Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
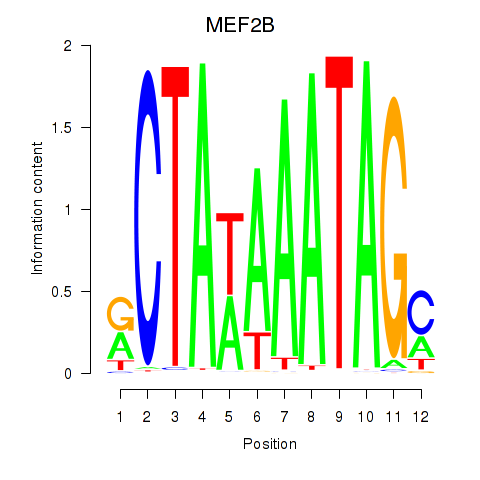
Results for MEF2B
Z-value: 1.10
Transcription factors associated with MEF2B
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MEF2B
|
ENSG00000213999.11 | myocyte enhancer factor 2B |
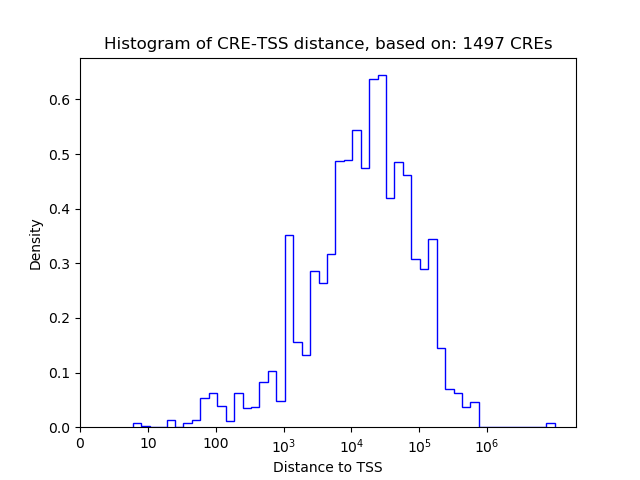
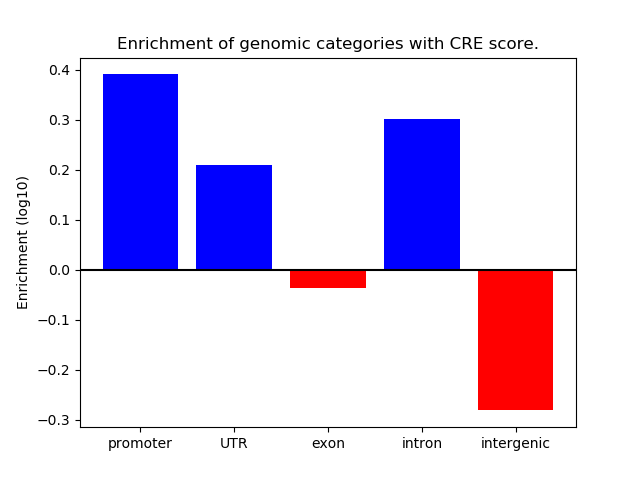
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr19_19271531_19271682 | MEF2B | 9448 | 0.106236 | 0.79 | 1.2e-02 | Click! |
| chr19_19266537_19266688 | MEF2B | 14442 | 0.102133 | 0.74 | 2.4e-02 | Click! |
| chr19_19270273_19270424 | MEF2B | 10706 | 0.105019 | 0.52 | 1.5e-01 | Click! |
| chr19_19282891_19283042 | MEF2B | 1868 | 0.198439 | 0.51 | 1.6e-01 | Click! |
| chr19_19271102_19271312 | MEF2B | 9847 | 0.105821 | 0.46 | 2.1e-01 | Click! |
Activity of the MEF2B motif across conditions
Conditions sorted by the z-value of the MEF2B motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
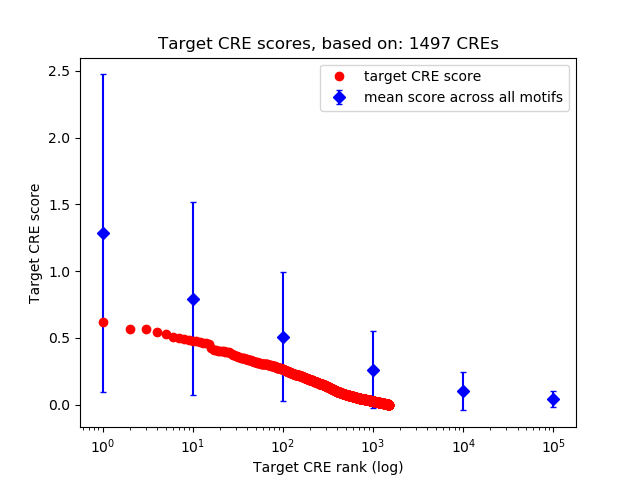
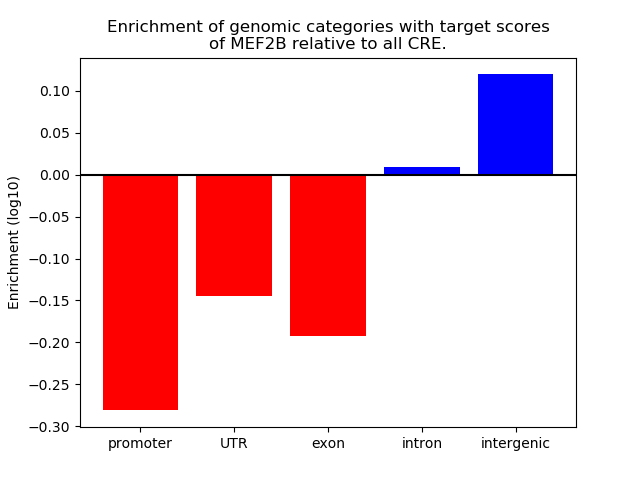
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
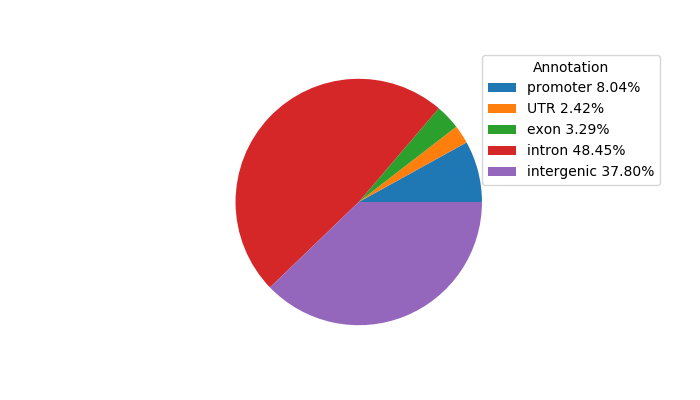
| chr1_83043773_83044235 | 0.62 |
LPHN2 |
latrophilin 2 |
598431 |
0.0 |
| chr10_114786502_114786745 | 0.57 |
RP11-57H14.2 |
|
74989 |
0.11 |
| chr6_85293592_85293821 | 0.57 |
RP11-132M7.3 |
|
105437 |
0.08 |
| chr3_177053679_177054041 | 0.55 |
TBL1XR1 |
transducin (beta)-like 1 X-linked receptor 1 |
138599 |
0.05 |
| chr13_101149154_101149445 | 0.53 |
PCCA-AS1 |
PCCA antisense RNA 1 |
15888 |
0.18 |
| chr20_23132592_23133523 | 0.51 |
ENSG00000201527 |
. |
8551 |
0.22 |
| chr13_30627332_30627483 | 0.50 |
LINC00572 |
long intergenic non-protein coding RNA 572 |
126619 |
0.06 |
| chr6_114130369_114130564 | 0.50 |
ENSG00000253091 |
. |
27799 |
0.16 |
| chr6_25179466_25179765 | 0.48 |
ENSG00000222373 |
. |
13026 |
0.19 |
| chr1_168467710_168467921 | 0.48 |
XCL2 |
chemokine (C motif) ligand 2 |
45420 |
0.16 |
| chr3_63908528_63908748 | 0.48 |
ATXN7 |
ataxin 7 |
10363 |
0.18 |
| chr4_86747445_86747851 | 0.47 |
ARHGAP24 |
Rho GTPase activating protein 24 |
1250 |
0.59 |
| chr1_184476924_184477075 | 0.46 |
GS1-115G20.1 |
|
99528 |
0.07 |
| chr5_148796784_148796935 | 0.46 |
ENSG00000208035 |
. |
11622 |
0.13 |
| chr1_50575855_50576371 | 0.45 |
ELAVL4 |
ELAV like neuron-specific RNA binding protein 4 |
735 |
0.74 |
| chr14_56777353_56777504 | 0.43 |
TMEM260 |
transmembrane protein 260 |
177644 |
0.03 |
| chr1_61299199_61299350 | 0.41 |
NFIA |
nuclear factor I/A |
31657 |
0.24 |
| chr5_124237918_124238155 | 0.41 |
ZNF608 |
zinc finger protein 608 |
153536 |
0.04 |
| chr1_234656705_234656856 | 0.41 |
TARBP1 |
TAR (HIV-1) RNA binding protein 1 |
41931 |
0.14 |
| chr3_48597793_48598316 | 0.40 |
PFKFB4 |
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 |
508 |
0.63 |
| chr2_173258743_173259031 | 0.40 |
ITGA6 |
integrin, alpha 6 |
33195 |
0.16 |
| chr16_73200279_73201088 | 0.40 |
C16orf47 |
chromosome 16 open reading frame 47 |
22337 |
0.26 |
| chr11_69264083_69264234 | 0.40 |
CCND1 |
cyclin D1 |
191697 |
0.03 |
| chr15_42651493_42651677 | 0.40 |
CAPN3 |
calpain 3, (p94) |
198 |
0.94 |
| chr6_37519897_37520048 | 0.40 |
ENSG00000263926 |
. |
3226 |
0.29 |
| chr16_75086975_75087126 | 0.39 |
ZNRF1 |
zinc and ring finger 1, E3 ubiquitin protein ligase |
50855 |
0.11 |
| chr20_3358805_3359063 | 0.38 |
ENSG00000201346 |
. |
5942 |
0.16 |
| chr8_17547050_17547201 | 0.37 |
ENSG00000265520 |
. |
7959 |
0.17 |
| chr2_208896310_208896486 | 0.37 |
PLEKHM3 |
pleckstrin homology domain containing, family M, member 3 |
6114 |
0.15 |
| chr21_39193991_39194257 | 0.37 |
KCNJ6 |
potassium inwardly-rectifying channel, subfamily J, member 6 |
91433 |
0.08 |
| chr8_120627310_120627516 | 0.36 |
ENPP2 |
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
22165 |
0.24 |
| chr8_108316239_108316473 | 0.36 |
ANGPT1 |
angiopoietin 1 |
32394 |
0.22 |
| chr2_158697443_158697594 | 0.35 |
ACVR1 |
activin A receptor, type I |
2679 |
0.28 |
| chr12_3764521_3764672 | 0.35 |
EFCAB4B |
EF-hand calcium binding domain 4B |
1207 |
0.6 |
| chr5_75367452_75367603 | 0.35 |
SV2C |
synaptic vesicle glycoprotein 2C |
11470 |
0.24 |
| chr14_105721504_105721672 | 0.35 |
BRF1 |
BRF1, RNA polymerase III transcription initiation factor 90 kDa subunit |
5847 |
0.17 |
| chr17_6503024_6503317 | 0.35 |
ENSG00000200914 |
. |
1201 |
0.36 |
| chr11_124595331_124595482 | 0.35 |
NRGN |
neurogranin (protein kinase C substrate, RC3) |
14336 |
0.1 |
| chr14_39317605_39317829 | 0.34 |
ENSG00000199285 |
. |
138768 |
0.05 |
| chr8_97803430_97803993 | 0.34 |
CPQ |
carboxypeptidase Q |
30243 |
0.25 |
| chr17_66315705_66315870 | 0.34 |
ARSG |
arylsulfatase G |
28128 |
0.15 |
| chr1_157745151_157745302 | 0.34 |
FCRL2 |
Fc receptor-like 2 |
1683 |
0.39 |
| chr15_39850731_39850882 | 0.33 |
THBS1 |
thrombospondin 1 |
22474 |
0.19 |
| chr12_12032698_12032849 | 0.33 |
ETV6 |
ets variant 6 |
6098 |
0.32 |
| chr1_68056079_68056230 | 0.33 |
ENSG00000207504 |
. |
49344 |
0.14 |
| chr9_101803098_101803249 | 0.33 |
TGFBR1 |
transforming growth factor, beta receptor 1 |
63147 |
0.11 |
| chr2_113429205_113429356 | 0.33 |
SLC20A1 |
solute carrier family 20 (phosphate transporter), member 1 |
24001 |
0.17 |
| chr12_15842783_15842934 | 0.32 |
ENSG00000200105 |
. |
18114 |
0.22 |
| chr7_29283267_29283581 | 0.32 |
AC004593.3 |
|
34838 |
0.16 |
| chr2_202819063_202819214 | 0.32 |
ENSG00000221489 |
. |
1079 |
0.52 |
| chr2_43443107_43443258 | 0.32 |
ZFP36L2 |
ZFP36 ring finger protein-like 2 |
10566 |
0.23 |
| chr22_27714638_27714789 | 0.32 |
ENSG00000200443 |
. |
282863 |
0.01 |
| chr16_84413717_84413987 | 0.31 |
ATP2C2 |
ATPase, Ca++ transporting, type 2C, member 2 |
11719 |
0.19 |
| chr5_75722311_75722532 | 0.31 |
IQGAP2 |
IQ motif containing GTPase activating protein 2 |
22172 |
0.25 |
| chr3_171038529_171038680 | 0.31 |
TNIK |
TRAF2 and NCK interacting kinase |
95104 |
0.08 |
| chr5_74615457_74615613 | 0.31 |
HMGCR |
3-hydroxy-3-methylglutaryl-CoA reductase |
16619 |
0.19 |
| chr2_155312519_155312670 | 0.31 |
AC009227.2 |
|
1356 |
0.52 |
| chr4_88832400_88832551 | 0.31 |
SPP1 |
secreted phosphoprotein 1 |
64344 |
0.1 |
| chr7_129630529_129630680 | 0.31 |
ENSG00000263557 |
. |
5017 |
0.14 |
| chr11_34469686_34469883 | 0.31 |
CAT |
catalase |
9312 |
0.22 |
| chr16_46689835_46690025 | 0.30 |
RP11-93O14.2 |
|
5573 |
0.18 |
| chr1_235051769_235052023 | 0.30 |
ENSG00000239690 |
. |
11963 |
0.27 |
| chr12_11932326_11932519 | 0.30 |
ETV6 |
ets variant 6 |
26987 |
0.25 |
| chr14_85775279_85775646 | 0.30 |
ENSG00000252529 |
. |
37186 |
0.24 |
| chr3_149160811_149161262 | 0.30 |
TM4SF4 |
transmembrane 4 L six family member 4 |
30725 |
0.16 |
| chr20_17649733_17649884 | 0.30 |
RRBP1 |
ribosome binding protein 1 |
8656 |
0.21 |
| chr19_19245450_19245992 | 0.30 |
TMEM161A |
transmembrane protein 161A |
2511 |
0.19 |
| chr3_148477195_148477534 | 0.30 |
AGTR1 |
angiotensin II receptor, type 1 |
19779 |
0.22 |
| chr9_124123057_124123675 | 0.30 |
STOM |
stomatin |
9117 |
0.15 |
| chr12_49626641_49626792 | 0.30 |
TUBA1C |
tubulin, alpha 1c |
5007 |
0.14 |
| chr2_55345739_55346346 | 0.29 |
RTN4 |
reticulon 4 |
6285 |
0.2 |
| chr13_91415783_91415934 | 0.29 |
ENSG00000251753 |
. |
18265 |
0.31 |
| chr14_73881705_73881856 | 0.29 |
NUMB |
numb homolog (Drosophila) |
5001 |
0.21 |
| chr2_218766852_218767096 | 0.29 |
TNS1 |
tensin 1 |
242 |
0.95 |
| chr3_140795767_140796225 | 0.29 |
SPSB4 |
splA/ryanodine receptor domain and SOCS box containing 4 |
25752 |
0.2 |
| chr12_392843_392994 | 0.29 |
RP11-283I3.6 |
|
6629 |
0.16 |
| chr7_70907526_70907677 | 0.29 |
ENSG00000266688 |
. |
132064 |
0.05 |
| chr14_56683663_56683814 | 0.29 |
PELI2 |
pellino E3 ubiquitin protein ligase family member 2 |
97911 |
0.08 |
| chr2_33682944_33683139 | 0.29 |
RASGRP3 |
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
72 |
0.98 |
| chr4_55555755_55555906 | 0.29 |
KIT |
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog |
31745 |
0.25 |
| chr1_201524755_201524906 | 0.29 |
CSRP1 |
cysteine and glycine-rich protein 1 |
46246 |
0.1 |
| chr20_52352229_52352983 | 0.29 |
ENSG00000238468 |
. |
67309 |
0.12 |
| chr22_24906519_24906756 | 0.29 |
AP000355.2 |
|
5839 |
0.15 |
| chr1_231242166_231242317 | 0.28 |
TRIM67 |
tripartite motif containing 67 |
55617 |
0.1 |
| chr15_25916381_25916532 | 0.28 |
ATP10A |
ATPase, class V, type 10A |
23516 |
0.19 |
| chr2_178332517_178332770 | 0.28 |
AGPS |
alkylglycerone phosphate synthase |
75147 |
0.07 |
| chr11_94502979_94503226 | 0.28 |
AMOTL1 |
angiomotin like 1 |
1565 |
0.45 |
| chr13_31774982_31775836 | 0.28 |
B3GALTL |
beta 1,3-galactosyltransferase-like |
1336 |
0.55 |
| chr8_56805555_56805843 | 0.28 |
RP11-318K15.2 |
|
455 |
0.77 |
| chr7_75340828_75341228 | 0.28 |
HIP1 |
huntingtin interacting protein 1 |
27231 |
0.18 |
| chr11_35936641_35936792 | 0.28 |
RP11-698N11.2 |
|
3552 |
0.24 |
| chr8_23830707_23830858 | 0.27 |
ENSG00000207201 |
. |
103889 |
0.07 |
| chr2_149372066_149372403 | 0.27 |
EPC2 |
enhancer of polycomb homolog 2 (Drosophila) |
29775 |
0.24 |
| chr6_108724802_108724953 | 0.27 |
ENSG00000200799 |
. |
11600 |
0.25 |
| chr2_216363831_216363982 | 0.27 |
AC012462.1 |
|
62930 |
0.13 |
| chr6_14639404_14639617 | 0.27 |
ENSG00000206960 |
. |
7256 |
0.34 |
| chr17_66576104_66576255 | 0.27 |
FAM20A |
family with sequence similarity 20, member A |
21351 |
0.19 |
| chr18_2068827_2069125 | 0.27 |
METTL4 |
methyltransferase like 4 |
485715 |
0.0 |
| chr11_3951087_3951238 | 0.27 |
ENSG00000206976 |
. |
7365 |
0.17 |
| chr12_66269655_66269806 | 0.27 |
RP11-366L20.2 |
Uncharacterized protein |
5628 |
0.2 |
| chr3_149050046_149050197 | 0.26 |
TM4SF18 |
transmembrane 4 L six family member 18 |
1148 |
0.5 |
| chr10_60494716_60495008 | 0.26 |
BICC1 |
bicaudal C homolog 1 (Drosophila) |
58433 |
0.16 |
| chrX_9057871_9058145 | 0.26 |
ENSG00000200852 |
. |
43861 |
0.18 |
| chr5_149424420_149424712 | 0.26 |
ENSG00000238369 |
. |
15551 |
0.14 |
| chr2_178603600_178603751 | 0.26 |
AC012499.1 |
|
40457 |
0.17 |
| chr6_73470213_73470364 | 0.26 |
KCNQ5-IT1 |
KCNQ5 intronic transcript 1 (non-protein coding) |
130065 |
0.05 |
| chr7_75341580_75341927 | 0.26 |
HIP1 |
huntingtin interacting protein 1 |
26506 |
0.18 |
| chr16_84770743_84770894 | 0.26 |
USP10 |
ubiquitin specific peptidase 10 |
31053 |
0.17 |
| chr17_74102777_74103137 | 0.25 |
EXOC7 |
exocyst complex component 7 |
3089 |
0.15 |
| chr12_70713258_70713711 | 0.25 |
CNOT2 |
CCR4-NOT transcription complex, subunit 2 |
12694 |
0.23 |
| chr4_186842803_186842954 | 0.25 |
ENSG00000239034 |
. |
24359 |
0.19 |
| chr13_78734998_78735287 | 0.25 |
RNF219-AS1 |
RNF219 antisense RNA 1 |
105852 |
0.08 |
| chr12_46867069_46867237 | 0.25 |
SLC38A2 |
solute carrier family 38, member 2 |
100503 |
0.08 |
| chr6_16903564_16904059 | 0.25 |
RP1-151F17.1 |
|
141668 |
0.05 |
| chr2_225812839_225813395 | 0.24 |
DOCK10 |
dedicator of cytokinesis 10 |
1335 |
0.59 |
| chr11_94661181_94661856 | 0.24 |
RP11-856F16.2 |
|
2832 |
0.29 |
| chr11_43855503_43855654 | 0.24 |
RP11-613D13.5 |
|
86 |
0.98 |
| chr7_114659768_114659936 | 0.24 |
MDFIC |
MyoD family inhibitor domain containing |
85928 |
0.11 |
| chr12_89340554_89340705 | 0.24 |
ENSG00000238302 |
. |
335433 |
0.01 |
| chr3_143676802_143676953 | 0.24 |
C3orf58 |
chromosome 3 open reading frame 58 |
13763 |
0.3 |
| chr1_239905020_239905171 | 0.24 |
ENSG00000233355 |
. |
11330 |
0.25 |
| chr21_17584166_17584317 | 0.24 |
ENSG00000201025 |
. |
72848 |
0.13 |
| chr11_129313202_129313353 | 0.24 |
BARX2 |
BARX homeobox 2 |
1301 |
0.61 |
| chr3_188139903_188140281 | 0.24 |
LPP-AS1 |
LPP antisense RNA 1 |
146362 |
0.04 |
| chr9_124122125_124122321 | 0.24 |
STOM |
stomatin |
10260 |
0.15 |
| chr22_30586930_30587243 | 0.23 |
RP3-438O4.4 |
|
16012 |
0.14 |
| chr8_131307946_131308498 | 0.23 |
ASAP1 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1 |
44961 |
0.19 |
| chr6_140204099_140204250 | 0.23 |
ENSG00000252107 |
. |
275657 |
0.02 |
| chr2_28556445_28556596 | 0.23 |
AC093690.1 |
|
23194 |
0.18 |
| chr2_30644578_30644729 | 0.23 |
ENSG00000221377 |
. |
10661 |
0.24 |
| chr8_82265328_82265556 | 0.23 |
RP11-363E6.3 |
|
71761 |
0.09 |
| chrX_38468479_38468630 | 0.23 |
ENSG00000238606 |
. |
1086 |
0.63 |
| chr2_169506366_169506517 | 0.23 |
ENSG00000199348 |
. |
43435 |
0.16 |
| chr15_42897406_42897557 | 0.23 |
STARD9 |
StAR-related lipid transfer (START) domain containing 9 |
29624 |
0.13 |
| chr1_55828154_55828305 | 0.23 |
ENSG00000199831 |
. |
14296 |
0.23 |
| chr1_161743421_161743663 | 0.23 |
ATF6 |
activating transcription factor 6 |
7458 |
0.13 |
| chr7_96467239_96467390 | 0.23 |
ENSG00000244318 |
. |
102362 |
0.07 |
| chr4_86629687_86629838 | 0.22 |
ENSG00000266421 |
. |
13859 |
0.27 |
| chr10_18498286_18498555 | 0.22 |
CACNB2 |
calcium channel, voltage-dependent, beta 2 subunit |
51278 |
0.15 |
| chr6_120007930_120008081 | 0.22 |
ENSG00000265725 |
. |
328320 |
0.01 |
| chr2_158163071_158163471 | 0.22 |
ENSG00000222404 |
. |
7601 |
0.22 |
| chr6_143023055_143023206 | 0.22 |
RP1-67K17.3 |
|
46450 |
0.19 |
| chr2_68543161_68543312 | 0.22 |
CNRIP1 |
cannabinoid receptor interacting protein 1 |
3296 |
0.21 |
| chr4_41180087_41180238 | 0.22 |
APBB2 |
amyloid beta (A4) precursor protein-binding, family B, member 2 |
36313 |
0.15 |
| chr21_34613920_34614547 | 0.22 |
AP000295.9 |
|
4846 |
0.16 |
| chr14_22950410_22950822 | 0.22 |
TRAJ55 |
T cell receptor alpha joining 55 (pseudogene) |
70 |
0.8 |
| chr20_2901732_2901883 | 0.22 |
PTPRA |
protein tyrosine phosphatase, receptor type, A |
2046 |
0.31 |
| chr4_71503415_71503566 | 0.22 |
ENAM |
enamelin |
9029 |
0.14 |
| chr10_34490797_34490948 | 0.22 |
ENSG00000199200 |
. |
130 |
0.98 |
| chr5_138542726_138542908 | 0.22 |
SIL1 |
SIL1 nucleotide exchange factor |
8730 |
0.16 |
| chr12_123568411_123568661 | 0.22 |
PITPNM2 |
phosphatidylinositol transfer protein, membrane-associated 2 |
2656 |
0.27 |
| chrX_64748269_64748420 | 0.22 |
LAS1L |
LAS1-like (S. cerevisiae) |
4847 |
0.28 |
| chr4_48207485_48207636 | 0.22 |
ENSG00000202014 |
. |
52109 |
0.12 |
| chr7_2843960_2844111 | 0.21 |
GNA12 |
guanine nucleotide binding protein (G protein) alpha 12 |
10857 |
0.26 |
| chr15_102040487_102040709 | 0.21 |
PCSK6 |
proprotein convertase subtilisin/kexin type 6 |
10411 |
0.26 |
| chr17_42247977_42248128 | 0.21 |
ASB16 |
ankyrin repeat and SOCS box containing 16 |
22 |
0.94 |
| chr4_9929319_9929470 | 0.21 |
RP13-560N11.1 |
|
4956 |
0.26 |
| chr2_217955017_217955405 | 0.21 |
ENSG00000251849 |
. |
86010 |
0.09 |
| chr1_183578719_183579088 | 0.21 |
NCF2 |
neutrophil cytosolic factor 2 |
18892 |
0.16 |
| chr2_118861943_118862094 | 0.21 |
INSIG2 |
insulin induced gene 2 |
15968 |
0.23 |
| chr22_50330984_50331151 | 0.21 |
CRELD2 |
cysteine-rich with EGF-like domains 2 |
18686 |
0.15 |
| chr16_88754480_88754631 | 0.21 |
SNAI3 |
snail family zinc finger 3 |
1654 |
0.16 |
| chr8_66507426_66507577 | 0.21 |
ENSG00000200714 |
. |
2535 |
0.33 |
| chr5_90410370_90410521 | 0.21 |
ENSG00000199643 |
. |
156099 |
0.04 |
| chr12_118784249_118784708 | 0.21 |
TAOK3 |
TAO kinase 3 |
12432 |
0.24 |
| chr16_88448984_88449499 | 0.21 |
ZNF469 |
zinc finger protein 469 |
44638 |
0.14 |
| chr16_69601308_69601964 | 0.21 |
NFAT5 |
nuclear factor of activated T-cells 5, tonicity-responsive |
1382 |
0.36 |
| chr2_65633649_65633902 | 0.21 |
SPRED2 |
sprouty-related, EVH1 domain containing 2 |
25536 |
0.19 |
| chr1_112462912_112463063 | 0.21 |
KCND3-AS1 |
KCND3 antisense RNA 1 |
10680 |
0.23 |
| chr16_48478901_48479413 | 0.21 |
RP11-44I10.6 |
|
3152 |
0.22 |
| chr10_71239313_71239464 | 0.20 |
TSPAN15 |
tetraspanin 15 |
4236 |
0.26 |
| chr16_11195081_11195232 | 0.20 |
CLEC16A |
C-type lectin domain family 16, member A |
22444 |
0.16 |
| chr6_16824855_16825024 | 0.20 |
RP1-151F17.1 |
|
62796 |
0.13 |
| chr22_24186722_24186873 | 0.20 |
DERL3 |
derlin 3 |
5598 |
0.1 |
| chr14_103121720_103121871 | 0.20 |
ENSG00000202459 |
. |
39930 |
0.14 |
| chr1_81677570_81677938 | 0.20 |
ENSG00000223026 |
. |
39720 |
0.21 |
| chr6_150131484_150131635 | 0.20 |
RP11-350J20.5 |
|
12251 |
0.13 |
| chr6_82970810_82970961 | 0.20 |
IBTK |
inhibitor of Bruton agammaglobulinemia tyrosine kinase |
13414 |
0.26 |
| chr1_25297951_25298189 | 0.20 |
RUNX3 |
runt-related transcription factor 3 |
6569 |
0.21 |
| chr2_65138467_65138618 | 0.20 |
ENSG00000238696 |
. |
2722 |
0.24 |
| chrX_117632578_117633370 | 0.20 |
DOCK11 |
dedicator of cytokinesis 11 |
3102 |
0.33 |
| chr14_75665184_75665335 | 0.20 |
ENSG00000252013 |
. |
3613 |
0.19 |
| chr10_98429524_98429700 | 0.20 |
PIK3AP1 |
phosphoinositide-3-kinase adaptor protein 1 |
244 |
0.94 |
| chr20_52715129_52715347 | 0.20 |
BCAS1 |
breast carcinoma amplified sequence 1 |
27934 |
0.18 |
| chr14_70921495_70921646 | 0.20 |
ADAM21 |
ADAM metallopeptidase domain 21 |
2647 |
0.27 |
| chr4_79578557_79578708 | 0.19 |
ENSG00000238816 |
. |
17378 |
0.2 |
| chr13_31381021_31381172 | 0.19 |
ALOX5AP |
arachidonate 5-lipoxygenase-activating protein |
71451 |
0.11 |
| chr5_74465276_74465427 | 0.19 |
ANKRD31 |
ankyrin repeat domain 31 |
67352 |
0.12 |
| chr2_171921049_171921298 | 0.19 |
TLK1 |
tousled-like kinase 1 |
2273 |
0.4 |
| chr5_160121300_160121451 | 0.19 |
CTC-529G1.1 |
|
9017 |
0.21 |
| chr3_143085241_143085392 | 0.19 |
SLC9A9-AS2 |
SLC9A9 antisense RNA 2 |
14864 |
0.21 |
| chr21_19260646_19260797 | 0.19 |
ENSG00000221335 |
. |
3307 |
0.27 |
| chr13_45553454_45553605 | 0.19 |
NUFIP1 |
nuclear fragile X mental retardation protein interacting protein 1 |
10089 |
0.18 |
| chr4_71814858_71815009 | 0.19 |
DCK |
deoxycytidine kinase |
43322 |
0.14 |
| chr12_9268627_9268778 | 0.19 |
A2M |
alpha-2-macroglobulin |
51 |
0.97 |
| chr22_51016890_51017529 | 0.19 |
CPT1B |
carnitine palmitoyltransferase 1B (muscle) |
113 |
0.91 |
| chr2_238570999_238571408 | 0.19 |
LRRFIP1 |
leucine rich repeat (in FLII) interacting protein 1 |
29585 |
0.17 |
| chr7_121012833_121012984 | 0.19 |
FAM3C |
family with sequence similarity 3, member C |
23454 |
0.22 |
| chr2_204553524_204553955 | 0.19 |
CD28 |
CD28 molecule |
17459 |
0.24 |
| chr12_105013205_105013356 | 0.19 |
ENSG00000264295 |
. |
27869 |
0.2 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) regulation of store-operated calcium channel activity(GO:1901339) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.1 | 0.3 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.1 | GO:0042637 | catagen(GO:0042637) |
| 0.0 | 0.2 | GO:0045916 | negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 0.0 | 0.5 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.0 | 0.1 | GO:0021801 | cerebral cortex radial glia guided migration(GO:0021801) telencephalon glial cell migration(GO:0022030) |
| 0.0 | 0.1 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.0 | 0.1 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.1 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.0 | 0.1 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.1 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.1 | GO:0003078 | obsolete regulation of natriuresis(GO:0003078) |
| 0.0 | 0.1 | GO:0007028 | cytoplasm organization(GO:0007028) |
| 0.0 | 0.2 | GO:1902603 | carnitine shuttle(GO:0006853) carnitine transmembrane transport(GO:1902603) |
| 0.0 | 0.1 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.0 | 0.1 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.1 | GO:0006573 | valine metabolic process(GO:0006573) |
| 0.0 | 0.1 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.1 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.1 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.0 | 0.1 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.0 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.0 | 0.0 | GO:0032898 | neurotrophin production(GO:0032898) |
| 0.0 | 0.1 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.0 | 0.0 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.0 | 0.1 | GO:0070874 | negative regulation of glycogen metabolic process(GO:0070874) |
| 0.0 | 0.0 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.0 | 0.1 | GO:0030239 | myofibril assembly(GO:0030239) |
| 0.0 | 0.1 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.0 | 0.1 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.0 | GO:0003056 | regulation of vascular smooth muscle contraction(GO:0003056) |
| 0.0 | 0.0 | GO:0090273 | regulation of somatostatin secretion(GO:0090273) |
| 0.0 | 0.0 | GO:0033131 | regulation of glucokinase activity(GO:0033131) positive regulation of glucokinase activity(GO:0033133) regulation of hexokinase activity(GO:1903299) positive regulation of hexokinase activity(GO:1903301) |
| 0.0 | 0.0 | GO:0014874 | response to stimulus involved in regulation of muscle adaptation(GO:0014874) |
| 0.0 | 0.0 | GO:0000492 | small nucleolar ribonucleoprotein complex assembly(GO:0000491) box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.0 | GO:0002538 | arachidonic acid metabolite production involved in inflammatory response(GO:0002538) leukotriene production involved in inflammatory response(GO:0002540) |
| 0.0 | 0.1 | GO:0046504 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.0 | GO:0002072 | optic cup morphogenesis involved in camera-type eye development(GO:0002072) |
| 0.0 | 0.0 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.1 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.0 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.0 | 0.0 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.0 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.1 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.1 | 0.2 | GO:0019958 | C-X-C chemokine binding(GO:0019958) interleukin-8 binding(GO:0019959) |
| 0.0 | 0.2 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.2 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.3 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.1 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.4 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.3 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.1 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.0 | 0.1 | GO:0004075 | biotin carboxylase activity(GO:0004075) |
| 0.0 | 0.0 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.0 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.0 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.1 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.0 | 0.0 | GO:1901474 | azole transporter activity(GO:0045118) azole transmembrane transporter activity(GO:1901474) |
| 0.0 | 0.1 | GO:0050543 | icosatetraenoic acid binding(GO:0050543) |
| 0.0 | 0.0 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.0 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.1 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.1 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 0.0 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.0 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.0 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.1 | GO:0001595 | angiotensin receptor activity(GO:0001595) angiotensin type II receptor activity(GO:0004945) |
| 0.0 | 0.2 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.0 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.0 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.0 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.1 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.2 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.2 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |