Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
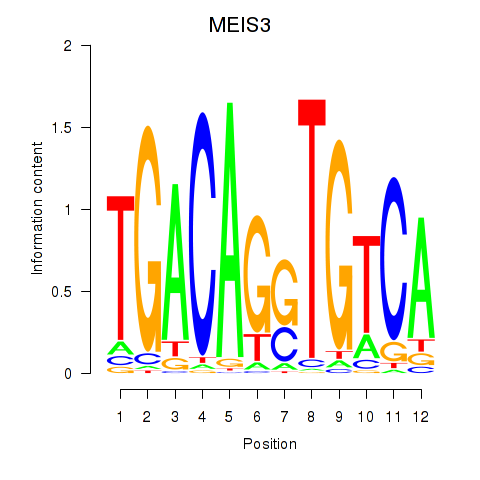
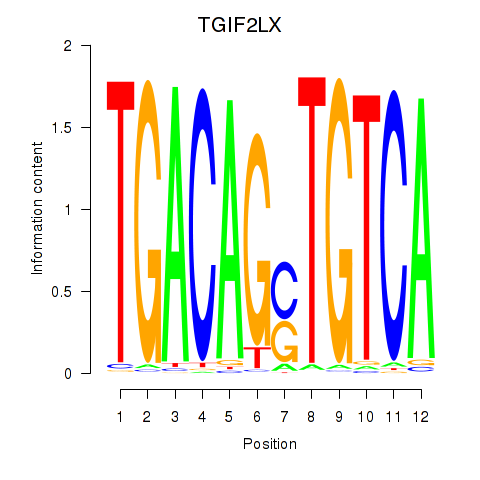
Results for MEIS3_TGIF2LX
Z-value: 1.06
Transcription factors associated with MEIS3_TGIF2LX
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MEIS3
|
ENSG00000105419.13 | Meis homeobox 3 |
|
TGIF2LX
|
ENSG00000153779.8 | TGFB induced factor homeobox 2 like X-linked |
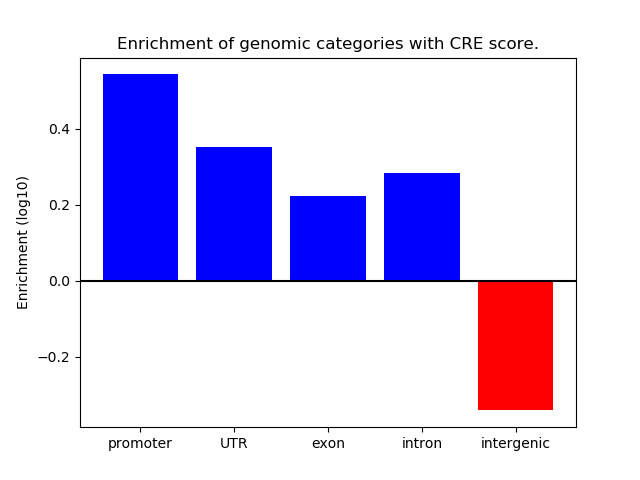
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr19_47930779_47930930 | MEIS3 | 8074 | 0.156354 | -0.55 | 1.2e-01 | Click! |
| chr19_47902354_47902505 | MEIS3 | 9980 | 0.157557 | 0.55 | 1.3e-01 | Click! |
| chr19_47936281_47936432 | MEIS3 | 13576 | 0.141825 | 0.54 | 1.3e-01 | Click! |
| chr19_47893667_47893818 | MEIS3 | 18667 | 0.139828 | 0.51 | 1.6e-01 | Click! |
| chr19_47932297_47932448 | MEIS3 | 9592 | 0.151645 | -0.51 | 1.6e-01 | Click! |
Activity of the MEIS3_TGIF2LX motif across conditions
Conditions sorted by the z-value of the MEIS3_TGIF2LX motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
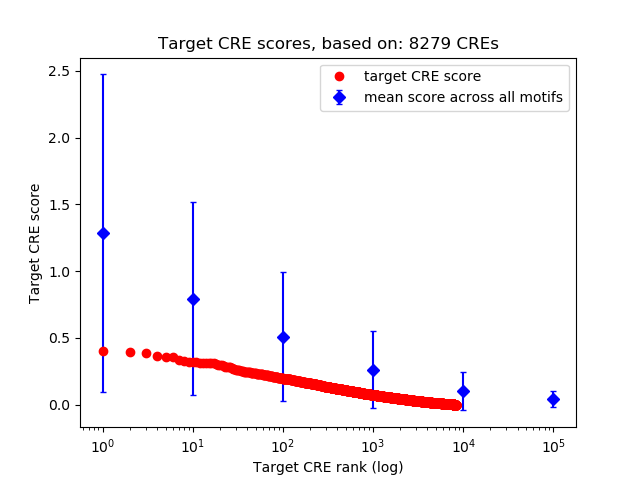
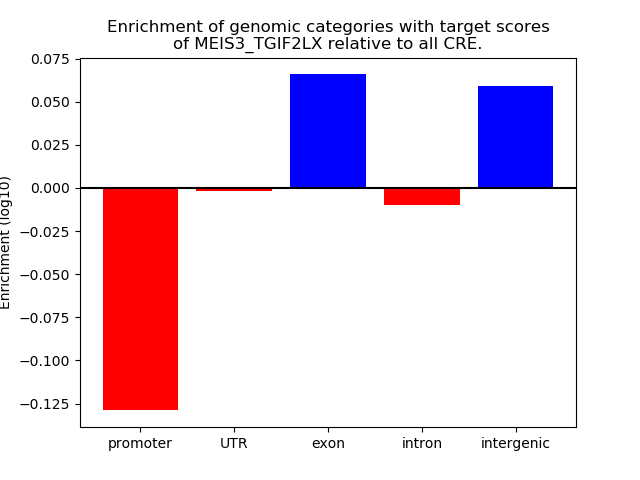
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr16_31364396_31364620 | 0.40 |
ITGAX |
integrin, alpha X (complement component 3 receptor 4 subunit) |
1947 |
0.22 |
| chr1_37195687_37195944 | 0.40 |
RP4-614N24.1 |
|
42926 |
0.19 |
| chr5_59090911_59091107 | 0.38 |
PDE4D |
phosphodiesterase 4D, cAMP-specific |
26538 |
0.25 |
| chr2_57825828_57826109 | 0.36 |
ENSG00000212168 |
. |
54298 |
0.18 |
| chr2_65132087_65132267 | 0.36 |
ENSG00000244534 |
. |
2095 |
0.28 |
| chr15_94406442_94406593 | 0.35 |
ENSG00000222409 |
. |
33739 |
0.25 |
| chr3_53206532_53206683 | 0.34 |
PRKCD |
protein kinase C, delta |
7462 |
0.18 |
| chr3_34304883_34305090 | 0.33 |
PDCD6IP |
programmed cell death 6 interacting protein |
464871 |
0.01 |
| chrX_123521860_123522011 | 0.32 |
SH2D1A |
SH2 domain containing 1A |
41496 |
0.21 |
| chr1_87771024_87771216 | 0.32 |
LMO4 |
LIM domain only 4 |
23031 |
0.26 |
| chr16_12449826_12449977 | 0.32 |
SNX29 |
sorting nexin 29 |
209 |
0.94 |
| chr9_81309923_81310074 | 0.32 |
PSAT1 |
phosphoserine aminotransferase 1 |
397939 |
0.01 |
| chr22_47080714_47080865 | 0.32 |
GRAMD4 |
GRAM domain containing 4 |
10284 |
0.22 |
| chr1_48711061_48711404 | 0.31 |
RP5-1024N4.4 |
|
15999 |
0.21 |
| chr16_89044159_89044400 | 0.31 |
CBFA2T3 |
core-binding factor, runt domain, alpha subunit 2; translocated to, 3 |
667 |
0.66 |
| chr9_95876458_95876701 | 0.31 |
C9orf89 |
chromosome 9 open reading frame 89 |
18079 |
0.14 |
| chr1_40529943_40530230 | 0.31 |
PPT1 |
palmitoyl-protein thioesterase 1 |
17907 |
0.17 |
| chr18_77269712_77269863 | 0.31 |
AC018445.1 |
Uncharacterized protein |
6270 |
0.3 |
| chr14_65519660_65519811 | 0.30 |
ENSG00000266531 |
. |
8329 |
0.14 |
| chr7_76139277_76139433 | 0.30 |
UPK3B |
uroplakin 3B |
390 |
0.81 |
| chr22_47017621_47018011 | 0.29 |
GRAMD4 |
GRAM domain containing 4 |
1517 |
0.48 |
| chr15_68077042_68077264 | 0.29 |
SKOR1 |
SKI family transcriptional corepressor 1 |
34889 |
0.2 |
| chr1_43765747_43766019 | 0.29 |
TIE1 |
tyrosine kinase with immunoglobulin-like and EGF-like domains 1 |
781 |
0.52 |
| chr16_11593490_11593706 | 0.28 |
CTD-3088G3.8 |
Protein LOC388210 |
7987 |
0.15 |
| chr6_34243280_34243535 | 0.28 |
C6orf1 |
chromosome 6 open reading frame 1 |
26160 |
0.18 |
| chr2_16844763_16844970 | 0.28 |
FAM49A |
family with sequence similarity 49, member A |
2230 |
0.47 |
| chr11_2697989_2698156 | 0.28 |
KCNQ1OT1 |
KCNQ1 opposite strand/antisense transcript 1 (non-protein coding) |
23152 |
0.22 |
| chr7_37429580_37430060 | 0.27 |
ENSG00000200113 |
. |
29411 |
0.16 |
| chr6_24951319_24951727 | 0.27 |
FAM65B |
family with sequence similarity 65, member B |
15335 |
0.22 |
| chr5_34865017_34865168 | 0.26 |
CTD-2517O10.6 |
|
25709 |
0.15 |
| chr3_194696258_194696409 | 0.26 |
XXYLT1-AS1 |
XXYLT1 antisense RNA 1 |
118984 |
0.05 |
| chr12_7060138_7060323 | 0.26 |
PTPN6 |
protein tyrosine phosphatase, non-receptor type 6 |
195 |
0.81 |
| chr11_76047608_76047759 | 0.26 |
PRKRIR |
protein-kinase, interferon-inducible double stranded RNA dependent inhibitor, repressor of (P58 repressor) |
44303 |
0.11 |
| chr15_42676695_42676925 | 0.25 |
CAPN3 |
calpain 3, (p94) |
17778 |
0.15 |
| chr11_105090744_105090960 | 0.25 |
CARD18 |
caspase recruitment domain family, member 18 |
80399 |
0.1 |
| chr10_131129546_131129832 | 0.25 |
MGMT |
O-6-methylguanine-DNA methyltransferase |
135759 |
0.05 |
| chr17_56692314_56692465 | 0.25 |
ENSG00000212195 |
. |
16808 |
0.12 |
| chr22_17582429_17582621 | 0.25 |
IL17RA |
interleukin 17 receptor A |
16676 |
0.14 |
| chr6_47203496_47203647 | 0.25 |
TNFRSF21 |
tumor necrosis factor receptor superfamily, member 21 |
74070 |
0.12 |
| chr4_16032933_16033084 | 0.24 |
ENSG00000251758 |
. |
22598 |
0.19 |
| chr16_81529220_81529493 | 0.24 |
CMIP |
c-Maf inducing protein |
402 |
0.91 |
| chr18_43904157_43904332 | 0.24 |
RNF165 |
ring finger protein 165 |
2528 |
0.4 |
| chr1_19235703_19235961 | 0.24 |
IFFO2 |
intermediate filament family orphan 2 |
2654 |
0.22 |
| chr9_21047360_21047561 | 0.24 |
PTPLAD2 |
protein tyrosine phosphatase-like A domain containing 2 |
15825 |
0.18 |
| chr9_84798219_84798630 | 0.24 |
SPATA31D1 |
SPATA31 subfamily D, member 1 |
194737 |
0.03 |
| chr5_78088517_78088668 | 0.24 |
LHFPL2 |
lipoma HMGIC fusion partner-like 2 |
143944 |
0.05 |
| chr1_206246898_206247049 | 0.24 |
ENSG00000252692 |
. |
14120 |
0.15 |
| chr7_29308125_29308409 | 0.24 |
AC004593.3 |
|
59681 |
0.12 |
| chr12_11939644_11939800 | 0.24 |
ETV6 |
ets variant 6 |
34287 |
0.22 |
| chr8_25095577_25095889 | 0.23 |
DOCK5 |
dedicator of cytokinesis 5 |
53353 |
0.15 |
| chr11_60939371_60939609 | 0.23 |
VPS37C |
vacuolar protein sorting 37 homolog C (S. cerevisiae) |
10401 |
0.15 |
| chr21_45595773_45596048 | 0.23 |
AP001055.1 |
|
2330 |
0.22 |
| chr8_92363748_92363899 | 0.23 |
SLC26A7 |
solute carrier family 26 (anion exchanger), member 7 |
55971 |
0.15 |
| chr1_175809524_175809675 | 0.23 |
TNR |
tenascin R |
96693 |
0.08 |
| chr19_50018940_50019106 | 0.23 |
FCGRT |
Fc fragment of IgG, receptor, transporter, alpha |
1708 |
0.13 |
| chr1_230413095_230413246 | 0.23 |
RP5-956O18.3 |
|
4469 |
0.26 |
| chr18_21695534_21696122 | 0.23 |
TTC39C |
tetratricopeptide repeat domain 39C |
1591 |
0.26 |
| chr15_31115309_31115461 | 0.23 |
ENSG00000221379 |
. |
21876 |
0.16 |
| chr6_12943954_12944105 | 0.23 |
PHACTR1 |
phosphatase and actin regulator 1 |
14185 |
0.3 |
| chr5_157661766_157661969 | 0.23 |
ENSG00000222626 |
. |
257903 |
0.02 |
| chr13_101325397_101325635 | 0.23 |
TMTC4 |
transmembrane and tetratricopeptide repeat containing 4 |
1512 |
0.47 |
| chr2_26232079_26232230 | 0.23 |
AC013449.1 |
Uncharacterized protein |
19327 |
0.15 |
| chr1_19686120_19686271 | 0.22 |
CAPZB |
capping protein (actin filament) muscle Z-line, beta |
26096 |
0.11 |
| chr15_28413886_28414277 | 0.22 |
HERC2 |
HECT and RLD domain containing E3 ubiquitin protein ligase 2 |
5488 |
0.29 |
| chr4_15190631_15190980 | 0.22 |
RP11-665G4.1 |
|
38779 |
0.19 |
| chr12_124228990_124229141 | 0.22 |
ATP6V0A2 |
ATPase, H+ transporting, lysosomal V0 subunit a2 |
5478 |
0.19 |
| chr17_27889238_27889394 | 0.22 |
RP11-68I3.2 |
|
1751 |
0.18 |
| chr9_111885537_111885688 | 0.22 |
TMEM245 |
transmembrane protein 245 |
3387 |
0.25 |
| chr21_34439493_34439682 | 0.22 |
OLIG1 |
oligodendrocyte transcription factor 1 |
2863 |
0.23 |
| chr10_134235955_134236340 | 0.22 |
RP11-432J24.3 |
|
18841 |
0.16 |
| chr12_27155870_27156021 | 0.22 |
TM7SF3 |
transmembrane 7 superfamily member 3 |
273 |
0.89 |
| chr2_60627914_60628065 | 0.22 |
ENSG00000266078 |
. |
13409 |
0.22 |
| chr11_70267723_70267874 | 0.21 |
CTTN |
cortactin |
1198 |
0.41 |
| chr15_57887157_57887308 | 0.21 |
GCOM1 |
GRINL1A complex locus 1 |
3001 |
0.22 |
| chr7_115625227_115625506 | 0.21 |
TFEC |
transcription factor EC |
17062 |
0.28 |
| chr17_48841397_48841548 | 0.21 |
LUC7L3 |
LUC7-like 3 (S. cerevisiae) |
17492 |
0.14 |
| chr19_47233988_47234360 | 0.21 |
CTB-174O21.2 |
|
2314 |
0.14 |
| chr22_18253018_18253331 | 0.21 |
BID |
BH3 interacting domain death agonist |
3608 |
0.22 |
| chr19_31765985_31766213 | 0.21 |
AC007796.1 |
|
73688 |
0.11 |
| chr7_93869278_93869476 | 0.21 |
COL1A2 |
collagen, type I, alpha 2 |
154496 |
0.04 |
| chr1_229137271_229137454 | 0.21 |
RP5-1061H20.5 |
|
225947 |
0.02 |
| chr1_9422560_9422755 | 0.21 |
SPSB1 |
splA/ryanodine receptor domain and SOCS box containing 1 |
6891 |
0.25 |
| chr4_40561931_40562177 | 0.21 |
RBM47 |
RNA binding motif protein 47 |
44064 |
0.15 |
| chr9_111597664_111597815 | 0.21 |
ACTL7B |
actin-like 7B |
21500 |
0.19 |
| chr2_225269583_225269734 | 0.21 |
FAM124B |
family with sequence similarity 124B |
2856 |
0.39 |
| chr1_245424145_245424296 | 0.20 |
RP11-62I21.1 |
|
26417 |
0.22 |
| chr19_44297367_44297518 | 0.20 |
LYPD5 |
LY6/PLAUR domain containing 5 |
6739 |
0.13 |
| chr21_45631442_45631593 | 0.20 |
AP001057.1 |
|
10344 |
0.12 |
| chrX_148713668_148713871 | 0.20 |
TMEM185A |
transmembrane protein 185A |
201 |
0.94 |
| chr2_64489091_64489290 | 0.20 |
AC074289.1 |
|
33588 |
0.2 |
| chr22_26564894_26565205 | 0.20 |
SEZ6L |
seizure related 6 homolog (mouse)-like |
391 |
0.87 |
| chr12_10186853_10187023 | 0.20 |
CLEC9A |
C-type lectin domain family 9, member A |
3662 |
0.14 |
| chr12_2639416_2639567 | 0.20 |
CACNA1C-AS3 |
CACNA1C antisense RNA 3 |
77115 |
0.1 |
| chr6_111897306_111897457 | 0.20 |
TRAF3IP2-AS1 |
TRAF3IP2 antisense RNA 1 |
1559 |
0.37 |
| chr4_86622619_86622800 | 0.20 |
ENSG00000266421 |
. |
20912 |
0.26 |
| chr10_94549884_94550135 | 0.20 |
EXOC6 |
exocyst complex component 6 |
40926 |
0.17 |
| chr21_39867625_39867922 | 0.20 |
ERG |
v-ets avian erythroblastosis virus E26 oncogene homolog |
2572 |
0.43 |
| chr6_20427872_20428026 | 0.20 |
ENSG00000252949 |
. |
5400 |
0.15 |
| chr1_234885846_234885997 | 0.20 |
ENSG00000201638 |
. |
87799 |
0.08 |
| chr10_23389093_23389313 | 0.20 |
MSRB2 |
methionine sulfoxide reductase B2 |
4768 |
0.23 |
| chr2_169482878_169483157 | 0.20 |
ENSG00000199348 |
. |
20011 |
0.22 |
| chr2_171218393_171218731 | 0.20 |
AC012594.1 |
|
10934 |
0.3 |
| chr12_117463237_117463436 | 0.20 |
TESC |
tescalcin |
21300 |
0.21 |
| chr6_42343133_42343301 | 0.19 |
ENSG00000221252 |
. |
36917 |
0.17 |
| chr2_233956975_233957215 | 0.19 |
INPP5D |
inositol polyphosphate-5-phosphatase, 145kDa |
31906 |
0.16 |
| chr19_52124679_52125032 | 0.19 |
SIGLEC5 |
sialic acid binding Ig-like lectin 5 |
8733 |
0.11 |
| chr3_4383206_4383357 | 0.19 |
SETMAR |
SET domain and mariner transposase fusion gene |
37989 |
0.21 |
| chr2_157327257_157327445 | 0.19 |
GPD2 |
glycerol-3-phosphate dehydrogenase 2 (mitochondrial) |
2733 |
0.29 |
| chr5_142596063_142596214 | 0.19 |
ARHGAP26 |
Rho GTPase activating protein 26 |
9373 |
0.25 |
| chr1_16827341_16827546 | 0.19 |
CROCCP3 |
ciliary rootlet coiled-coil, rootletin pseudogene 3 |
9641 |
0.13 |
| chr12_111587807_111587958 | 0.19 |
CUX2 |
cut-like homeobox 2 |
50632 |
0.15 |
| chr19_18090395_18090751 | 0.19 |
ENSG00000252591 |
. |
7739 |
0.14 |
| chr11_3188344_3188495 | 0.19 |
OSBPL5 |
oxysterol binding protein-like 5 |
1419 |
0.35 |
| chr10_81698298_81698654 | 0.19 |
SFTPD |
surfactant protein D |
10383 |
0.26 |
| chr17_61753634_61753814 | 0.19 |
LIMD2 |
LIM domain containing 2 |
22798 |
0.13 |
| chr10_50507704_50507855 | 0.19 |
C10orf71 |
chromosome 10 open reading frame 71 |
541 |
0.55 |
| chr5_135347344_135347609 | 0.19 |
TGFBI |
transforming growth factor, beta-induced, 68kDa |
17108 |
0.2 |
| chr6_37017148_37017362 | 0.19 |
COX6A1P2 |
cytochrome c oxidase subunit VIa polypeptide 1 pseudogene 2 |
4648 |
0.23 |
| chr4_174202594_174202800 | 0.19 |
GALNT7 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 7 (GalNAc-T7) |
10585 |
0.17 |
| chr22_26972149_26972362 | 0.19 |
TPST2 |
tyrosylprotein sulfotransferase 2 |
10563 |
0.15 |
| chr18_74769125_74769276 | 0.19 |
MBP |
myelin basic protein |
39466 |
0.19 |
| chr4_109901475_109901626 | 0.19 |
COL25A1 |
collagen, type XXV, alpha 1 |
156175 |
0.04 |
| chr2_64694768_64695032 | 0.19 |
LGALSL |
lectin, galactoside-binding-like |
13224 |
0.19 |
| chr7_50251693_50252080 | 0.19 |
AC020743.2 |
|
69467 |
0.11 |
| chr10_88117437_88117588 | 0.19 |
GRID1 |
glutamate receptor, ionotropic, delta 1 |
8723 |
0.24 |
| chr4_55557239_55557390 | 0.19 |
KIT |
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog |
33229 |
0.24 |
| chr7_87944172_87944366 | 0.18 |
STEAP4 |
STEAP family member 4 |
8063 |
0.23 |
| chr20_20037098_20037366 | 0.18 |
C20orf26 |
chromosome 20 open reading frame 26 |
58 |
0.9 |
| chr1_21510914_21511065 | 0.18 |
EIF4G3 |
eukaryotic translation initiation factor 4 gamma, 3 |
7612 |
0.21 |
| chr6_44585641_44585792 | 0.18 |
ENSG00000266619 |
. |
182338 |
0.03 |
| chr6_159461301_159461469 | 0.18 |
TAGAP |
T-cell activation RhoGTPase activating protein |
4665 |
0.21 |
| chr11_33952668_33952907 | 0.18 |
LMO2 |
LIM domain only 2 (rhombotin-like 1) |
38951 |
0.16 |
| chrX_128913033_128913442 | 0.18 |
SASH3 |
SAM and SH3 domain containing 3 |
723 |
0.69 |
| chr10_101940587_101940738 | 0.18 |
ERLIN1 |
ER lipid raft associated 1 |
5124 |
0.15 |
| chr3_126248151_126248302 | 0.18 |
CHST13 |
carbohydrate (chondroitin 4) sulfotransferase 13 |
5100 |
0.16 |
| chr17_72329157_72329308 | 0.18 |
KIF19 |
kinesin family member 19 |
6881 |
0.11 |
| chr4_55529764_55530043 | 0.18 |
KIT |
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog |
5818 |
0.34 |
| chr9_135757349_135757570 | 0.18 |
C9orf9 |
chromosome 9 open reading frame 9 |
3174 |
0.17 |
| chr13_32691700_32692027 | 0.18 |
FRY |
furry homolog (Drosophila) |
56862 |
0.14 |
| chr21_46954664_46954827 | 0.18 |
SLC19A1 |
solute carrier family 19 (folate transporter), member 1 |
216 |
0.95 |
| chr11_75091839_75092030 | 0.18 |
RPS3 |
ribosomal protein S3 |
18596 |
0.11 |
| chr18_42882859_42883010 | 0.18 |
SLC14A2 |
solute carrier family 14 (urea transporter), member 2 |
89974 |
0.09 |
| chr12_111618066_111618305 | 0.18 |
CUX2 |
cut-like homeobox 2 |
80935 |
0.09 |
| chr2_127818917_127819068 | 0.18 |
ENSG00000238788 |
. |
43995 |
0.16 |
| chr10_11155205_11156000 | 0.18 |
CELF2-AS2 |
CELF2 antisense RNA 2 |
15166 |
0.21 |
| chr7_139585204_139585374 | 0.18 |
TBXAS1 |
thromboxane A synthase 1 (platelet) |
56179 |
0.14 |
| chr8_70591137_70591370 | 0.18 |
ENSG00000222889 |
. |
21206 |
0.21 |
| chr11_44938433_44938696 | 0.18 |
TSPAN18 |
tetraspanin 18 |
10606 |
0.25 |
| chr11_65084227_65084378 | 0.18 |
CDC42EP2 |
CDC42 effector protein (Rho GTPase binding) 2 |
211 |
0.9 |
| chr16_57712273_57712424 | 0.18 |
GPR97 |
G protein-coupled receptor 97 |
10128 |
0.13 |
| chr5_132644386_132644537 | 0.18 |
CTB-49A3.4 |
|
42184 |
0.15 |
| chr6_12659751_12659902 | 0.18 |
PHACTR1 |
phosphatase and actin regulator 1 |
58067 |
0.15 |
| chr19_35950537_35950782 | 0.17 |
FFAR2 |
free fatty acid receptor 2 |
10042 |
0.11 |
| chr9_135685079_135685230 | 0.17 |
RP11-295G24.5 |
|
39555 |
0.12 |
| chr4_144278414_144278571 | 0.17 |
ENSG00000265623 |
. |
13879 |
0.2 |
| chr20_58566781_58566961 | 0.17 |
CDH26 |
cadherin 26 |
2710 |
0.28 |
| chr6_53600725_53601075 | 0.17 |
RP13-476E20.1 |
|
58172 |
0.11 |
| chr4_40310355_40310506 | 0.17 |
CHRNA9 |
cholinergic receptor, nicotinic, alpha 9 (neuronal) |
26916 |
0.19 |
| chr5_108994298_108994449 | 0.17 |
ENSG00000266090 |
. |
26908 |
0.19 |
| chr19_19653103_19653257 | 0.17 |
CILP2 |
cartilage intermediate layer protein 2 |
4106 |
0.13 |
| chr17_397150_397301 | 0.17 |
RP5-1029F21.3 |
|
9186 |
0.19 |
| chr2_238828434_238828585 | 0.17 |
UBE2F |
ubiquitin-conjugating enzyme E2F (putative) |
47088 |
0.12 |
| chr8_10053593_10053832 | 0.17 |
MSRA |
methionine sulfoxide reductase A |
100475 |
0.07 |
| chr16_68775300_68775451 | 0.17 |
ENSG00000200558 |
. |
1008 |
0.48 |
| chr2_143909839_143910067 | 0.17 |
RP11-190J23.1 |
|
19788 |
0.22 |
| chr17_62032388_62032539 | 0.17 |
SCN4A |
sodium channel, voltage-gated, type IV, alpha subunit |
17815 |
0.11 |
| chr19_54885476_54885627 | 0.17 |
LAIR1 |
leukocyte-associated immunoglobulin-like receptor 1 |
3386 |
0.14 |
| chr2_220074664_220075050 | 0.17 |
ZFAND2B |
zinc finger, AN1-type domain 2B |
1677 |
0.16 |
| chr1_44889322_44889516 | 0.17 |
RNF220 |
ring finger protein 220 |
293 |
0.91 |
| chr2_10815795_10815988 | 0.17 |
NOL10 |
nucleolar protein 10 |
4075 |
0.21 |
| chr7_29288155_29288306 | 0.17 |
AC004593.3 |
|
39644 |
0.16 |
| chr8_96087280_96087459 | 0.17 |
MIR3150A |
microRNA 3150a |
1959 |
0.28 |
| chr6_116784453_116784778 | 0.17 |
RP1-93H18.6 |
|
240 |
0.83 |
| chr6_6637501_6637686 | 0.17 |
LY86-AS1 |
LY86 antisense RNA 1 |
14589 |
0.26 |
| chr14_75152469_75152620 | 0.17 |
AREL1 |
apoptosis resistant E3 ubiquitin protein ligase 1 |
9626 |
0.15 |
| chr5_78204185_78204336 | 0.17 |
ARSB |
arylsulfatase B |
77348 |
0.11 |
| chr5_59560361_59560512 | 0.17 |
ENSG00000202017 |
. |
39542 |
0.2 |
| chr19_46179501_46179652 | 0.17 |
ENSG00000207773 |
. |
1390 |
0.21 |
| chr21_43845315_43845466 | 0.17 |
ENSG00000252619 |
. |
7688 |
0.14 |
| chr4_82421002_82421153 | 0.17 |
RASGEF1B |
RasGEF domain family, member 1B |
28008 |
0.26 |
| chr1_45819749_45819900 | 0.17 |
ENSG00000238945 |
. |
4596 |
0.17 |
| chrX_22015726_22015877 | 0.17 |
PHEX |
phosphate regulating endopeptidase homolog, X-linked |
34758 |
0.17 |
| chr5_148148648_148148804 | 0.17 |
ADRB2 |
adrenoceptor beta 2, surface |
57430 |
0.14 |
| chr19_46005187_46005379 | 0.16 |
PPM1N |
protein phosphatase, Mg2+/Mn2+ dependent, 1N (putative) |
2064 |
0.19 |
| chr16_81787336_81787487 | 0.16 |
PLCG2 |
phospholipase C, gamma 2 (phosphatidylinositol-specific) |
14709 |
0.27 |
| chr10_114502069_114502220 | 0.16 |
RP11-25C19.3 |
|
64566 |
0.12 |
| chr3_32447849_32448023 | 0.16 |
CMTM7 |
CKLF-like MARVEL transmembrane domain containing 7 |
14405 |
0.24 |
| chr7_41506541_41506692 | 0.16 |
INHBA-AS1 |
INHBA antisense RNA 1 |
226898 |
0.02 |
| chr20_2271241_2271392 | 0.16 |
TGM3 |
transglutaminase 3 |
5331 |
0.27 |
| chr2_221996968_221997119 | 0.16 |
EPHA4 |
EPH receptor A4 |
370237 |
0.01 |
| chr22_24525026_24525177 | 0.16 |
CABIN1 |
calcineurin binding protein 1 |
26705 |
0.14 |
| chr2_173998078_173998369 | 0.16 |
MLTK |
Mitogen-activated protein kinase kinase kinase MLT |
42876 |
0.16 |
| chr8_82030046_82030197 | 0.16 |
PAG1 |
phosphoprotein associated with glycosphingolipid microdomains 1 |
5818 |
0.31 |
| chr20_58653485_58653636 | 0.16 |
C20orf197 |
chromosome 20 open reading frame 197 |
22580 |
0.23 |
| chr2_232164549_232164700 | 0.16 |
ARMC9 |
armadillo repeat containing 9 |
29379 |
0.17 |
| chr6_5403263_5403414 | 0.16 |
FARS2 |
phenylalanyl-tRNA synthetase 2, mitochondrial |
1437 |
0.53 |
| chr22_22889208_22889359 | 0.16 |
PRAME |
preferentially expressed antigen in melanoma |
7320 |
0.07 |
| chr10_99126345_99126553 | 0.16 |
RRP12 |
ribosomal RNA processing 12 homolog (S. cerevisiae) |
24153 |
0.09 |
| chr3_140732457_140732639 | 0.16 |
SPSB4 |
splA/ryanodine receptor domain and SOCS box containing 4 |
37696 |
0.16 |
| chr5_66338060_66338433 | 0.16 |
MAST4 |
microtubule associated serine/threonine kinase family member 4 |
37763 |
0.19 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0032959 | inositol trisphosphate biosynthetic process(GO:0032959) |
| 0.1 | 0.2 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.0 | 0.1 | GO:0045906 | negative regulation of vasoconstriction(GO:0045906) |
| 0.0 | 0.1 | GO:0000101 | sulfur amino acid transport(GO:0000101) L-cystine transport(GO:0015811) |
| 0.0 | 0.2 | GO:0031272 | pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.1 | GO:0010193 | response to ozone(GO:0010193) |
| 0.0 | 0.1 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 0.0 | 0.1 | GO:0031223 | response to auditory stimulus(GO:0010996) auditory behavior(GO:0031223) |
| 0.0 | 0.1 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) positive regulation of hematopoietic progenitor cell differentiation(GO:1901534) |
| 0.0 | 0.1 | GO:0014041 | regulation of neuron maturation(GO:0014041) regulation of cell maturation(GO:1903429) |
| 0.0 | 0.1 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.0 | 0.2 | GO:0015884 | folic acid transport(GO:0015884) |
| 0.0 | 0.1 | GO:0043320 | natural killer cell activation involved in immune response(GO:0002323) natural killer cell degranulation(GO:0043320) |
| 0.0 | 0.1 | GO:0006154 | adenosine catabolic process(GO:0006154) |
| 0.0 | 0.1 | GO:0033622 | integrin activation(GO:0033622) |
| 0.0 | 0.1 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.1 | GO:0010886 | positive regulation of cholesterol storage(GO:0010886) |
| 0.0 | 0.1 | GO:1903224 | endodermal cell fate specification(GO:0001714) regulation of endodermal cell fate specification(GO:0042663) regulation of endodermal cell differentiation(GO:1903224) |
| 0.0 | 0.0 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.0 | 0.0 | GO:0072194 | kidney smooth muscle tissue development(GO:0072194) |
| 0.0 | 0.1 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.1 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 | 0.1 | GO:1901503 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.0 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.0 | 0.1 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.2 | GO:0051155 | positive regulation of striated muscle cell differentiation(GO:0051155) |
| 0.0 | 0.0 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.0 | 0.1 | GO:0007028 | cytoplasm organization(GO:0007028) |
| 0.0 | 0.1 | GO:0006734 | NADH metabolic process(GO:0006734) |
| 0.0 | 0.0 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.1 | GO:0035751 | lysosomal lumen acidification(GO:0007042) regulation of lysosomal lumen pH(GO:0035751) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.2 | GO:0030851 | granulocyte differentiation(GO:0030851) |
| 0.0 | 0.1 | GO:0060123 | regulation of growth hormone secretion(GO:0060123) |
| 0.0 | 0.3 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.0 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.0 | 0.1 | GO:0009912 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.0 | 0.0 | GO:0032632 | interleukin-3 production(GO:0032632) |
| 0.0 | 0.0 | GO:0046931 | pore complex assembly(GO:0046931) |
| 0.0 | 0.0 | GO:0070340 | detection of bacterial lipoprotein(GO:0042494) detection of bacterial lipopeptide(GO:0070340) |
| 0.0 | 0.1 | GO:0034204 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.0 | 0.2 | GO:0050732 | negative regulation of peptidyl-tyrosine phosphorylation(GO:0050732) |
| 0.0 | 0.1 | GO:0032849 | positive regulation of cellular pH reduction(GO:0032849) |
| 0.0 | 0.1 | GO:0045356 | positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.0 | 0.0 | GO:0045002 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) double-strand break repair via single-strand annealing(GO:0045002) |
| 0.0 | 0.1 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.1 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.0 | 0.0 | GO:0008215 | spermine metabolic process(GO:0008215) |
| 0.0 | 0.0 | GO:0015959 | diadenosine polyphosphate metabolic process(GO:0015959) |
| 0.0 | 0.0 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 0.1 | GO:0098876 | Golgi to plasma membrane transport(GO:0006893) vesicle-mediated transport to the plasma membrane(GO:0098876) |
| 0.0 | 0.1 | GO:0033197 | response to vitamin E(GO:0033197) |
| 0.0 | 0.1 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.0 | GO:0043129 | surfactant homeostasis(GO:0043129) chemical homeostasis within a tissue(GO:0048875) |
| 0.0 | 0.0 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.1 | GO:0001562 | response to protozoan(GO:0001562) |
| 0.0 | 0.0 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.0 | GO:0052805 | histidine catabolic process(GO:0006548) imidazole-containing compound catabolic process(GO:0052805) |
| 0.0 | 0.1 | GO:0034331 | cell junction maintenance(GO:0034331) cell-cell junction maintenance(GO:0045217) |
| 0.0 | 0.1 | GO:0046874 | quinolinate metabolic process(GO:0046874) |
| 0.0 | 0.0 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.0 | 0.0 | GO:0009296 | obsolete flagellum assembly(GO:0009296) |
| 0.0 | 0.0 | GO:0033194 | response to hydroperoxide(GO:0033194) |
| 0.0 | 0.0 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 0.0 | 0.0 | GO:0097035 | regulation of membrane lipid distribution(GO:0097035) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.1 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.1 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.1 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.0 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.0 | GO:0033150 | cytoskeletal calyx(GO:0033150) |
| 0.0 | 0.1 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.0 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.1 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.1 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.0 | 0.1 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.1 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.0 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.2 | GO:0008518 | reduced folate carrier activity(GO:0008518) |
| 0.0 | 0.1 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.0 | 0.1 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.0 | 0.1 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.1 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.1 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.1 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.0 | 0.1 | GO:0001846 | opsonin binding(GO:0001846) |
| 0.0 | 0.1 | GO:0052659 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) inositol-1,3,4,5-tetrakisphosphate 5-phosphatase activity(GO:0052659) |
| 0.0 | 0.1 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.1 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.1 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.1 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.0 | 0.1 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 0.0 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.0 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.0 | 0.0 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.0 | GO:0017060 | 3-galactosyl-N-acetylglucosaminide 4-alpha-L-fucosyltransferase activity(GO:0017060) |
| 0.0 | 0.0 | GO:0001099 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.0 | 0.0 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.0 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.0 | 0.1 | GO:0045569 | TRAIL binding(GO:0045569) |
| 0.0 | 0.0 | GO:0004803 | transposase activity(GO:0004803) |
| 0.0 | 0.2 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 0.1 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.0 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.0 | 0.0 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.1 | GO:0042285 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.0 | GO:0043175 | RNA polymerase core enzyme binding(GO:0043175) |
| 0.0 | 0.0 | GO:0032450 | maltose alpha-glucosidase activity(GO:0032450) |
| 0.0 | 0.2 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 0.0 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.0 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.0 | 0.1 | GO:0016520 | growth hormone-releasing hormone receptor activity(GO:0016520) |
| 0.0 | 0.1 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.3 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 0.0 | 0.0 | GO:0019958 | C-X-C chemokine binding(GO:0019958) interleukin-8 binding(GO:0019959) |
| 0.0 | 0.0 | GO:0050542 | icosanoid binding(GO:0050542) fatty acid derivative binding(GO:1901567) |
| 0.0 | 0.0 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.1 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.0 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0051635 | obsolete bacterial cell surface binding(GO:0051635) |
| 0.0 | 0.0 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.0 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.0 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 0.0 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.0 | 0.0 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 0.0 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.0 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.2 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.1 | GO:0042895 | antibiotic transporter activity(GO:0042895) |
| 0.0 | 0.1 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.1 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.0 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.0 | GO:0071723 | lipopeptide binding(GO:0071723) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.0 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 0.4 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 0.1 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.1 | PID TRAIL PATHWAY | TRAIL signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME DOWNSTREAM SIGNALING EVENTS OF B CELL RECEPTOR BCR | Genes involved in Downstream Signaling Events Of B Cell Receptor (BCR) |
| 0.0 | 0.4 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.1 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.2 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.0 | REACTOME DAG AND IP3 SIGNALING | Genes involved in DAG and IP3 signaling |
| 0.0 | 0.2 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.1 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |