Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
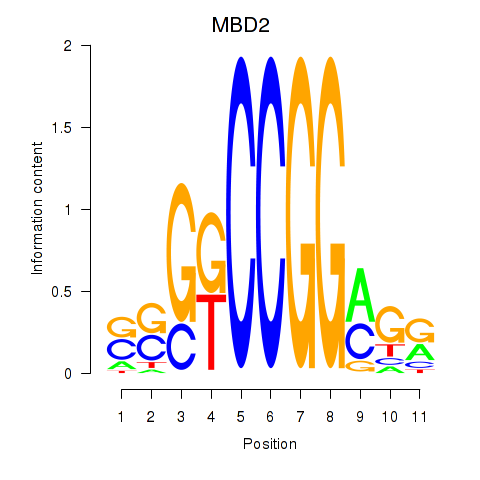
Results for MBD2
Z-value: 0.83
Transcription factors associated with MBD2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MBD2
|
ENSG00000134046.7 | methyl-CpG binding domain protein 2 |
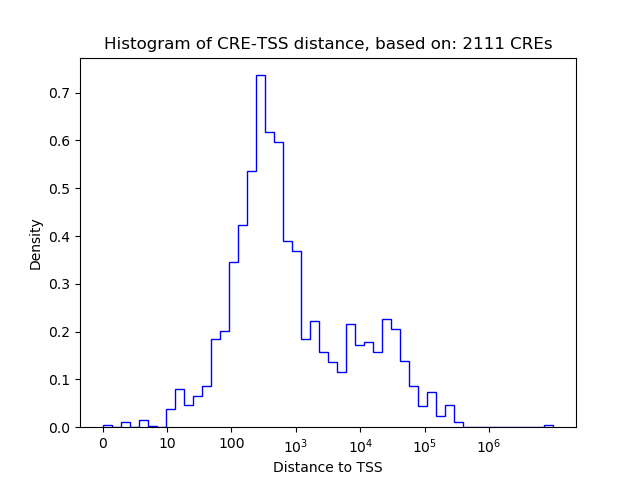
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr18_51750110_51750261 | MBD2 | 802 | 0.554668 | 0.76 | 1.7e-02 | Click! |
| chr18_51749861_51750099 | MBD2 | 1007 | 0.430976 | 0.65 | 5.6e-02 | Click! |
| chr18_51765765_51765916 | MBD2 | 14682 | 0.186836 | 0.54 | 1.4e-01 | Click! |
| chr18_51766674_51767013 | MBD2 | 15685 | 0.184618 | 0.36 | 3.4e-01 | Click! |
| chr18_51765483_51765634 | MBD2 | 14400 | 0.187460 | 0.34 | 3.7e-01 | Click! |
Activity of the MBD2 motif across conditions
Conditions sorted by the z-value of the MBD2 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
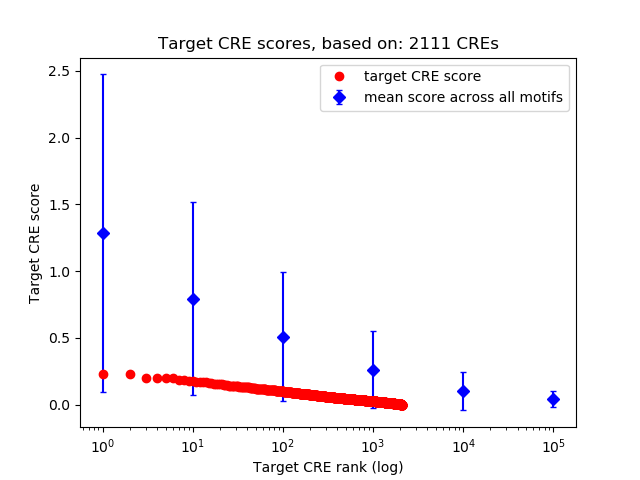
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr5_158525970_158526185 | 0.23 |
EBF1 |
early B-cell factor 1 |
624 |
0.8 |
| chr17_4047225_4047376 | 0.23 |
CYB5D2 |
cytochrome b5 domain containing 2 |
304 |
0.77 |
| chr2_227664099_227664289 | 0.20 |
IRS1 |
insulin receptor substrate 1 |
281 |
0.92 |
| chr1_36923322_36923556 | 0.20 |
MRPS15 |
mitochondrial ribosomal protein S15 |
6599 |
0.14 |
| chr2_28616465_28616616 | 0.20 |
FOSL2 |
FOS-like antigen 2 |
815 |
0.43 |
| chr12_106641074_106641225 | 0.20 |
RP11-651L5.2 |
|
415 |
0.6 |
| chr4_129731338_129731616 | 0.19 |
JADE1 |
jade family PHD finger 1 |
291 |
0.94 |
| chr1_200962213_200962376 | 0.19 |
KIF21B |
kinesin family member 21B |
30242 |
0.15 |
| chr1_28586799_28587133 | 0.18 |
SESN2 |
sestrin 2 |
928 |
0.45 |
| chr3_55521069_55521220 | 0.18 |
WNT5A |
wingless-type MMTV integration site family, member 5A |
187 |
0.84 |
| chr19_47222599_47222782 | 0.17 |
PRKD2 |
protein kinase D2 |
2306 |
0.14 |
| chr4_77871771_77872019 | 0.17 |
SEPT11 |
septin 11 |
880 |
0.67 |
| chr14_67826123_67826274 | 0.17 |
ATP6V1D |
ATPase, H+ transporting, lysosomal 34kDa, V1 subunit D |
306 |
0.65 |
| chr15_40211997_40212191 | 0.17 |
GPR176 |
G protein-coupled receptor 176 |
332 |
0.85 |
| chr10_70320624_70320793 | 0.17 |
TET1 |
tet methylcytosine dioxygenase 1 |
295 |
0.9 |
| chr6_70576954_70577105 | 0.16 |
COL19A1 |
collagen, type XIX, alpha 1 |
566 |
0.85 |
| chr11_62313253_62313404 | 0.16 |
RP11-864I4.4 |
|
143 |
0.52 |
| chr12_57483024_57483175 | 0.16 |
NAB2 |
NGFI-A binding protein 2 (EGR1 binding protein 2) |
209 |
0.88 |
| chr7_139875650_139875801 | 0.16 |
KDM7A |
lysine (K)-specific demethylase 7A |
1016 |
0.41 |
| chr8_49833855_49834028 | 0.15 |
SNAI2 |
snail family zinc finger 2 |
47 |
0.99 |
| chr9_131182814_131182965 | 0.15 |
CERCAM |
cerebral endothelial cell adhesion molecule |
129 |
0.93 |
| chr9_114659468_114659619 | 0.15 |
UGCG |
UDP-glucose ceramide glucosyltransferase |
497 |
0.81 |
| chr17_76129423_76129727 | 0.15 |
TMC6 |
transmembrane channel-like 6 |
1087 |
0.35 |
| chr2_71693709_71693860 | 0.15 |
DYSF |
dysferlin |
48 |
0.98 |
| chr5_87564263_87564414 | 0.14 |
TMEM161B |
transmembrane protein 161B |
301 |
0.56 |
| chr10_126107594_126107745 | 0.14 |
OAT |
ornithine aminotransferase |
124 |
0.96 |
| chr14_61124038_61124189 | 0.14 |
SIX1 |
SIX homeobox 1 |
864 |
0.65 |
| chr14_50778528_50778679 | 0.14 |
L2HGDH |
L-2-hydroxyglutarate dehydrogenase |
269 |
0.65 |
| chr1_169075983_169076134 | 0.14 |
ATP1B1 |
ATPase, Na+/K+ transporting, beta 1 polypeptide |
497 |
0.77 |
| chr3_52739027_52739199 | 0.14 |
SPCS1 |
signal peptidase complex subunit 1 homolog (S. cerevisiae) |
142 |
0.81 |
| chr6_139308621_139308772 | 0.14 |
REPS1 |
RALBP1 associated Eps domain containing 1 |
81 |
0.98 |
| chr17_1552892_1553043 | 0.14 |
RILP |
Rab interacting lysosomal protein |
404 |
0.69 |
| chr6_36646575_36646743 | 0.14 |
CDKN1A |
cyclin-dependent kinase inhibitor 1A (p21, Cip1) |
161 |
0.94 |
| chr5_175665590_175665741 | 0.14 |
SIMC1 |
SUMO-interacting motifs containing 1 |
262 |
0.93 |
| chr12_15941961_15942112 | 0.14 |
EPS8 |
epidermal growth factor receptor pathway substrate 8 |
279 |
0.95 |
| chr3_62304846_62304997 | 0.14 |
C3orf14 |
chromosome 3 open reading frame 14 |
203 |
0.62 |
| chr18_59561361_59561512 | 0.13 |
RNF152 |
ring finger protein 152 |
28 |
0.99 |
| chr2_37898161_37898436 | 0.13 |
CDC42EP3 |
CDC42 effector protein (Rho GTPase binding) 3 |
1036 |
0.66 |
| chr3_149689144_149689375 | 0.13 |
AC117395.1 |
LOC646903 protein; Uncharacterized protein |
193 |
0.57 |
| chr8_144329183_144329334 | 0.13 |
ZFP41 |
|
22 |
0.96 |
| chr7_42276182_42276486 | 0.13 |
GLI3 |
GLI family zinc finger 3 |
278 |
0.96 |
| chr17_48133834_48134023 | 0.13 |
ITGA3 |
integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor) |
124 |
0.95 |
| chr17_8025338_8025489 | 0.13 |
HES7 |
hes family bHLH transcription factor 7 |
1989 |
0.15 |
| chr10_12390683_12390842 | 0.13 |
CAMK1D |
calcium/calmodulin-dependent protein kinase ID |
719 |
0.67 |
| chr10_96162263_96162513 | 0.13 |
TBC1D12 |
TBC1 domain family, member 12 |
127 |
0.97 |
| chr13_95254337_95254566 | 0.13 |
GPR180 |
G protein-coupled receptor 180 |
294 |
0.91 |
| chr22_38349909_38350064 | 0.13 |
POLR2F |
polymerase (RNA) II (DNA directed) polypeptide F |
252 |
0.53 |
| chr7_99097504_99097655 | 0.12 |
ZNF394 |
zinc finger protein 394 |
308 |
0.8 |
| chr1_44878601_44878847 | 0.12 |
RNF220 |
ring finger protein 220 |
954 |
0.56 |
| chr13_37493848_37494116 | 0.12 |
SMAD9 |
SMAD family member 9 |
393 |
0.88 |
| chr1_17764676_17764827 | 0.12 |
RCC2 |
regulator of chromosome condensation 2 |
306 |
0.88 |
| chr9_139895229_139895380 | 0.12 |
CLIC3 |
chloride intracellular channel 3 |
4049 |
0.07 |
| chr16_88533909_88534060 | 0.12 |
ENSG00000263456 |
. |
1342 |
0.4 |
| chr12_124155798_124155949 | 0.12 |
TCTN2 |
tectonic family member 2 |
213 |
0.91 |
| chr8_61564342_61564493 | 0.12 |
CHD7 |
chromodomain helicase DNA binding protein 7 |
26920 |
0.22 |
| chr17_75842366_75842622 | 0.12 |
FLJ45079 |
|
36165 |
0.19 |
| chr2_97535201_97535472 | 0.12 |
SEMA4C |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4C |
372 |
0.81 |
| chr3_118959156_118959307 | 0.12 |
B4GALT4 |
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 4 |
227 |
0.92 |
| chr11_73716058_73716371 | 0.12 |
UCP3 |
uncoupling protein 3 (mitochondrial, proton carrier) |
2135 |
0.25 |
| chr12_66219604_66219755 | 0.12 |
HMGA2 |
high mobility group AT-hook 2 |
776 |
0.69 |
| chr8_128806386_128806537 | 0.12 |
ENSG00000249859 |
. |
1747 |
0.46 |
| chr20_50721480_50721631 | 0.12 |
ZFP64 |
ZFP64 zinc finger protein |
93 |
0.98 |
| chr17_73149465_73149616 | 0.11 |
HN1 |
hematological and neurological expressed 1 |
152 |
0.9 |
| chr2_232329699_232329872 | 0.11 |
NCL |
nucleolin |
480 |
0.67 |
| chr6_3157928_3158079 | 0.11 |
TUBB2A |
tubulin, beta 2A class IIa |
243 |
0.9 |
| chr2_12627750_12628043 | 0.11 |
ENSG00000207183 |
. |
76269 |
0.12 |
| chr1_12206631_12206794 | 0.11 |
TNFRSF1B |
tumor necrosis factor receptor superfamily, member 1B |
20348 |
0.14 |
| chr1_117113459_117113610 | 0.11 |
CD58 |
CD58 molecule |
114 |
0.96 |
| chr16_1664631_1664782 | 0.11 |
CRAMP1L |
Crm, cramped-like (Drosophila) |
65 |
0.94 |
| chr17_72920305_72920516 | 0.11 |
OTOP2 |
otopetrin 2 |
40 |
0.93 |
| chr8_143592312_143592463 | 0.11 |
BAI1 |
brain-specific angiogenesis inhibitor 1 |
47010 |
0.13 |
| chr14_105531218_105531594 | 0.11 |
GPR132 |
G protein-coupled receptor 132 |
361 |
0.87 |
| chr11_27721665_27721841 | 0.11 |
BDNF |
brain-derived neurotrophic factor |
282 |
0.94 |
| chr1_247267190_247267341 | 0.11 |
ZNF669 |
zinc finger protein 669 |
315 |
0.87 |
| chr1_204253357_204253508 | 0.11 |
RP11-203F10.5 |
|
7299 |
0.2 |
| chr22_50625070_50625221 | 0.11 |
TRABD |
TraB domain containing |
799 |
0.4 |
| chr15_70755158_70755354 | 0.11 |
UACA |
uveal autoantigen with coiled-coil domains and ankyrin repeats |
239364 |
0.02 |
| chr7_2079893_2080044 | 0.11 |
MAD1L1 |
MAD1 mitotic arrest deficient-like 1 (yeast) |
39680 |
0.19 |
| chr17_17307764_17307974 | 0.11 |
ENSG00000201741 |
. |
55889 |
0.1 |
| chr16_23860249_23860431 | 0.11 |
PRKCB |
protein kinase C, beta |
11796 |
0.24 |
| chr9_4297910_4298061 | 0.11 |
GLIS3 |
GLIS family zinc finger 3 |
511 |
0.74 |
| chr8_142427631_142427782 | 0.11 |
PTP4A3 |
protein tyrosine phosphatase type IVA, member 3 |
4301 |
0.15 |
| chr4_174255283_174255434 | 0.11 |
HMGB2 |
high mobility group box 2 |
42 |
0.97 |
| chr2_45236571_45236881 | 0.11 |
SIX2 |
SIX homeobox 2 |
157 |
0.97 |
| chr17_42298747_42298947 | 0.11 |
RP5-882C2.2 |
|
78 |
0.58 |
| chr16_29164996_29165147 | 0.11 |
CTB-134H23.3 |
|
46305 |
0.11 |
| chr5_98109480_98109631 | 0.10 |
RGMB |
repulsive guidance molecule family member b |
216 |
0.69 |
| chr1_9242870_9243036 | 0.10 |
ENSG00000207865 |
. |
31117 |
0.15 |
| chr1_221174763_221174914 | 0.10 |
HLX |
H2.0-like homeobox |
120254 |
0.06 |
| chr10_88516958_88517218 | 0.10 |
BMPR1A |
bone morphogenetic protein receptor, type IA |
681 |
0.52 |
| chr10_3109354_3109505 | 0.10 |
PFKP |
phosphofructokinase, platelet |
283 |
0.94 |
| chr7_139875894_139876045 | 0.10 |
KDM7A |
lysine (K)-specific demethylase 7A |
772 |
0.49 |
| chr17_27169488_27169639 | 0.10 |
FAM222B |
family with sequence similarity 222, member B |
182 |
0.87 |
| chr7_130418710_130418861 | 0.10 |
KLF14 |
Kruppel-like factor 14 |
103 |
0.97 |
| chr6_166854947_166855218 | 0.10 |
RPS6KA2-IT1 |
RPS6KA2 intronic transcript 1 (non-protein coding) |
23789 |
0.17 |
| chr1_840236_840387 | 0.10 |
SAMD11 |
sterile alpha motif domain containing 11 |
19949 |
0.1 |
| chr11_63535181_63535583 | 0.10 |
ENSG00000264519 |
. |
30161 |
0.13 |
| chr19_46102764_46102973 | 0.10 |
GPR4 |
G protein-coupled receptor 4 |
2598 |
0.14 |
| chr10_129845544_129845695 | 0.10 |
PTPRE |
protein tyrosine phosphatase, receptor type, E |
51 |
0.99 |
| chr3_9885266_9885439 | 0.10 |
RPUSD3 |
RNA pseudouridylate synthase domain containing 3 |
274 |
0.82 |
| chr8_27491503_27491654 | 0.10 |
SCARA3 |
scavenger receptor class A, member 3 |
120 |
0.96 |
| chr3_192635854_192636005 | 0.10 |
MB21D2 |
Mab-21 domain containing 2 |
21 |
0.99 |
| chr19_2841960_2842118 | 0.10 |
ZNF555 |
zinc finger protein 555 |
597 |
0.58 |
| chr3_39425233_39425481 | 0.10 |
SLC25A38 |
solute carrier family 25, member 38 |
477 |
0.75 |
| chr11_57405830_57405981 | 0.10 |
AP000662.4 |
|
56 |
0.95 |
| chr7_40174611_40174762 | 0.10 |
SUGCT |
succinylCoA:glutarate-CoA transferase |
67 |
0.86 |
| chr12_49760368_49760519 | 0.10 |
SPATS2 |
spermatogenesis associated, serine-rich 2 |
76 |
0.96 |
| chr17_79361175_79361326 | 0.10 |
RP11-1055B8.6 |
Uncharacterized protein |
8025 |
0.12 |
| chr2_239335294_239335445 | 0.10 |
ASB1 |
ankyrin repeat and SOCS box containing 1 |
96 |
0.94 |
| chr12_115121630_115121887 | 0.10 |
TBX3 |
T-box 3 |
211 |
0.95 |
| chr7_135347682_135347918 | 0.10 |
C7orf73 |
chromosome 7 open reading frame 73 |
531 |
0.73 |
| chr10_119135171_119135322 | 0.09 |
PDZD8 |
PDZ domain containing 8 |
268 |
0.93 |
| chr6_34112547_34112722 | 0.09 |
GRM4 |
glutamate receptor, metabotropic 4 |
1223 |
0.55 |
| chr1_228401023_228401174 | 0.09 |
C1orf145 |
chromosome 1 open reading frame 145 |
263 |
0.87 |
| chr13_113344676_113344827 | 0.09 |
ATP11A |
ATPase, class VI, type 11A |
108 |
0.97 |
| chr1_45808097_45808295 | 0.09 |
MUTYH |
mutY homolog |
2054 |
0.23 |
| chr7_43909566_43909799 | 0.09 |
MRPS24 |
mitochondrial ribosomal protein S24 |
526 |
0.74 |
| chr13_50017891_50018071 | 0.09 |
CAB39L |
calcium binding protein 39-like |
208 |
0.73 |
| chr8_67581847_67582257 | 0.09 |
C8orf44 |
chromosome 8 open reading frame 44 |
2165 |
0.2 |
| chr8_79578462_79578613 | 0.09 |
ZC2HC1A |
zinc finger, C2HC-type containing 1A |
255 |
0.94 |
| chr4_140477398_140477823 | 0.09 |
SETD7 |
SET domain containing (lysine methyltransferase) 7 |
218 |
0.64 |
| chr7_79764645_79764797 | 0.09 |
GNAI1 |
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 |
116 |
0.98 |
| chr12_113314531_113314682 | 0.09 |
OAS1 |
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
29976 |
0.16 |
| chrX_39864924_39865126 | 0.09 |
BCOR |
BCL6 corepressor |
57165 |
0.16 |
| chr4_140098673_140098889 | 0.09 |
ELF2 |
E74-like factor 2 (ets domain transcription factor) |
409 |
0.8 |
| chr17_71188126_71188355 | 0.09 |
COG1 |
component of oligomeric golgi complex 1 |
889 |
0.54 |
| chr17_36811230_36811381 | 0.09 |
C17orf96 |
chromosome 17 open reading frame 96 |
19882 |
0.09 |
| chr20_61604608_61604759 | 0.09 |
SLC17A9 |
solute carrier family 17 (vesicular nucleotide transporter), member 9 |
20285 |
0.14 |
| chr17_79849821_79850029 | 0.09 |
ANAPC11 |
anaphase promoting complex subunit 11 |
14 |
0.85 |
| chr2_27580084_27580240 | 0.09 |
AC074117.10 |
|
107 |
0.72 |
| chr17_3814962_3815143 | 0.09 |
P2RX1 |
purinergic receptor P2X, ligand-gated ion channel, 1 |
4742 |
0.18 |
| chr15_31619854_31620040 | 0.09 |
KLF13 |
Kruppel-like factor 13 |
889 |
0.73 |
| chr3_57581237_57581439 | 0.09 |
ARF4 |
ADP-ribosylation factor 4 |
1714 |
0.21 |
| chr13_42622874_42623025 | 0.09 |
DGKH |
diacylglycerol kinase, eta |
60 |
0.98 |
| chr6_21598054_21598402 | 0.09 |
SOX4 |
SRY (sex determining region Y)-box 4 |
4157 |
0.38 |
| chr12_42982787_42982938 | 0.09 |
PRICKLE1 |
prickle homolog 1 (Drosophila) |
616 |
0.82 |
| chr19_695715_695866 | 0.09 |
PRSS57 |
protease, serine, 57 |
330 |
0.79 |
| chr22_37628846_37629004 | 0.09 |
RAC2 |
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2) |
11363 |
0.13 |
| chr17_27920798_27921030 | 0.09 |
GIT1 |
G protein-coupled receptor kinase interacting ArfGAP 1 |
158 |
0.73 |
| chr9_133814267_133814515 | 0.09 |
FIBCD1 |
fibrinogen C domain containing 1 |
64 |
0.97 |
| chr1_45008474_45008671 | 0.09 |
ENSG00000263381 |
. |
2593 |
0.31 |
| chr1_190448073_190448224 | 0.09 |
BRINP3 |
bone morphogenetic protein/retinoic acid inducible neural-specific 3 |
1389 |
0.62 |
| chr21_47790160_47790311 | 0.09 |
PCNT |
pericentrin |
46199 |
0.1 |
| chr20_3073916_3074245 | 0.09 |
ENSG00000263905 |
. |
781 |
0.5 |
| chr3_193721076_193721316 | 0.09 |
RP11-135A1.2 |
|
127794 |
0.05 |
| chr10_105110241_105110401 | 0.09 |
PCGF6 |
polycomb group ring finger 6 |
515 |
0.7 |
| chr7_150757151_150757478 | 0.09 |
SLC4A2 |
solute carrier family 4 (anion exchanger), member 2 |
157 |
0.88 |
| chr4_109089309_109089460 | 0.09 |
LEF1 |
lymphoid enhancer-binding factor 1 |
194 |
0.86 |
| chr1_115212366_115212517 | 0.09 |
DENND2C |
DENN/MADD domain containing 2C |
215 |
0.92 |
| chr8_8243228_8243507 | 0.09 |
SGK223 |
Tyrosine-protein kinase SgK223 |
641 |
0.8 |
| chr2_8534822_8535050 | 0.08 |
AC011747.7 |
|
280960 |
0.01 |
| chr4_174292034_174292185 | 0.08 |
SAP30 |
Sin3A-associated protein, 30kDa |
15 |
0.9 |
| chr5_177580113_177580362 | 0.08 |
NHP2 |
NHP2 ribonucleoprotein |
343 |
0.87 |
| chr5_74907731_74907925 | 0.08 |
ANKDD1B |
ankyrin repeat and death domain containing 1B |
544 |
0.77 |
| chr3_142682236_142682387 | 0.08 |
PAQR9 |
progestin and adipoQ receptor family member IX |
133 |
0.82 |
| chr17_43226535_43226808 | 0.08 |
HEXIM1 |
hexamethylene bis-acetamide inducible 1 |
1987 |
0.18 |
| chr19_47231631_47231782 | 0.08 |
CTB-174O21.2 |
|
154 |
0.78 |
| chr3_55522055_55522206 | 0.08 |
WNT5A-AS1 |
WNT5A antisense RNA 1 |
403 |
0.72 |
| chr9_132312469_132312650 | 0.08 |
RP11-492E3.2 |
|
25129 |
0.14 |
| chr5_133984728_133984879 | 0.08 |
SAR1B |
SAR1 homolog B (S. cerevisiae) |
158 |
0.66 |
| chr17_44271140_44271291 | 0.08 |
KANSL1 |
KAT8 regulatory NSL complex subunit 1 |
30 |
0.73 |
| chr5_57755480_57755679 | 0.08 |
PLK2 |
polo-like kinase 2 |
508 |
0.83 |
| chr5_39424907_39425058 | 0.08 |
DAB2 |
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
12 |
0.99 |
| chr1_151131154_151131481 | 0.08 |
SCNM1 |
sodium channel modifier 1 |
2177 |
0.13 |
| chr15_70767188_70767491 | 0.08 |
UACA |
uveal autoantigen with coiled-coil domains and ankyrin repeats |
227281 |
0.02 |
| chr9_130742404_130742642 | 0.08 |
FAM102A |
family with sequence similarity 102, member A |
269 |
0.86 |
| chr11_72301686_72301842 | 0.08 |
PDE2A |
phosphodiesterase 2A, cGMP-stimulated |
301 |
0.86 |
| chr11_10328178_10328394 | 0.08 |
ADM |
adrenomedullin |
1362 |
0.36 |
| chr1_86046251_86046504 | 0.08 |
CYR61 |
cysteine-rich, angiogenic inducer, 61 |
67 |
0.97 |
| chr7_140773539_140773706 | 0.08 |
TMEM178B |
transmembrane protein 178B |
410 |
0.87 |
| chr11_74460077_74460228 | 0.08 |
RNF169 |
ring finger protein 169 |
239 |
0.89 |
| chr20_633536_633687 | 0.08 |
SRXN1 |
sulfiredoxin 1 |
403 |
0.83 |
| chr6_150263305_150263552 | 0.08 |
ULBP2 |
UL16 binding protein 2 |
292 |
0.86 |
| chr5_16935760_16935953 | 0.08 |
MYO10 |
myosin X |
191 |
0.95 |
| chr5_64776318_64776785 | 0.08 |
ADAMTS6 |
ADAM metallopeptidase with thrombospondin type 1 motif, 6 |
1153 |
0.57 |
| chr12_9949678_9949885 | 0.08 |
KLRF1 |
killer cell lectin-like receptor subfamily F, member 1 |
30296 |
0.12 |
| chr2_8451327_8451593 | 0.08 |
AC011747.7 |
|
364436 |
0.01 |
| chr14_89020572_89020999 | 0.08 |
PTPN21 |
protein tyrosine phosphatase, non-receptor type 21 |
292 |
0.87 |
| chr1_25756426_25756577 | 0.08 |
RHCE |
Rh blood group, CcEe antigens |
182 |
0.88 |
| chr5_2739627_2739778 | 0.08 |
IRX2 |
iroquois homeobox 2 |
12067 |
0.25 |
| chr7_27162967_27163166 | 0.08 |
HOXA-AS2 |
HOXA cluster antisense RNA 2 |
279 |
0.75 |
| chr3_27524774_27525101 | 0.08 |
SLC4A7 |
solute carrier family 4, sodium bicarbonate cotransporter, member 7 |
901 |
0.62 |
| chr5_1495230_1495381 | 0.08 |
LPCAT1 |
lysophosphatidylcholine acyltransferase 1 |
28787 |
0.18 |
| chr4_183839114_183839265 | 0.08 |
DCTD |
dCMP deaminase |
100 |
0.98 |
| chr1_25466873_25467076 | 0.08 |
RP4-706G24.1 |
|
67656 |
0.1 |
| chr21_45209574_45209725 | 0.08 |
RRP1 |
ribosomal RNA processing 1 |
255 |
0.9 |
| chr11_61596533_61596684 | 0.08 |
FADS1 |
fatty acid desaturase 1 |
173 |
0.86 |
| chr18_60903891_60904042 | 0.08 |
ENSG00000238988 |
. |
42068 |
0.15 |
| chrX_9983834_9983985 | 0.08 |
WWC3 |
WWC family member 3 |
307 |
0.9 |
| chr3_150263594_150263745 | 0.08 |
SERP1 |
stress-associated endoplasmic reticulum protein 1 |
617 |
0.53 |
| chr3_42814583_42814734 | 0.08 |
CCDC13 |
coiled-coil domain containing 13 |
87 |
0.94 |
| chr1_183604255_183604406 | 0.08 |
ARPC5 |
actin related protein 2/3 complex, subunit 5, 16kDa |
487 |
0.63 |
| chr4_77342405_77342556 | 0.08 |
SHROOM3 |
shroom family member 3 |
13773 |
0.15 |
| chr14_55033500_55033670 | 0.08 |
SAMD4A |
sterile alpha motif domain containing 4A |
230 |
0.95 |
| chr19_4809653_4809804 | 0.08 |
AC005523.2 |
|
14157 |
0.1 |
| chr14_58618488_58618681 | 0.08 |
C14orf37 |
chromosome 14 open reading frame 37 |
347 |
0.88 |
| chr21_47600208_47600359 | 0.08 |
SPATC1L |
spermatogenesis and centriole associated 1-like |
4090 |
0.11 |
| chr9_114361618_114361849 | 0.08 |
PTGR1 |
prostaglandin reductase 1 |
46 |
0.97 |
| chr7_50256424_50256575 | 0.08 |
AC020743.2 |
|
74080 |
0.1 |
| chr20_19997607_19997892 | 0.08 |
NAA20 |
N(alpha)-acetyltransferase 20, NatB catalytic subunit |
11 |
0.98 |
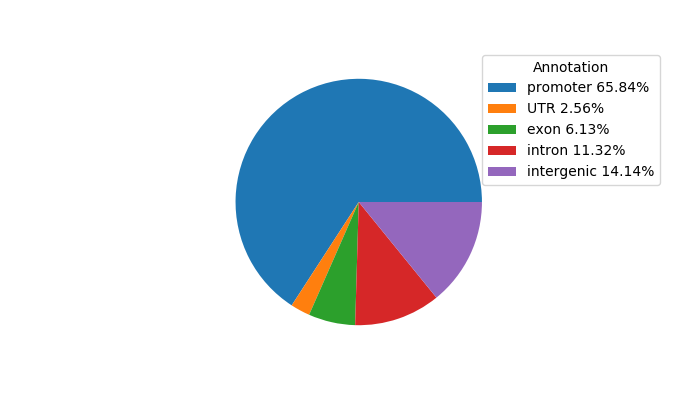
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0090273 | regulation of somatostatin secretion(GO:0090273) |
| 0.1 | 0.2 | GO:0072201 | negative regulation of mesenchymal cell proliferation(GO:0072201) |
| 0.0 | 0.1 | GO:0060831 | smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:0060831) |
| 0.0 | 0.2 | GO:0090400 | stress-induced premature senescence(GO:0090400) |
| 0.0 | 0.1 | GO:0046476 | glycosylceramide biosynthetic process(GO:0046476) |
| 0.0 | 0.1 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.0 | 0.1 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) positive regulation of hematopoietic progenitor cell differentiation(GO:1901534) |
| 0.0 | 0.1 | GO:0060596 | mammary placode formation(GO:0060596) |
| 0.0 | 0.1 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.1 | GO:0080111 | DNA demethylation(GO:0080111) |
| 0.0 | 0.1 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.1 | GO:0070296 | sarcoplasmic reticulum calcium ion transport(GO:0070296) |
| 0.0 | 0.1 | GO:0032714 | negative regulation of interleukin-5 production(GO:0032714) |
| 0.0 | 0.1 | GO:0045906 | negative regulation of vasoconstriction(GO:0045906) |
| 0.0 | 0.1 | GO:0009103 | lipopolysaccharide biosynthetic process(GO:0009103) |
| 0.0 | 0.0 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.0 | 0.1 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.1 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.1 | GO:0043570 | maintenance of DNA repeat elements(GO:0043570) |
| 0.0 | 0.1 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.0 | 0.0 | GO:0060231 | mesenchymal to epithelial transition(GO:0060231) |
| 0.0 | 0.1 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.0 | 0.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.1 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.0 | 0.1 | GO:0072033 | renal vesicle formation(GO:0072033) |
| 0.0 | 0.1 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.1 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.1 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.0 | 0.0 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.0 | 0.0 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.1 | GO:0060363 | cranial suture morphogenesis(GO:0060363) craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.1 | GO:0072273 | metanephric nephron morphogenesis(GO:0072273) |
| 0.0 | 0.0 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.0 | GO:0021631 | optic nerve morphogenesis(GO:0021631) |
| 0.0 | 0.1 | GO:0031017 | exocrine pancreas development(GO:0031017) |
| 0.0 | 0.1 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.1 | GO:2000095 | regulation of non-canonical Wnt signaling pathway(GO:2000050) regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.0 | 0.0 | GO:0006565 | L-serine catabolic process(GO:0006565) |
| 0.0 | 0.1 | GO:0021535 | cell migration in hindbrain(GO:0021535) |
| 0.0 | 0.0 | GO:0060206 | estrous cycle phase(GO:0060206) |
| 0.0 | 0.1 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.0 | 0.0 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.0 | 0.1 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) telomere maintenance in response to DNA damage(GO:0043247) |
| 0.0 | 0.1 | GO:0017085 | response to insecticide(GO:0017085) |
| 0.0 | 0.0 | GO:0048669 | collateral sprouting in absence of injury(GO:0048669) |
| 0.0 | 0.1 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 0.1 | GO:0048207 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) COPII-coated vesicle budding(GO:0090114) |
| 0.0 | 0.0 | GO:0021801 | cerebral cortex radial glia guided migration(GO:0021801) telencephalon glial cell migration(GO:0022030) |
| 0.0 | 0.0 | GO:0046398 | UDP-glucuronate biosynthetic process(GO:0006065) UDP-glucuronate metabolic process(GO:0046398) |
| 0.0 | 0.0 | GO:0046469 | platelet activating factor biosynthetic process(GO:0006663) platelet activating factor metabolic process(GO:0046469) |
| 0.0 | 0.0 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.1 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.0 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.0 | 0.1 | GO:0009186 | deoxyribonucleoside diphosphate metabolic process(GO:0009186) |
| 0.0 | 0.0 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 0.1 | GO:0009650 | UV protection(GO:0009650) |
| 0.0 | 0.1 | GO:0034643 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.0 | 0.1 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.1 | GO:0003214 | cardiac left ventricle morphogenesis(GO:0003214) |
| 0.0 | 0.0 | GO:0046606 | negative regulation of centrosome duplication(GO:0010826) negative regulation of centrosome cycle(GO:0046606) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.1 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.1 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.1 | GO:0005954 | calcium- and calmodulin-dependent protein kinase complex(GO:0005954) |
| 0.0 | 0.1 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.1 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.1 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.1 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.1 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.1 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) ribonuclease P complex(GO:0030677) multimeric ribonuclease P complex(GO:0030681) |
| 0.0 | 0.1 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.0 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.0 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.1 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.1 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.0 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.1 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.0 | 0.1 | GO:0031105 | septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.2 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.1 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.1 | GO:0008061 | chitin binding(GO:0008061) |
| 0.0 | 0.1 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.3 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.1 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.1 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.0 | 0.1 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.1 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.2 | GO:0016565 | obsolete general transcriptional repressor activity(GO:0016565) |
| 0.0 | 0.2 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.0 | 0.1 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.0 | 0.1 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.1 | GO:0016717 | oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water(GO:0016717) |
| 0.0 | 0.1 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.1 | GO:0003701 | obsolete RNA polymerase I transcription factor activity(GO:0003701) |
| 0.0 | 0.1 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.0 | 0.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.1 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.1 | GO:0032357 | single guanine insertion binding(GO:0032142) oxidized DNA binding(GO:0032356) oxidized purine DNA binding(GO:0032357) |
| 0.0 | 0.1 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.0 | 0.1 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.1 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 0.0 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.1 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.1 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.1 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 0.0 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.0 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.0 | 0.1 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.0 | 0.1 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 0.1 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.0 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.0 | 0.0 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.0 | GO:0055103 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.1 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
| 0.0 | 0.1 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.0 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.1 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.0 | 0.0 | GO:0070735 | protein-glycine ligase activity(GO:0070735) |
| 0.0 | 0.0 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.1 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.0 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.0 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.0 | GO:0031544 | procollagen-proline 3-dioxygenase activity(GO:0019797) peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.0 | 0.0 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.1 | GO:0004526 | ribonuclease P activity(GO:0004526) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.3 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 0.2 | PID ALK2 PATHWAY | ALK2 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.2 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.2 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.1 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.3 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.1 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |