 |
16945 |
0.19 |
| chr10_77492995_77493165 | 0.24 |
RP11-367B6.2 |
|
9418 |
0.24 |
| chr3_69245956_69246107 | 0.24 |
FRMD4B |
FERM domain containing 4B |
2390 |
0.37 |
| chr1_12185179_12185361 | 0.24 |
TNFRSF8 |
tumor necrosis factor receptor superfamily, member 8 |
686 |
0.66 |
| chr14_78219731_78219882 | 0.24 |
C14orf178 |
chromosome 14 open reading frame 178 |
7367 |
0.14 |
| chr2_242285300_242285489 | 0.24 |
SEPT2 |
septin 2 |
4108 |
0.17 |
| chr16_58913891_58914042 | 0.24 |
ENSG00000244003 |
. |
109538 |
0.06 |
| chr3_142557847_142557998 | 0.24 |
ENSG00000251787 |
. |
37873 |
0.14 |
| chr9_71399265_71399416 | 0.24 |
FAM122A |
family with sequence similarity 122A |
4376 |
0.27 |
| chrX_38666948_38667099 | 0.24 |
MID1IP1-AS1 |
MID1IP1 antisense RNA 1 |
3887 |
0.27 |
| chr10_121237439_121237906 | 0.24 |
RGS10 |
regulator of G-protein signaling 10 |
49317 |
0.14 |
| chr15_59146843_59146994 | 0.24 |
RP11-30K9.5 |
|
10344 |
0.14 |
| chr14_61836268_61836532 | 0.23 |
PRKCH |
protein kinase C, eta |
8608 |
0.24 |
| chr11_60610377_60610615 | 0.23 |
RP11-804A23.2 |
|
58 |
0.92 |
| chr13_41132128_41132279 | 0.23 |
AL133318.1 |
Uncharacterized protein |
20880 |
0.24 |
| chr6_137081513_137081694 | 0.23 |
MAP3K5 |
mitogen-activated protein kinase kinase kinase 5 |
32053 |
0.18 |
| chr12_65090896_65091133 | 0.23 |
AC025262.1 |
Mesenchymal stem cell protein DSC96; Uncharacterized protein |
685 |
0.64 |
| chr9_21024355_21024653 | 0.23 |
PTPLAD2 |
protein tyrosine phosphatase-like A domain containing 2 |
7104 |
0.22 |
| chr11_94954464_94954615 | 0.23 |
RP11-712B9.2 |
|
8811 |
0.23 |
| chr4_109031503_109031852 | 0.23 |
LEF1 |
lymphoid enhancer-binding factor 1 |
55780 |
0.13 |
| chr2_162229850_162230001 | 0.23 |
ENSG00000221590 |
. |
23054 |
0.15 |
| chr20_33732912_33733126 | 0.23 |
EDEM2 |
ER degradation enhancer, mannosidase alpha-like 2 |
60 |
0.96 |
| chr5_60639238_60639389 | 0.23 |
ZSWIM6 |
zinc finger, SWIM-type containing 6 |
11213 |
0.29 |
| chrX_17813136_17813287 | 0.23 |
RAI2 |
retinoic acid induced 2 |
42639 |
0.18 |
| chr3_113950179_113950372 | 0.23 |
ZNF80 |
zinc finger protein 80 |
6150 |
0.18 |
| chr6_12184707_12184858 | 0.23 |
HIVEP1 |
human immunodeficiency virus type I enhancer binding protein 1 |
58803 |
0.15 |
| chr20_43657355_43657581 | 0.23 |
ENSG00000252021 |
. |
24479 |
0.14 |
| chr11_3760141_3760292 | 0.23 |
NUP98 |
nucleoporin 98kDa |
21608 |
0.1 |
| chr4_15256524_15256675 | 0.23 |
RP11-665G4.1 |
|
27015 |
0.23 |
| chr3_4949233_4949520 | 0.22 |
BHLHE40-AS1 |
BHLHE40 antisense RNA 1 |
1190 |
0.49 |
| chr17_29143963_29144169 | 0.22 |
CRLF3 |
cytokine receptor-like factor 3 |
7623 |
0.13 |
| chr9_82247341_82247492 | 0.22 |
TLE4 |
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
20092 |
0.3 |
| chr1_36015652_36015803 | 0.22 |
KIAA0319L |
KIAA0319-like |
4861 |
0.17 |
| chr21_38809303_38809462 | 0.22 |
DYRK1A |
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A |
16705 |
0.23 |
| chr4_109065027_109065178 | 0.22 |
LEF1 |
lymphoid enhancer-binding factor 1 |
22355 |
0.2 |
| chr8_108830160_108830311 | 0.22 |
ENSG00000200806 |
. |
66487 |
0.14 |
| chr20_3972793_3973048 | 0.22 |
RNF24 |
ring finger protein 24 |
17886 |
0.19 |
| chr6_129766976_129767127 | 0.22 |
ENSG00000252554 |
. |
8926 |
0.27 |
| chr14_39287950_39288101 | 0.22 |
ENSG00000199285 |
. |
168460 |
0.04 |
| chr12_9805985_9806454 | 0.22 |
RP11-705C15.2 |
|
60 |
0.96 |
| chr2_225443234_225443385 | 0.22 |
CUL3 |
cullin 3 |
6756 |
0.33 |
| chr9_129163662_129163813 | 0.22 |
ENSG00000253079 |
. |
6952 |
0.21 |
| chr12_102453949_102454100 | 0.22 |
CCDC53 |
coiled-coil domain containing 53 |
1833 |
0.31 |
| chr21_15911267_15911507 | 0.22 |
SAMSN1 |
SAM domain, SH3 domain and nuclear localization signals 1 |
7275 |
0.26 |
| chr5_142500061_142500271 | 0.22 |
ARHGAP26 |
Rho GTPase activating protein 26 |
64518 |
0.13 |
| chr17_46485231_46485382 | 0.22 |
SKAP1 |
src kinase associated phosphoprotein 1 |
22246 |
0.12 |
| chr3_131089789_131089940 | 0.22 |
NUDT16P1 |
nudix (nucleoside diphosphate linked moiety X)-type motif 16 pseudogene 1 |
9090 |
0.17 |
| chr10_24910488_24910681 | 0.22 |
ARHGAP21 |
Rho GTPase activating protein 21 |
1179 |
0.56 |
| chrX_138672209_138672360 | 0.22 |
ENSG00000216114 |
. |
9178 |
0.21 |
| chr4_90232073_90232224 | 0.22 |
GPRIN3 |
GPRIN family member 3 |
2987 |
0.4 |
| chrX_48586382_48586533 | 0.21 |
ENSG00000206723 |
. |
3491 |
0.14 |
| chr8_82065263_82065697 | 0.21 |
PAG1 |
phosphoprotein associated with glycosphingolipid microdomains 1 |
41177 |
0.19 |
| chr10_104868083_104868234 | 0.21 |
NT5C2 |
5'-nucleotidase, cytosolic II |
1751 |
0.44 |
| chr4_101961293_101961465 | 0.21 |
EMCN-IT1 |
EMCN intronic transcript 1 (non-protein coding) |
225942 |
0.02 |
| chr4_153738011_153738162 | 0.21 |
ARFIP1 |
ADP-ribosylation factor interacting protein 1 |
36949 |
0.16 |
| chrX_47556921_47557072 | 0.21 |
CXXC1P1 |
CXXC finger protein 1 pseudogene 1 |
24734 |
0.13 |
| chr11_68504611_68504762 | 0.21 |
MTL5 |
metallothionein-like 5, testis-specific (tesmin) |
14242 |
0.2 |
| chr11_17976069_17976220 | 0.21 |
SERGEF |
secretion regulating guanine nucleotide exchange factor |
34160 |
0.17 |
| chr15_38902764_38903045 | 0.21 |
RASGRP1 |
RAS guanyl releasing protein 1 (calcium and DAG-regulated) |
45128 |
0.15 |
| chr12_4058973_4059124 | 0.21 |
RP11-664D1.1 |
|
44662 |
0.18 |
| chr10_28338660_28338811 | 0.21 |
ARMC4 |
armadillo repeat containing 4 |
50758 |
0.17 |
| chr5_142182149_142182300 | 0.21 |
ARHGAP26 |
Rho GTPase activating protein 26 |
29942 |
0.21 |
| chr17_1365840_1366319 | 0.21 |
CRK |
v-crk avian sarcoma virus CT10 oncogene homolog |
6527 |
0.15 |
| chr8_126657314_126657465 | 0.21 |
ENSG00000266452 |
. |
200582 |
0.03 |
| chr3_114028090_114028374 | 0.21 |
ENSG00000207770 |
. |
7184 |
0.18 |
| chrX_67401924_67402162 | 0.21 |
ENSG00000252145 |
. |
80069 |
0.11 |
| chrX_20229909_20230060 | 0.21 |
RPS6KA3 |
ribosomal protein S6 kinase, 90kDa, polypeptide 3 |
6995 |
0.22 |
| chr11_46513342_46513546 | 0.21 |
ENSG00000265014 |
. |
40005 |
0.14 |
| chr8_42000592_42000772 | 0.21 |
RP11-589C21.5 |
|
9599 |
0.17 |
| chr4_106081664_106081815 | 0.21 |
TET2 |
tet methylcytosine dioxygenase 2 |
13125 |
0.21 |
| chr12_29318198_29318373 | 0.21 |
FAR2 |
fatty acyl CoA reductase 2 |
16152 |
0.24 |
| chr18_29622386_29622822 | 0.21 |
ENSG00000265063 |
. |
18348 |
0.13 |
| chr6_41986819_41986970 | 0.21 |
ENSG00000206875 |
. |
642 |
0.68 |
| chr16_11991411_11991562 | 0.21 |
GSPT1 |
G1 to S phase transition 1 |
252 |
0.86 |
| chr2_235367929_235368080 | 0.21 |
ARL4C |
ADP-ribosylation factor-like 4C |
37240 |
0.23 |
| chr5_17438596_17438747 | 0.21 |
ENSG00000201715 |
. |
92946 |
0.09 |
| chr8_71132477_71132643 | 0.21 |
NCOA2 |
nuclear receptor coactivator 2 |
25050 |
0.23 |
| chr7_132979014_132979165 | 0.21 |
ENSG00000238844 |
. |
31604 |
0.21 |
| chr5_169698045_169698343 | 0.21 |
LCP2 |
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa) |
3863 |
0.27 |
| chr5_142789348_142789499 | 0.21 |
NR3C1 |
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
4535 |
0.34 |
| chr1_109202283_109202603 | 0.21 |
HENMT1 |
HEN1 methyltransferase homolog 1 (Arabidopsis) |
1234 |
0.47 |
| chr21_39623507_39623658 | 0.21 |
KCNJ15 |
potassium inwardly-rectifying channel, subfamily J, member 15 |
5081 |
0.29 |
| chr6_41048012_41048396 | 0.21 |
NFYA |
nuclear transcription factor Y, alpha |
7482 |
0.13 |
| chr18_72372150_72372301 | 0.21 |
ZNF407 |
zinc finger protein 407 |
29249 |
0.24 |
| chr13_74598508_74598659 | 0.21 |
KLF12 |
Kruppel-like factor 12 |
29397 |
0.26 |
| chr16_23879975_23880302 | 0.21 |
PRKCB |
protein kinase C, beta |
31594 |
0.2 |
| chr3_9434517_9434896 | 0.21 |
SETD5-AS1 |
SETD5 antisense RNA 1 |
1200 |
0.39 |
| chr3_27575150_27575759 | 0.21 |
ENSG00000238912 |
. |
13895 |
0.2 |
| chr2_181641800_181641951 | 0.21 |
ENSG00000264976 |
. |
159942 |
0.04 |
| chr5_101632941_101633136 | 0.21 |
SLCO4C1 |
solute carrier organic anion transporter family, member 4C1 |
785 |
0.67 |
| chr4_4462962_4463617 | 0.21 |
STX18 |
syntaxin 18 |
392 |
0.87 |
| chr12_14548937_14549088 | 0.21 |
ATF7IP |
activating transcription factor 7 interacting protein |
1215 |
0.56 |
| chr9_110897638_110897789 | 0.21 |
ENSG00000222459 |
. |
216454 |
0.02 |
| chr14_97586844_97587256 | 0.21 |
VRK1 |
vaccinia related kinase 1 |
244684 |
0.02 |
| chr3_43371875_43372026 | 0.21 |
ENSG00000241939 |
. |
15507 |
0.16 |
| chr10_63944095_63944273 | 0.20 |
RTKN2 |
rhotekin 2 |
51838 |
0.15 |
| chr7_139249827_139249978 | 0.20 |
CLEC2L |
C-type lectin domain family 2, member L |
41074 |
0.17 |
| chr12_30835807_30835958 | 0.20 |
IPO8 |
importin 8 |
5704 |
0.3 |
| chr18_2988877_2989028 | 0.20 |
LPIN2 |
lipin 2 |
6081 |
0.17 |
| chrX_96026603_96026754 | 0.20 |
ENSG00000207426 |
. |
70060 |
0.12 |
| chrX_73067879_73068030 | 0.20 |
RP13-216E22.5 |
|
121366 |
0.06 |
| chr5_50046144_50046322 | 0.20 |
PARP8 |
poly (ADP-ribose) polymerase family, member 8 |
82842 |
0.11 |
| chr3_71447838_71447989 | 0.20 |
FOXP1 |
forkhead box P1 |
94002 |
0.08 |
| chr10_111975212_111975532 | 0.20 |
MXI1 |
MAX interactor 1, dimerization protein |
5383 |
0.23 |
| chr20_51990277_51990428 | 0.20 |
RP4-669H2.1 |
|
39096 |
0.17 |
| chr14_95563807_95563958 | 0.20 |
DICER1 |
dicer 1, ribonuclease type III |
5885 |
0.23 |
| chr8_61859867_61860032 | 0.20 |
CLVS1 |
clavesin 1 |
109768 |
0.07 |
| chr10_20639964_20640115 | 0.20 |
ENSG00000265372 |
. |
200860 |
0.03 |
| chr4_79322016_79322167 | 0.20 |
FRAS1 |
Fraser syndrome 1 |
18101 |
0.28 |
| chr14_23494825_23495047 | 0.20 |
ENSG00000207765 |
. |
7911 |
0.08 |
| chr3_49298618_49298769 | 0.20 |
RP11-3B7.1 |
Uncharacterized protein |
1175 |
0.31 |
| chr3_71820514_71820665 | 0.20 |
PROK2 |
prokineticin 2 |
13623 |
0.18 |
| chr2_84879868_84880019 | 0.20 |
DNAH6 |
dynein, axonemal, heavy chain 6 |
5488 |
0.31 |
| chr2_208802616_208802767 | 0.20 |
PLEKHM3 |
pleckstrin homology domain containing, family M, member 3 |
39483 |
0.14 |
| chr8_103896614_103896765 | 0.20 |
KB-1507C5.3 |
|
7415 |
0.17 |
| chr6_149669103_149669254 | 0.20 |
TAB2 |
TGF-beta activated kinase 1/MAP3K7 binding protein 2 |
21866 |
0.18 |
| chr1_224010673_224010824 | 0.20 |
TP53BP2 |
tumor protein p53 binding protein, 2 |
8718 |
0.27 |
| chr2_26058872_26059023 | 0.20 |
ASXL2 |
additional sex combs like 2 (Drosophila) |
42388 |
0.17 |
| chr2_136719996_136720195 | 0.20 |
AC093391.2 |
|
22651 |
0.18 |
| chr13_110302972_110303123 | 0.20 |
LINC00676 |
long intergenic non-protein coding RNA 676 |
77582 |
0.12 |
| chr12_14557948_14558281 | 0.20 |
ATF7IP |
activating transcription factor 7 interacting protein |
3310 |
0.31 |
| chr1_40630962_40631113 | 0.20 |
RLF |
rearranged L-myc fusion |
3992 |
0.2 |
| chr18_21823884_21824035 | 0.20 |
ENSG00000206766 |
. |
25695 |
0.13 |
| chr22_40338171_40338362 | 0.20 |
GRAP2 |
GRB2-related adaptor protein 2 |
4555 |
0.2 |
| chr11_9183093_9183244 | 0.19 |
DENND5A |
DENN/MADD domain containing 5A |
9214 |
0.2 |
| chr10_49537749_49537900 | 0.19 |
MAPK8 |
mitogen-activated protein kinase 8 |
22755 |
0.22 |
| chr5_156969848_156970007 | 0.19 |
AC106801.1 |
|
23045 |
0.14 |
| chr7_72482355_72482506 | 0.19 |
ENSG00000201282 |
. |
765 |
0.47 |
| chr5_17453794_17453945 | 0.19 |
ENSG00000201715 |
. |
108144 |
0.07 |
| chr3_29196189_29196340 | 0.19 |
RBMS3 |
RNA binding motif, single stranded interacting protein 3 |
126209 |
0.06 |
| chr6_37769563_37769803 | 0.19 |
RP3-441A12.1 |
|
17311 |
0.18 |
| chr15_55533028_55533179 | 0.19 |
RAB27A |
RAB27A, member RAS oncogene family |
8130 |
0.22 |
| chr17_28194872_28195023 | 0.19 |
RP11-338L22.2 |
|
4527 |
0.17 |
| chr8_73988156_73988307 | 0.19 |
RP11-956J14.1 |
|
9883 |
0.16 |
| chr7_41695372_41695523 | 0.19 |
INHBA-AS1 |
INHBA antisense RNA 1 |
38067 |
0.17 |
| chr14_45426633_45426853 | 0.19 |
KLHL28 |
kelch-like family member 28 |
3592 |
0.2 |
| chr6_144029827_144029978 | 0.19 |
PHACTR2 |
phosphatase and actin regulator 2 |
30697 |
0.21 |
| chr1_111727139_111727290 | 0.19 |
RP5-1180E21.5 |
|
469 |
0.71 |
| chr5_55981642_55982013 | 0.19 |
AC022431.2 |
Homo sapiens uncharacterized LOC101928448 (LOC101928448), mRNA. |
79768 |
0.09 |
| chr9_75205109_75205260 | 0.19 |
TMC1 |
transmembrane channel-like 1 |
24432 |
0.24 |
| chr13_30893918_30894069 | 0.19 |
KATNAL1 |
katanin p60 subunit A-like 1 |
12372 |
0.24 |
| chr12_96617227_96617404 | 0.19 |
RP11-394J1.2 |
|
436 |
0.85 |
| chr3_156020631_156020782 | 0.19 |
KCNAB1 |
potassium voltage-gated channel, shaker-related subfamily, beta member 1 |
11009 |
0.27 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.1 | 0.3 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.1 | 0.2 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.1 | GO:0061299 | retina vasculature development in camera-type eye(GO:0061298) retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 | 0.1 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 0.0 | 0.1 | GO:0032071 | regulation of endodeoxyribonuclease activity(GO:0032071) |
| 0.0 | 0.1 | GO:0071436 | sodium ion export(GO:0071436) |
| 0.0 | 0.2 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000794) |
| 0.0 | 0.1 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.0 | 0.1 | GO:0042996 | regulation of Golgi to plasma membrane protein transport(GO:0042996) |
| 0.0 | 0.1 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) regulation of store-operated calcium channel activity(GO:1901339) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.1 | GO:0001911 | negative regulation of leukocyte mediated cytotoxicity(GO:0001911) |
| 0.0 | 0.2 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) negative regulation of signal transduction by p53 class mediator(GO:1901797) |
| 0.0 | 0.0 | GO:0070669 | response to interleukin-2(GO:0070669) |
| 0.0 | 0.1 | GO:0033629 | negative regulation of cell adhesion mediated by integrin(GO:0033629) |
| 0.0 | 0.1 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.1 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 | 0.3 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.0 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.0 | 0.1 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.0 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.0 | 0.0 | GO:0002513 | tolerance induction to self antigen(GO:0002513) |
| 0.0 | 0.1 | GO:0061029 | eyelid development in camera-type eye(GO:0061029) |
| 0.0 | 0.1 | GO:0009048 | dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 0.1 | GO:0032933 | SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.1 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.0 | 0.1 | GO:0030540 | female genitalia development(GO:0030540) |
| 0.0 | 0.2 | GO:0060122 | inner ear receptor stereocilium organization(GO:0060122) |
| 0.0 | 0.1 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.1 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.0 | 0.1 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.1 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.0 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.0 | 0.1 | GO:0019987 | obsolete negative regulation of anti-apoptosis(GO:0019987) |
| 0.0 | 0.1 | GO:0002328 | pro-B cell differentiation(GO:0002328) |
| 0.0 | 0.1 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.1 | GO:1903429 | regulation of neuron maturation(GO:0014041) regulation of cell maturation(GO:1903429) |
| 0.0 | 0.1 | GO:0008611 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.0 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 0.1 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.1 | GO:0045040 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.1 | GO:0031641 | regulation of myelination(GO:0031641) |
| 0.0 | 0.1 | GO:0050651 | dermatan sulfate proteoglycan biosynthetic process(GO:0050651) |
| 0.0 | 0.1 | GO:0002634 | regulation of germinal center formation(GO:0002634) |
| 0.0 | 0.1 | GO:0042339 | keratan sulfate metabolic process(GO:0042339) |
| 0.0 | 0.1 | GO:0002313 | mature B cell differentiation involved in immune response(GO:0002313) |
| 0.0 | 0.1 | GO:0051459 | corticotropin secretion(GO:0051458) regulation of corticotropin secretion(GO:0051459) positive regulation of corticotropin secretion(GO:0051461) |
| 0.0 | 0.1 | GO:0042023 | regulation of DNA endoreduplication(GO:0032875) negative regulation of DNA endoreduplication(GO:0032876) DNA endoreduplication(GO:0042023) |
| 0.0 | 0.0 | GO:0032070 | regulation of deoxyribonuclease activity(GO:0032070) |
| 0.0 | 0.0 | GO:0002072 | optic cup morphogenesis involved in camera-type eye development(GO:0002072) |
| 0.0 | 0.0 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.0 | 0.0 | GO:0021860 | pyramidal neuron differentiation(GO:0021859) pyramidal neuron development(GO:0021860) |
| 0.0 | 0.1 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.0 | 0.1 | GO:0043368 | positive T cell selection(GO:0043368) |
| 0.0 | 0.1 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.1 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.0 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.0 | 0.0 | GO:0045627 | positive regulation of T-helper 1 cell differentiation(GO:0045627) |
| 0.0 | 0.0 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 | 0.1 | GO:0045356 | positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.0 | 0.0 | GO:0090047 | obsolete positive regulation of transcription regulator activity(GO:0090047) |
| 0.0 | 0.1 | GO:0009138 | pyrimidine nucleoside diphosphate metabolic process(GO:0009138) |
| 0.0 | 0.1 | GO:0043578 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.0 | GO:0045341 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) |
| 0.0 | 0.0 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.2 | GO:0032733 | positive regulation of interleukin-10 production(GO:0032733) |
| 0.0 | 0.1 | GO:0006498 | N-terminal protein lipidation(GO:0006498) |
| 0.0 | 0.0 | GO:0045636 | positive regulation of melanocyte differentiation(GO:0045636) positive regulation of pigment cell differentiation(GO:0050942) |
| 0.0 | 0.1 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.0 | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.0 | 0.1 | GO:0072531 | pyrimidine-containing compound transmembrane transport(GO:0072531) |
| 0.0 | 0.0 | GO:0021819 | layer formation in cerebral cortex(GO:0021819) |
| 0.0 | 0.0 | GO:0050765 | negative regulation of phagocytosis(GO:0050765) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.0 | GO:0015917 | aminophospholipid transport(GO:0015917) |
| 0.0 | 0.0 | GO:0032703 | negative regulation of interleukin-2 production(GO:0032703) |
| 0.0 | 0.0 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.0 | GO:0031946 | regulation of glucocorticoid biosynthetic process(GO:0031946) |
| 0.0 | 0.1 | GO:0042535 | positive regulation of tumor necrosis factor biosynthetic process(GO:0042535) |
| 0.0 | 0.1 | GO:0006911 | phagocytosis, engulfment(GO:0006911) membrane invagination(GO:0010324) |
| 0.0 | 0.1 | GO:0006978 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) |
| 0.0 | 0.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.1 | GO:0033089 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.0 | 0.0 | GO:0034776 | response to histamine(GO:0034776) |
| 0.0 | 0.0 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.1 | GO:0070498 | interleukin-1-mediated signaling pathway(GO:0070498) |
| 0.0 | 0.1 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.0 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.0 | GO:0043320 | natural killer cell activation involved in immune response(GO:0002323) natural killer cell degranulation(GO:0043320) |
| 0.0 | 0.1 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.0 | GO:0039656 | modulation by virus of host gene expression(GO:0039656) |
| 0.0 | 0.1 | GO:0031033 | myosin filament organization(GO:0031033) |
| 0.0 | 0.1 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 0.1 | GO:0070670 | response to interleukin-4(GO:0070670) |
| 0.0 | 0.0 | GO:0033024 | mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) |
| 0.0 | 0.4 | GO:1903038 | negative regulation of T cell activation(GO:0050868) negative regulation of leukocyte cell-cell adhesion(GO:1903038) |
| 0.0 | 0.0 | GO:0045060 | negative thymic T cell selection(GO:0045060) |
| 0.0 | 0.1 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.1 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.0 | 0.0 | GO:0031943 | regulation of glucocorticoid metabolic process(GO:0031943) |
| 0.0 | 0.1 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.0 | 0.1 | GO:0009649 | entrainment of circadian clock(GO:0009649) |
| 0.0 | 0.0 | GO:0033689 | negative regulation of osteoblast proliferation(GO:0033689) |
| 0.0 | 0.2 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.0 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.0 | 0.0 | GO:0072216 | positive regulation of metanephros development(GO:0072216) |
| 0.0 | 0.1 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 0.1 | GO:0002483 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 0.1 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 | 0.0 | GO:0032455 | nerve growth factor processing(GO:0032455) |
| 0.0 | 0.0 | GO:0060972 | left/right pattern formation(GO:0060972) |
| 0.0 | 0.0 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.0 | 0.1 | GO:0045723 | positive regulation of fatty acid biosynthetic process(GO:0045723) |
| 0.0 | 0.0 | GO:0046469 | platelet activating factor metabolic process(GO:0046469) |
| 0.0 | 0.0 | GO:0042501 | serine phosphorylation of STAT protein(GO:0042501) |
| 0.0 | 0.0 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 0.0 | 0.0 | GO:0060437 | lung growth(GO:0060437) |
| 0.0 | 0.2 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.0 | 0.1 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 | 0.2 | GO:0034199 | activation of protein kinase A activity(GO:0034199) |
| 0.0 | 0.0 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.0 | 0.0 | GO:0002283 | neutrophil activation involved in immune response(GO:0002283) neutrophil degranulation(GO:0043312) |
| 0.0 | 0.1 | GO:0034643 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.0 | 0.0 | GO:1903053 | regulation of extracellular matrix disassembly(GO:0010715) negative regulation of extracellular matrix disassembly(GO:0010716) regulation of extracellular matrix organization(GO:1903053) negative regulation of extracellular matrix organization(GO:1903054) |
| 0.0 | 0.2 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.0 | 0.0 | GO:0045061 | thymic T cell selection(GO:0045061) |
| 0.0 | 0.1 | GO:0071044 | histone mRNA catabolic process(GO:0071044) |
| 0.0 | 0.0 | GO:0001779 | natural killer cell differentiation(GO:0001779) |
| 0.0 | 0.0 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 | 0.1 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.0 | GO:0035066 | positive regulation of histone acetylation(GO:0035066) positive regulation of protein acetylation(GO:1901985) positive regulation of peptidyl-lysine acetylation(GO:2000758) |
| 0.0 | 0.0 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.0 | GO:0090400 | stress-induced premature senescence(GO:0090400) |
| 0.0 | 0.0 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.0 | 0.0 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.0 | 0.0 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.1 | GO:0042362 | fat-soluble vitamin biosynthetic process(GO:0042362) |
| 0.0 | 0.1 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 | 0.0 | GO:0071636 | positive regulation of transforming growth factor beta production(GO:0071636) |
| 0.0 | 0.1 | GO:0007350 | blastoderm segmentation(GO:0007350) |
| 0.0 | 0.0 | GO:0002068 | glandular epithelial cell development(GO:0002068) |
| 0.0 | 0.0 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.0 | 0.0 | GO:0042159 | lipoprotein catabolic process(GO:0042159) |
| 0.0 | 0.0 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.0 | 0.1 | GO:0032528 | microvillus assembly(GO:0030033) microvillus organization(GO:0032528) |
| 0.0 | 0.0 | GO:0034139 | regulation of toll-like receptor 3 signaling pathway(GO:0034139) positive regulation of toll-like receptor 4 signaling pathway(GO:0034145) |
| 0.0 | 0.0 | GO:0021780 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) oligodendrocyte cell fate specification(GO:0021778) glial cell fate specification(GO:0021780) |
| 0.0 | 0.1 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.0 | GO:0032814 | regulation of natural killer cell activation(GO:0032814) |
| 0.0 | 0.0 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) positive regulation of lamellipodium organization(GO:1902745) |
| 0.0 | 0.1 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.0 | 0.1 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.1 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.1 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.1 | GO:0001740 | Barr body(GO:0001740) |
| 0.0 | 0.1 | GO:0002139 | stereocilia coupling link(GO:0002139) stereocilia ankle link(GO:0002141) stereocilia ankle link complex(GO:0002142) |
| 0.0 | 0.1 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.4 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.0 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.1 | GO:0031332 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.1 | GO:0031313 | extrinsic component of vacuolar membrane(GO:0000306) extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.1 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.2 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.0 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.1 | GO:0070688 | MLL5-L complex(GO:0070688) |
| 0.0 | 0.1 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.2 | GO:0045495 | P granule(GO:0043186) pole plasm(GO:0045495) germ plasm(GO:0060293) |
| 0.0 | 0.1 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.0 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.1 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.1 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.0 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.0 | 0.1 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.2 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.1 | GO:0098984 | asymmetric synapse(GO:0032279) neuron to neuron synapse(GO:0098984) |
| 0.0 | 0.0 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.1 | GO:0005845 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.0 | 0.1 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.0 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.1 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.0 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.1 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.1 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.0 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.0 | GO:0000800 | lateral element(GO:0000800) |
| 0.0 | 0.1 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.1 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.1 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.1 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.1 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.0 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.0 | 0.2 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.0 | GO:0042584 | chromaffin granule(GO:0042583) chromaffin granule membrane(GO:0042584) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0032296 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.1 | 0.2 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.1 | 0.2 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.1 | 0.2 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.2 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.2 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.1 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.0 | 0.2 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.2 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.1 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.1 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.1 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.1 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.1 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.1 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.1 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.2 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.1 | GO:0015065 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.0 | 0.1 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.1 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.1 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.2 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.1 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.0 | 0.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.1 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.2 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.1 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.0 | 0.1 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.0 | 0.0 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 0.1 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.1 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.0 | 0.3 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.2 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) |
| 0.0 | 0.0 | GO:0019958 | C-X-C chemokine binding(GO:0019958) interleukin-8 binding(GO:0019959) |
| 0.0 | 0.1 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.1 | GO:0008518 | reduced folate carrier activity(GO:0008518) |
| 0.0 | 0.1 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.1 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.1 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.0 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.3 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.1 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.0 | 0.1 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.1 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.1 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.0 | GO:0032450 | maltose alpha-glucosidase activity(GO:0032450) |
| 0.0 | 0.0 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.0 | 0.0 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.3 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.2 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.1 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.1 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.0 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.0 | 0.1 | GO:0015385 | sodium:proton antiporter activity(GO:0015385) |
| 0.0 | 0.1 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.0 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 0.1 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.0 | GO:0099602 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.0 | 0.1 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.1 | GO:0045309 | protein phosphorylated amino acid binding(GO:0045309) |
| 0.0 | 0.1 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.1 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.0 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.0 | 0.0 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.0 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.0 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.0 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.1 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.0 | 0.0 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.0 | GO:0090482 | vitamin transmembrane transporter activity(GO:0090482) |
| 0.0 | 0.1 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.0 | 0.0 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.0 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.1 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.0 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.4 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.3 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.0 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.1 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.2 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.8 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.0 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 0.0 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.0 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 0.1 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 0.3 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 0.0 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.0 | 0.0 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 0.1 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.0 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.0 | 0.0 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.4 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.3 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.3 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.0 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.2 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.2 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.2 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.1 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.0 | 0.0 | REACTOME CDT1 ASSOCIATION WITH THE CDC6 ORC ORIGIN COMPLEX | Genes involved in CDT1 association with the CDC6:ORC:origin complex |
| 0.0 | 0.2 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.3 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 0.2 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.3 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.2 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.1 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.0 | 0.0 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 0.4 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 0.2 | REACTOME BOTULINUM NEUROTOXICITY | Genes involved in Botulinum neurotoxicity |
| 0.0 | 0.1 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.1 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.2 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.0 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.0 | 0.0 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.0 | 0.1 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |