Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
Results for LMX1B_MNX1_RAX2
Z-value: 0.50
Transcription factors associated with LMX1B_MNX1_RAX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
LMX1B
|
ENSG00000136944.13 | LIM homeobox transcription factor 1 beta |
|
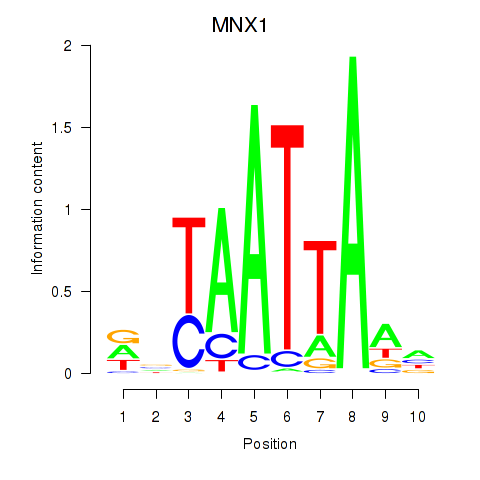
MNX1
|
ENSG00000130675.10 | motor neuron and pancreas homeobox 1 |
|
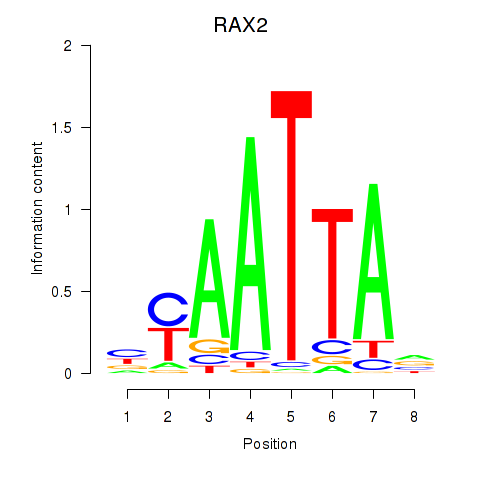
RAX2
|
ENSG00000173976.11 | retina and anterior neural fold homeobox 2 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr9_129388786_129388937 | LMX1B | 12113 | 0.157468 | -0.52 | 1.5e-01 | Click! |
| chr9_129387667_129387818 | LMX1B | 10994 | 0.159314 | -0.50 | 1.7e-01 | Click! |
| chr9_129388528_129388727 | LMX1B | 11879 | 0.157845 | -0.41 | 2.7e-01 | Click! |
| chr9_129386356_129386507 | LMX1B | 9683 | 0.161683 | -0.38 | 3.2e-01 | Click! |
| chr9_129378843_129378994 | LMX1B | 2170 | 0.252632 | -0.36 | 3.4e-01 | Click! |
| chr7_156801782_156801933 | MNX1 | 88 | 0.952809 | 0.35 | 3.6e-01 | Click! |
| chr7_156801023_156801174 | MNX1 | 671 | 0.600026 | 0.25 | 5.1e-01 | Click! |
| chr7_156800821_156800972 | MNX1 | 873 | 0.486004 | 0.21 | 5.8e-01 | Click! |
| chr7_156801980_156802430 | MNX1 | 76 | 0.950727 | 0.12 | 7.7e-01 | Click! |
| chr7_156801401_156801562 | MNX1 | 288 | 0.861126 | -0.05 | 9.0e-01 | Click! |
Activity of the LMX1B_MNX1_RAX2 motif across conditions
Conditions sorted by the z-value of the LMX1B_MNX1_RAX2 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
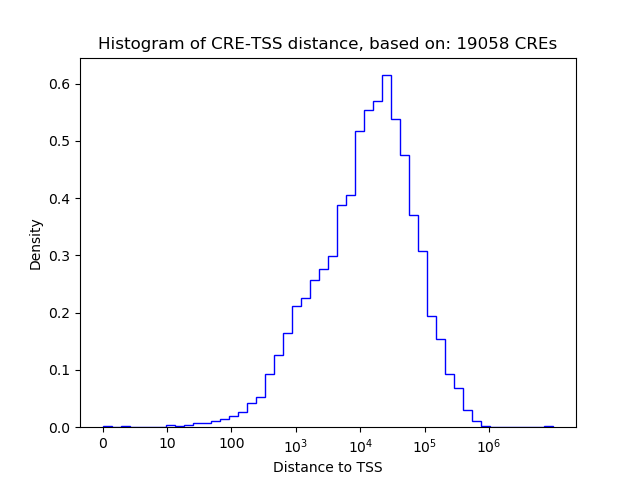
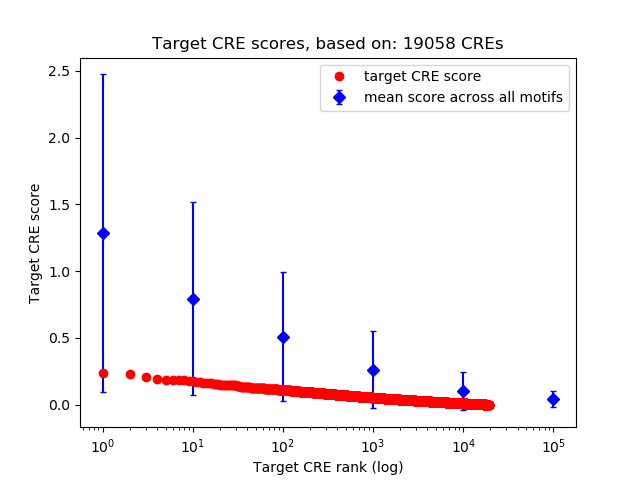
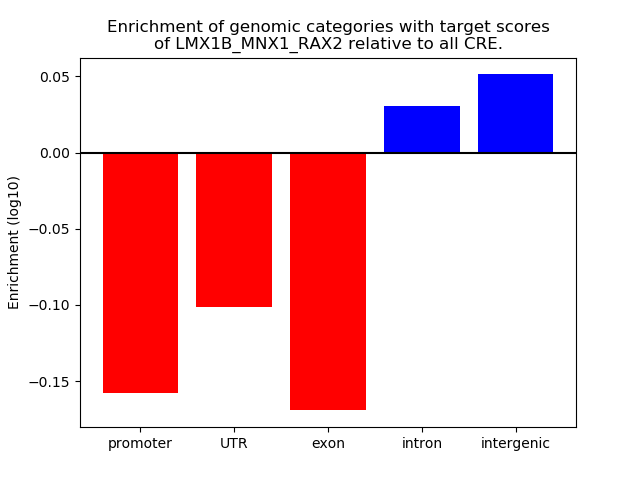
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr5_148365723_148365874 | 0.24 |
RP11-44B19.1 |
|
15895 |
0.18 |
| chr17_28275117_28275440 | 0.23 |
EFCAB5 |
EF-hand calcium binding domain 5 |
6655 |
0.17 |
| chr3_154690294_154690445 | 0.21 |
MME |
membrane metallo-endopeptidase |
51544 |
0.19 |
| chrX_55741083_55741234 | 0.19 |
RRAGB |
Ras-related GTP binding B |
3014 |
0.33 |
| chr7_105903512_105903663 | 0.19 |
NAMPT |
nicotinamide phosphoribosyltransferase |
21780 |
0.19 |
| chr5_169227490_169227641 | 0.18 |
CTB-37A13.1 |
|
21196 |
0.23 |
| chr2_8007405_8007668 | 0.18 |
ENSG00000221255 |
. |
290564 |
0.01 |
| chrX_96151518_96151669 | 0.18 |
RPA4 |
replication protein A4, 30kDa |
12686 |
0.31 |
| chr8_62569308_62569459 | 0.18 |
ASPH |
aspartate beta-hydroxylase |
10017 |
0.23 |
| chr6_2509288_2509439 | 0.18 |
ENSG00000266252 |
. |
99494 |
0.08 |
| chr9_123478112_123478263 | 0.17 |
MEGF9 |
multiple EGF-like-domains 9 |
1439 |
0.51 |
| chr11_122897468_122897619 | 0.17 |
ENSG00000200879 |
. |
31326 |
0.12 |
| chr6_17972496_17972680 | 0.16 |
KIF13A |
kinesin family member 13A |
15106 |
0.27 |
| chr9_28922506_28922753 | 0.16 |
ENSG00000215939 |
. |
33676 |
0.22 |
| chr16_4998738_4998889 | 0.16 |
ENSG00000200980 |
. |
3079 |
0.19 |
| chr9_94178983_94179134 | 0.16 |
NFIL3 |
nuclear factor, interleukin 3 regulated |
7086 |
0.3 |
| chr9_38042251_38042402 | 0.16 |
SHB |
Src homology 2 domain containing adaptor protein B |
26882 |
0.21 |
| chr12_81262405_81262556 | 0.15 |
LIN7A |
lin-7 homolog A (C. elegans) |
20647 |
0.19 |
| chr15_101143578_101144308 | 0.15 |
ASB7 |
ankyrin repeat and SOCS box containing 7 |
1129 |
0.36 |
| chr5_17282736_17283060 | 0.15 |
ENSG00000252908 |
. |
42178 |
0.15 |
| chr2_134912163_134912443 | 0.15 |
ENSG00000263813 |
. |
27607 |
0.2 |
| chr9_41085505_41085742 | 0.15 |
RP11-95K23.5 |
|
199734 |
0.03 |
| chr2_124505371_124505522 | 0.15 |
ENSG00000221689 |
. |
65655 |
0.14 |
| chr5_127457688_127457839 | 0.15 |
SLC12A2 |
solute carrier family 12 (sodium/potassium/chloride transporter), member 2 |
38223 |
0.23 |
| chr12_80333289_80333440 | 0.15 |
PPP1R12A |
protein phosphatase 1, regulatory subunit 12A |
4124 |
0.23 |
| chr5_66255925_66256427 | 0.15 |
MAST4 |
microtubule associated serine/threonine kinase family member 4 |
1360 |
0.56 |
| chr18_22012413_22012586 | 0.15 |
RP11-178F10.1 |
|
2829 |
0.18 |
| chrX_7163379_7163530 | 0.15 |
STS |
steroid sulfatase (microsomal), isozyme S |
25957 |
0.25 |
| chr7_25345986_25346137 | 0.15 |
ENSG00000202233 |
. |
41786 |
0.18 |
| chr6_17906953_17907342 | 0.14 |
KIF13A |
kinesin family member 13A |
80547 |
0.1 |
| chr20_46413000_46413151 | 0.14 |
SULF2 |
sulfatase 2 |
1158 |
0.56 |
| chr10_129077210_129077361 | 0.14 |
FAM196A |
family with sequence similarity 196, member A |
82863 |
0.11 |
| chr13_72218273_72218424 | 0.14 |
DACH1 |
dachshund homolog 1 (Drosophila) |
222559 |
0.02 |
| chr5_40608102_40608403 | 0.14 |
ENSG00000199552 |
. |
46813 |
0.16 |
| chr6_25005285_25005494 | 0.14 |
ENSG00000244618 |
. |
26121 |
0.17 |
| chr1_109632204_109632387 | 0.13 |
TMEM167B |
transmembrane protein 167B |
130 |
0.91 |
| chr13_30472600_30472751 | 0.13 |
LINC00572 |
long intergenic non-protein coding RNA 572 |
28113 |
0.22 |
| chr1_180279497_180279775 | 0.13 |
RP5-1180C10.2 |
|
34277 |
0.19 |
| chr17_74601770_74601921 | 0.13 |
ST6GALNAC2 |
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 2 |
19635 |
0.08 |
| chr5_115823878_115824029 | 0.13 |
SEMA6A |
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
9580 |
0.25 |
| chr20_39135706_39135857 | 0.13 |
ENSG00000252434 |
. |
90266 |
0.1 |
| chr12_104200199_104200350 | 0.13 |
RP11-650K20.2 |
|
13114 |
0.14 |
| chr7_8328595_8328746 | 0.13 |
ICA1 |
islet cell autoantigen 1, 69kDa |
26353 |
0.17 |
| chrX_120811108_120811259 | 0.13 |
ENSG00000221217 |
. |
191537 |
0.03 |
| chr2_210868461_210869072 | 0.13 |
RPE |
ribulose-5-phosphate-3-epimerase |
1380 |
0.42 |
| chr5_53987308_53987459 | 0.13 |
ENSG00000221073 |
. |
161393 |
0.04 |
| chr9_21383132_21383283 | 0.13 |
IFNA2 |
interferon, alpha 2 |
2189 |
0.22 |
| chr3_177443249_177443545 | 0.13 |
ENSG00000200288 |
. |
101200 |
0.08 |
| chr4_83969657_83969808 | 0.13 |
COPS4 |
COP9 signalosome subunit 4 |
13354 |
0.17 |
| chr7_36108267_36108418 | 0.13 |
PP13004 |
|
10352 |
0.22 |
| chr13_49475290_49475441 | 0.13 |
ENSG00000265585 |
. |
28983 |
0.2 |
| chr2_240340277_240340428 | 0.13 |
AC062017.1 |
Uncharacterized protein |
16853 |
0.19 |
| chr2_152510948_152511099 | 0.13 |
NEB |
nebulin |
70890 |
0.13 |
| chr2_37576208_37576507 | 0.12 |
QPCT |
glutaminyl-peptide cyclotransferase |
4505 |
0.2 |
| chr3_12242557_12242708 | 0.12 |
TIMP4 |
TIMP metallopeptidase inhibitor 4 |
41781 |
0.18 |
| chr16_69775894_69776045 | 0.12 |
NOB1 |
NIN1/RPN12 binding protein 1 homolog (S. cerevisiae) |
12874 |
0.1 |
| chr13_32715851_32716002 | 0.12 |
FRY |
furry homolog (Drosophila) |
80925 |
0.1 |
| chr9_107830314_107830790 | 0.12 |
ENSG00000201583 |
. |
28485 |
0.23 |
| chr8_90851049_90851200 | 0.12 |
ENSG00000207359 |
. |
61928 |
0.12 |
| chr6_35002148_35002432 | 0.12 |
TCP11 |
t-complex 11, testis-specific |
86532 |
0.08 |
| chr2_14212942_14213093 | 0.12 |
ENSG00000251859 |
. |
201832 |
0.03 |
| chr13_110651559_110651710 | 0.12 |
ENSG00000201161 |
. |
84191 |
0.1 |
| chr7_37760403_37760554 | 0.12 |
GPR141 |
G protein-coupled receptor 141 |
19518 |
0.23 |
| chr14_103106612_103106763 | 0.12 |
RCOR1 |
REST corepressor 1 |
47454 |
0.12 |
| chr9_137251018_137251277 | 0.12 |
ENSG00000263897 |
. |
20110 |
0.22 |
| chr8_9331082_9331233 | 0.12 |
TNKS |
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase |
82267 |
0.09 |
| chr8_97536479_97536735 | 0.12 |
SDC2 |
syndecan 2 |
30371 |
0.22 |
| chr20_6888062_6888352 | 0.12 |
ENSG00000251833 |
. |
46546 |
0.2 |
| chr4_123694862_123695013 | 0.12 |
BBS12 |
Bardet-Biedl syndrome 12 |
40985 |
0.14 |
| chr9_590173_590324 | 0.12 |
ENSG00000202172 |
. |
31422 |
0.15 |
| chr3_23930377_23930528 | 0.12 |
RPL15 |
ribosomal protein L15 |
27584 |
0.16 |
| chr2_8012045_8012196 | 0.12 |
ENSG00000221255 |
. |
295148 |
0.01 |
| chr22_36793445_36793596 | 0.12 |
MYH9 |
myosin, heavy chain 9, non-muscle |
9457 |
0.18 |
| chr7_80350905_80351077 | 0.12 |
CD36 |
CD36 molecule (thrombospondin receptor) |
48299 |
0.19 |
| chr18_10213675_10214158 | 0.12 |
ENSG00000239031 |
. |
176081 |
0.03 |
| chr4_10009068_10009219 | 0.12 |
RP11-448G15.1 |
|
1037 |
0.53 |
| chr4_39152584_39152984 | 0.12 |
RP11-360F5.1 |
|
24346 |
0.17 |
| chr21_39659541_39659821 | 0.12 |
KCNJ15 |
potassium inwardly-rectifying channel, subfamily J, member 15 |
8798 |
0.29 |
| chr10_64252258_64252409 | 0.12 |
ZNF365 |
zinc finger protein 365 |
27874 |
0.24 |
| chr13_107573389_107573540 | 0.12 |
ENSG00000239050 |
. |
53910 |
0.18 |
| chr4_101903241_101903392 | 0.12 |
EMCN-IT1 |
EMCN intronic transcript 1 (non-protein coding) |
167879 |
0.04 |
| chr11_33934050_33934291 | 0.12 |
LMO2 |
LIM domain only 2 (rhombotin-like 1) |
20334 |
0.2 |
| chr17_65372251_65373170 | 0.12 |
PITPNC1 |
phosphatidylinositol transfer protein, cytoplasmic 1 |
865 |
0.53 |
| chr8_28272280_28272431 | 0.12 |
ENSG00000207361 |
. |
1610 |
0.28 |
| chr11_32843513_32843664 | 0.11 |
PRRG4 |
proline rich Gla (G-carboxyglutamic acid) 4 (transmembrane) |
7901 |
0.23 |
| chr19_18239230_18239381 | 0.11 |
AC007192.6 |
|
23196 |
0.1 |
| chr4_101813891_101814042 | 0.11 |
EMCN-IT1 |
EMCN intronic transcript 1 (non-protein coding) |
78529 |
0.11 |
| chr18_2154076_2154227 | 0.11 |
METTL4 |
methyltransferase like 4 |
400540 |
0.01 |
| chr2_197857511_197857662 | 0.11 |
ANKRD44 |
ankyrin repeat domain 44 |
7640 |
0.27 |
| chr3_48800674_48800825 | 0.11 |
PRKAR2A |
protein kinase, cAMP-dependent, regulatory, type II, alpha |
2034 |
0.27 |
| chr20_4811132_4811283 | 0.11 |
RASSF2 |
Ras association (RalGDS/AF-6) domain family member 2 |
6916 |
0.2 |
| chr2_100664977_100665128 | 0.11 |
AFF3 |
AF4/FMR2 family, member 3 |
55940 |
0.14 |
| chr14_70110903_70111120 | 0.11 |
KIAA0247 |
KIAA0247 |
32698 |
0.18 |
| chr5_102181314_102181465 | 0.11 |
PAM |
peptidylglycine alpha-amidating monooxygenase |
18852 |
0.29 |
| chr4_65272983_65273134 | 0.11 |
TECRL |
trans-2,3-enoyl-CoA reductase-like |
2058 |
0.4 |
| chr1_88995690_88995841 | 0.11 |
RP11-76N22.2 |
|
40778 |
0.18 |
| chr4_74569730_74569881 | 0.11 |
IL8 |
interleukin 8 |
36418 |
0.18 |
| chr6_87895108_87895259 | 0.11 |
ZNF292 |
zinc finger protein 292 |
29844 |
0.16 |
| chr8_93968938_93969089 | 0.11 |
TRIQK |
triple QxxK/R motif containing |
8804 |
0.25 |
| chr15_78594610_78594888 | 0.11 |
WDR61 |
WD repeat domain 61 |
2613 |
0.18 |
| chr3_187492287_187492506 | 0.11 |
BCL6 |
B-cell CLL/lymphoma 6 |
28881 |
0.2 |
| chrX_147724893_147725044 | 0.11 |
AFF2 |
AF4/FMR2 family, member 2 |
75647 |
0.11 |
| chr10_31273280_31273431 | 0.11 |
ZNF438 |
zinc finger protein 438 |
0 |
0.99 |
| chr7_26143414_26143565 | 0.11 |
ENSG00000266430 |
. |
43530 |
0.14 |
| chr7_33973779_33973930 | 0.11 |
BMPER |
BMP binding endothelial regulator |
28709 |
0.26 |
| chr11_69414414_69414565 | 0.11 |
CCND1 |
cyclin D1 |
41366 |
0.17 |
| chr1_87801300_87801913 | 0.11 |
LMO4 |
LIM domain only 4 |
4255 |
0.35 |
| chr10_31272970_31273270 | 0.11 |
ZNF438 |
zinc finger protein 438 |
235 |
0.96 |
| chr14_33625609_33625904 | 0.11 |
NPAS3 |
neuronal PAS domain protein 3 |
57128 |
0.18 |
| chr1_31870163_31870721 | 0.11 |
SERINC2 |
serine incorporator 2 |
11970 |
0.15 |
| chr2_201958936_201959087 | 0.11 |
ENSG00000252148 |
. |
14446 |
0.12 |
| chr7_155382227_155382378 | 0.11 |
AC009403.2 |
Protein LOC100506302 |
54793 |
0.11 |
| chr19_19539763_19539914 | 0.11 |
GATAD2A |
GATA zinc finger domain containing 2A |
1502 |
0.3 |
| chr4_26727880_26728031 | 0.11 |
ENSG00000200999 |
. |
24024 |
0.23 |
| chr12_27197460_27197611 | 0.11 |
MED21 |
mediator complex subunit 21 |
22052 |
0.15 |
| chr4_16034494_16034749 | 0.11 |
ENSG00000251758 |
. |
24211 |
0.19 |
| chr3_39256267_39256418 | 0.11 |
XIRP1 |
xin actin-binding repeat containing 1 |
22255 |
0.18 |
| chr4_101993104_101993255 | 0.11 |
EMCN-IT1 |
EMCN intronic transcript 1 (non-protein coding) |
257742 |
0.02 |
| chr14_23583904_23584167 | 0.11 |
CEBPE |
CCAAT/enhancer binding protein (C/EBP), epsilon |
4790 |
0.11 |
| chr7_127763401_127763552 | 0.11 |
ENSG00000207588 |
. |
41563 |
0.16 |
| chrX_14818705_14818856 | 0.11 |
ENSG00000221278 |
. |
35541 |
0.19 |
| chr6_138820271_138821054 | 0.11 |
NHSL1 |
NHS-like 1 |
31 |
0.98 |
| chr2_33309027_33309178 | 0.11 |
LTBP1 |
latent transforming growth factor beta binding protein 1 |
50371 |
0.16 |
| chr14_103586056_103586207 | 0.11 |
TNFAIP2 |
tumor necrosis factor, alpha-induced protein 2 |
3667 |
0.2 |
| chr7_21979875_21980026 | 0.11 |
CDCA7L |
cell division cycle associated 7-like |
4541 |
0.34 |
| chr12_117472876_117473027 | 0.10 |
TESC |
tescalcin |
11685 |
0.24 |
| chr1_100293238_100293389 | 0.10 |
ENSG00000252226 |
. |
1708 |
0.32 |
| chr8_80718909_80719060 | 0.10 |
ENSG00000238884 |
. |
6554 |
0.27 |
| chr6_97580328_97580479 | 0.10 |
KLHL32 |
kelch-like family member 32 |
67899 |
0.13 |
| chr7_37748458_37748609 | 0.10 |
GPR141 |
G protein-coupled receptor 141 |
25058 |
0.2 |
| chr20_44601777_44602076 | 0.10 |
ZNF335 |
zinc finger protein 335 |
1093 |
0.38 |
| chr14_51406161_51406312 | 0.10 |
PYGL |
phosphorylase, glycogen, liver |
4938 |
0.21 |
| chr6_166670086_166670237 | 0.10 |
ENSG00000201292 |
. |
47270 |
0.13 |
| chr5_109078948_109079099 | 0.10 |
ENSG00000202512 |
. |
43488 |
0.16 |
| chr4_85836939_85837233 | 0.10 |
ENSG00000252062 |
. |
29447 |
0.21 |
| chr1_180382000_180382151 | 0.10 |
ENSG00000265435 |
. |
25450 |
0.21 |
| chr7_26351236_26351387 | 0.10 |
SNX10 |
sorting nexin 10 |
18637 |
0.22 |
| chr2_165699907_165700058 | 0.10 |
COBLL1 |
cordon-bleu WH2 repeat protein-like 1 |
207 |
0.93 |
| chr18_61561537_61561693 | 0.10 |
SERPINB10 |
serpin peptidase inhibitor, clade B (ovalbumin), member 10 |
2793 |
0.24 |
| chr19_10891817_10891968 | 0.10 |
ENSG00000265879 |
. |
962 |
0.4 |
| chr15_77646528_77646679 | 0.10 |
PEAK1 |
pseudopodium-enriched atypical kinase 1 |
65839 |
0.12 |
| chr5_139968834_139969062 | 0.10 |
ENSG00000200235 |
. |
9696 |
0.08 |
| chr2_190637879_190638030 | 0.10 |
AC013468.1 |
|
5203 |
0.12 |
| chr2_144454482_144454633 | 0.10 |
RP11-434H14.1 |
|
14049 |
0.22 |
| chr18_46195874_46196025 | 0.10 |
ENSG00000266276 |
. |
1022 |
0.6 |
| chr19_43853396_43853679 | 0.10 |
CD177 |
CD177 molecule |
4288 |
0.2 |
| chr9_129190475_129190626 | 0.10 |
ENSG00000252985 |
. |
1378 |
0.46 |
| chr18_456798_457125 | 0.10 |
RP11-720L2.2 |
|
32545 |
0.16 |
| chr3_11734918_11735069 | 0.10 |
VGLL4 |
vestigial like 4 (Drosophila) |
17018 |
0.25 |
| chr7_156836903_156837054 | 0.10 |
MNX1-AS1 |
MNX1 antisense RNA 1 (head to head) |
33479 |
0.13 |
| chr3_158546491_158546642 | 0.10 |
MFSD1 |
major facilitator superfamily domain containing 1 |
19540 |
0.17 |
| chr3_81783533_81783684 | 0.10 |
GBE1 |
glucan (1,4-alpha-), branching enzyme 1 |
9172 |
0.33 |
| chr14_59803635_59803786 | 0.10 |
ENSG00000252869 |
. |
63031 |
0.12 |
| chr1_33793881_33794032 | 0.10 |
ENSG00000266239 |
. |
4137 |
0.14 |
| chr15_69730252_69730403 | 0.10 |
RPLP1 |
ribosomal protein, large, P1 |
14796 |
0.12 |
| chr11_3748512_3748663 | 0.10 |
ENSG00000251934 |
. |
12482 |
0.11 |
| chr9_114557676_114557827 | 0.10 |
C9orf84 |
chromosome 9 open reading frame 84 |
463 |
0.84 |
| chr5_57134689_57134840 | 0.10 |
ENSG00000266864 |
. |
34109 |
0.24 |
| chr22_40574959_40575808 | 0.10 |
TNRC6B |
trinucleotide repeat containing 6B |
1437 |
0.53 |
| chr20_45754256_45754424 | 0.10 |
ENSG00000264901 |
. |
41269 |
0.18 |
| chr13_49838164_49838315 | 0.10 |
CDADC1 |
cytidine and dCMP deaminase domain containing 1 |
16055 |
0.21 |
| chr8_38264429_38264741 | 0.10 |
RP11-350N15.6 |
|
1675 |
0.23 |
| chr2_64074407_64074558 | 0.10 |
UGP2 |
UDP-glucose pyrophosphorylase 2 |
1274 |
0.55 |
| chr10_78792555_78792706 | 0.10 |
KCNMA1 |
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
46652 |
0.17 |
| chr7_8290002_8290153 | 0.10 |
ICA1 |
islet cell autoantigen 1, 69kDa |
11605 |
0.18 |
| chr1_21067705_21067856 | 0.10 |
RP5-930J4.4 |
|
1700 |
0.27 |
| chr1_76545726_76545919 | 0.10 |
ST6GALNAC3 |
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 3 |
5418 |
0.32 |
| chr1_234667803_234667954 | 0.10 |
TARBP1 |
TAR (HIV-1) RNA binding protein 1 |
53029 |
0.12 |
| chr1_200865216_200865405 | 0.10 |
C1orf106 |
chromosome 1 open reading frame 106 |
1361 |
0.42 |
| chr2_153267355_153267720 | 0.10 |
FMNL2 |
formin-like 2 |
75786 |
0.12 |
| chr5_158647032_158647183 | 0.10 |
RNF145 |
ring finger protein 145 |
10046 |
0.16 |
| chr3_47123059_47123210 | 0.10 |
ENSG00000251938 |
. |
6964 |
0.22 |
| chr12_42228024_42228175 | 0.10 |
ENSG00000252364 |
. |
175621 |
0.03 |
| chr3_168752701_168752852 | 0.10 |
MECOM |
MDS1 and EVI1 complex locus |
93046 |
0.1 |
| chr11_46479636_46479787 | 0.10 |
ENSG00000265014 |
. |
6272 |
0.19 |
| chr3_138595962_138596113 | 0.10 |
PIK3CB |
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit beta |
42257 |
0.15 |
| chr1_83043773_83044235 | 0.10 |
LPHN2 |
latrophilin 2 |
598431 |
0.0 |
| chr2_225451534_225451729 | 0.10 |
CUL3 |
cullin 3 |
1521 |
0.57 |
| chr2_163197962_163198113 | 0.10 |
GCA |
grancalcin, EF-hand calcium binding protein |
2561 |
0.33 |
| chr3_122029022_122029173 | 0.10 |
CSTA |
cystatin A (stefin A) |
14994 |
0.16 |
| chr3_148706459_148706610 | 0.10 |
GYG1 |
glycogenin 1 |
2594 |
0.29 |
| chrX_16661091_16661259 | 0.10 |
S100G |
S100 calcium binding protein G |
7106 |
0.18 |
| chr4_185186357_185186631 | 0.09 |
ENSG00000221523 |
. |
2090 |
0.34 |
| chr2_163263039_163263190 | 0.09 |
GCA |
grancalcin, EF-hand calcium binding protein |
50166 |
0.15 |
| chr10_120480954_120481105 | 0.09 |
CACUL1 |
CDK2-associated, cullin domain 1 |
33729 |
0.18 |
| chr7_37726411_37726562 | 0.09 |
GPR141 |
G protein-coupled receptor 141 |
3011 |
0.28 |
| chr22_30257161_30257312 | 0.09 |
MTMR3 |
myotubularin related protein 3 |
21908 |
0.14 |
| chr5_115627433_115627584 | 0.09 |
ENSG00000212457 |
. |
103259 |
0.07 |
| chr17_29906221_29906372 | 0.09 |
ENSG00000199187 |
. |
3866 |
0.14 |
| chr16_3081576_3081727 | 0.09 |
RP11-473M20.5 |
|
831 |
0.27 |
| chr17_2285977_2286170 | 0.09 |
RP1-59D14.1 |
|
2068 |
0.18 |
| chr4_15662221_15662454 | 0.09 |
FBXL5 |
F-box and leucine-rich repeat protein 5 |
847 |
0.63 |
| chrX_38576524_38576675 | 0.09 |
MID1IP1 |
MID1 interacting protein 1 |
84086 |
0.09 |
| chr2_223688143_223688294 | 0.09 |
ACSL3 |
acyl-CoA synthetase long-chain family member 3 |
37434 |
0.18 |
| chr6_51167339_51167725 | 0.09 |
ENSG00000212532 |
. |
161956 |
0.04 |
| chr1_233375664_233375815 | 0.09 |
PCNXL2 |
pecanex-like 2 (Drosophila) |
12724 |
0.29 |
| chr9_90215657_90216062 | 0.09 |
DAPK1-IT1 |
DAPK1 intronic transcript 1 (non-protein coding) |
47490 |
0.16 |
| chr17_66307289_66307440 | 0.09 |
ARSG |
arylsulfatase G |
19705 |
0.16 |
| chr1_64697428_64697579 | 0.09 |
UBE2U |
ubiquitin-conjugating enzyme E2U (putative) |
10956 |
0.25 |
| chr4_48382974_48383176 | 0.09 |
SLAIN2 |
SLAIN motif family, member 2 |
2620 |
0.36 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0071436 | sodium ion export(GO:0071436) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.2 | GO:0061001 | dendritic spine morphogenesis(GO:0060997) regulation of dendritic spine morphogenesis(GO:0061001) dendritic spine organization(GO:0097061) |
| 0.0 | 0.1 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.0 | 0.1 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.1 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.1 | GO:0006065 | UDP-glucuronate biosynthetic process(GO:0006065) |
| 0.0 | 0.0 | GO:0060129 | thyroid-stimulating hormone-secreting cell differentiation(GO:0060129) |
| 0.0 | 0.1 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.1 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) |
| 0.0 | 0.0 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.1 | GO:0045091 | regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045091) |
| 0.0 | 0.0 | GO:0034139 | regulation of toll-like receptor 3 signaling pathway(GO:0034139) |
| 0.0 | 0.1 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 0.0 | GO:0060585 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.0 | 0.1 | GO:0001821 | histamine secretion(GO:0001821) |
| 0.0 | 0.0 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.0 | 0.1 | GO:0060083 | smooth muscle contraction involved in micturition(GO:0060083) |
| 0.0 | 0.1 | GO:0032493 | response to bacterial lipoprotein(GO:0032493) response to bacterial lipopeptide(GO:0070339) cellular response to bacterial lipoprotein(GO:0071220) cellular response to bacterial lipopeptide(GO:0071221) |
| 0.0 | 0.1 | GO:0072112 | renal filtration cell differentiation(GO:0061318) glomerular epithelium development(GO:0072010) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.0 | 0.1 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.1 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.0 | 0.1 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.0 | 0.0 | GO:0022605 | oogenesis stage(GO:0022605) |
| 0.0 | 0.0 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.0 | 0.0 | GO:0043320 | natural killer cell activation involved in immune response(GO:0002323) natural killer cell degranulation(GO:0043320) |
| 0.0 | 0.0 | GO:1903579 | negative regulation of nucleoside metabolic process(GO:0045978) negative regulation of ATP metabolic process(GO:1903579) |
| 0.0 | 0.0 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.0 | 0.0 | GO:0048631 | regulation of skeletal muscle tissue growth(GO:0048631) |
| 0.0 | 0.1 | GO:0051770 | positive regulation of nitric-oxide synthase biosynthetic process(GO:0051770) |
| 0.0 | 0.0 | GO:0010193 | response to ozone(GO:0010193) |
| 0.0 | 0.1 | GO:0060287 | epithelial cilium movement(GO:0003351) epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.0 | GO:0031946 | regulation of glucocorticoid biosynthetic process(GO:0031946) |
| 0.0 | 0.0 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.0 | 0.1 | GO:0010659 | striated muscle cell apoptotic process(GO:0010658) cardiac muscle cell apoptotic process(GO:0010659) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) regulation of cardiac muscle cell apoptotic process(GO:0010665) negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.0 | 0.1 | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.0 | 0.1 | GO:2000171 | negative regulation of dendrite development(GO:2000171) |
| 0.0 | 0.0 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.1 | GO:0009414 | response to water deprivation(GO:0009414) response to water(GO:0009415) |
| 0.0 | 0.1 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.0 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.0 | 0.0 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.0 | 0.1 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.0 | GO:0009215 | purine deoxyribonucleoside triphosphate metabolic process(GO:0009215) |
| 0.0 | 0.0 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.0 | 0.0 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.0 | 0.0 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 0.0 | GO:0046882 | negative regulation of follicle-stimulating hormone secretion(GO:0046882) |
| 0.0 | 0.0 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.1 | GO:0002903 | negative regulation of B cell apoptotic process(GO:0002903) |
| 0.0 | 0.1 | GO:0009629 | response to gravity(GO:0009629) |
| 0.0 | 0.0 | GO:0060449 | bud elongation involved in lung branching(GO:0060449) |
| 0.0 | 0.1 | GO:0006534 | cysteine metabolic process(GO:0006534) |
| 0.0 | 0.1 | GO:0010591 | regulation of lamellipodium assembly(GO:0010591) |
| 0.0 | 0.0 | GO:2000095 | regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.0 | 0.0 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.0 | GO:0016102 | retinoic acid biosynthetic process(GO:0002138) diterpenoid biosynthetic process(GO:0016102) |
| 0.0 | 0.0 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 | 0.0 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.0 | 0.1 | GO:0046835 | carbohydrate phosphorylation(GO:0046835) |
| 0.0 | 0.0 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.0 | 0.1 | GO:0042119 | neutrophil activation(GO:0042119) |
| 0.0 | 0.1 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.0 | GO:0071692 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.1 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0031313 | extrinsic component of vacuolar membrane(GO:0000306) extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.1 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.1 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.0 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.2 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.0 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.1 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.0 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.0 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.1 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.1 | GO:0005350 | pyrimidine nucleobase transmembrane transporter activity(GO:0005350) |
| 0.0 | 0.1 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.1 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.1 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.1 | GO:0019958 | C-X-C chemokine binding(GO:0019958) interleukin-8 binding(GO:0019959) |
| 0.0 | 0.1 | GO:0004516 | nicotinate phosphoribosyltransferase activity(GO:0004516) |
| 0.0 | 0.2 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.1 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.1 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.1 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.1 | GO:0042808 | obsolete neuronal Cdc2-like kinase binding(GO:0042808) |
| 0.0 | 0.1 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.1 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.0 | 0.0 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.0 | GO:0000987 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.0 | 0.1 | GO:0016565 | obsolete general transcriptional repressor activity(GO:0016565) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.1 | GO:0001846 | opsonin binding(GO:0001846) |
| 0.0 | 0.1 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.1 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.0 | 0.0 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.1 | GO:0051637 | obsolete Gram-positive bacterial cell surface binding(GO:0051637) |
| 0.0 | 0.1 | GO:0004339 | glucan 1,4-alpha-glucosidase activity(GO:0004339) |
| 0.0 | 0.2 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 0.0 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.0 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.0 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.1 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.1 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.1 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.0 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.0 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 0.1 | GO:0005536 | glucose binding(GO:0005536) |
| 0.0 | 0.0 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.0 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.0 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.0 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.0 | 0.0 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.0 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.0 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.0 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 0.0 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.0 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.1 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.0 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 0.0 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.0 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.0 | 0.0 | REACTOME PI3K EVENTS IN ERBB2 SIGNALING | Genes involved in PI3K events in ERBB2 signaling |
| 0.0 | 0.0 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.1 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.0 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |