Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
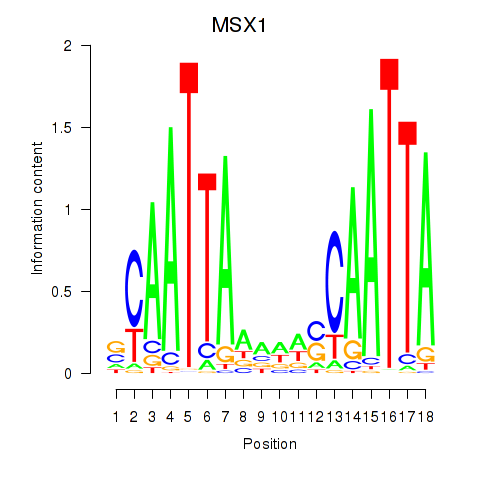
Results for MSX1
Z-value: 1.28
Transcription factors associated with MSX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MSX1
|
ENSG00000163132.6 | msh homeobox 1 |
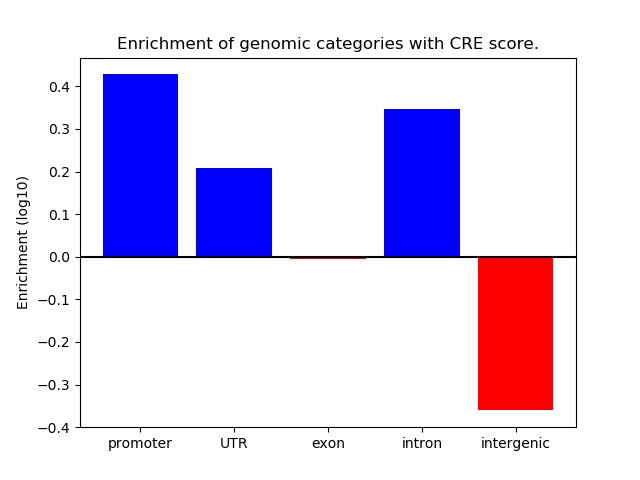
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr4_4862740_4862954 | MSX1 | 1454 | 0.530254 | -0.81 | 7.9e-03 | Click! |
| chr4_4860648_4860832 | MSX1 | 653 | 0.801189 | -0.80 | 8.9e-03 | Click! |
| chr4_4857303_4857507 | MSX1 | 3988 | 0.321058 | -0.75 | 1.9e-02 | Click! |
| chr4_4793701_4793852 | MSX1 | 67617 | 0.125212 | 0.72 | 2.7e-02 | Click! |
| chr4_4862254_4862686 | MSX1 | 1077 | 0.636089 | -0.60 | 8.5e-02 | Click! |
Activity of the MSX1 motif across conditions
Conditions sorted by the z-value of the MSX1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
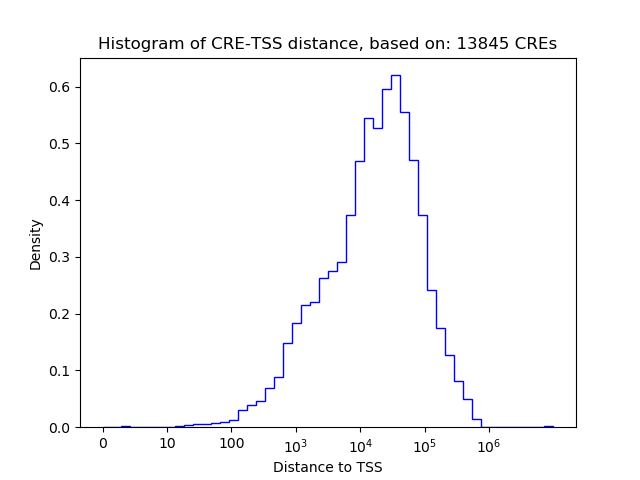
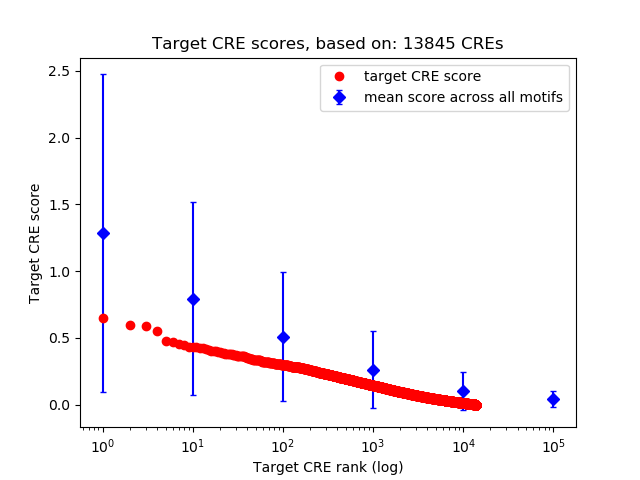
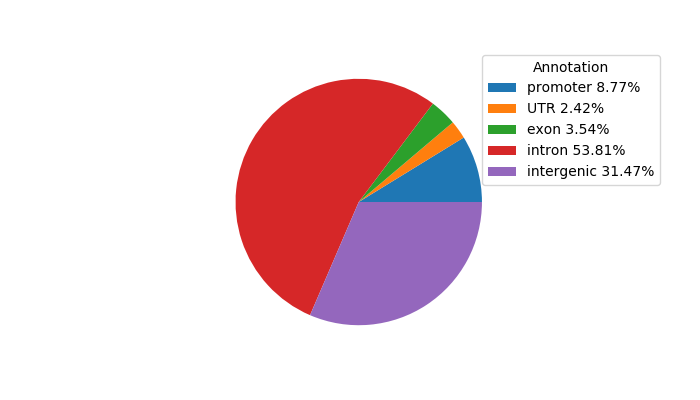
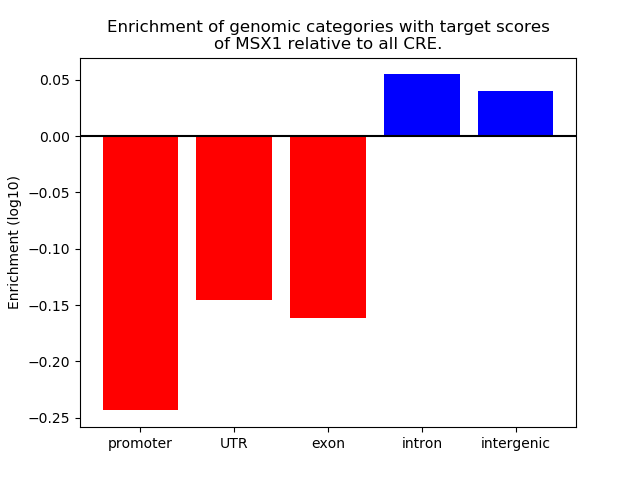
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr10_93221945_93222246 | 0.65 |
HECTD2 |
HECT domain containing E3 ubiquitin protein ligase 2 |
198 |
0.97 |
| chr7_116416400_116416551 | 0.60 |
MET |
met proto-oncogene |
754 |
0.71 |
| chr1_206593311_206593758 | 0.59 |
SRGAP2 |
SLIT-ROBO Rho GTPase activating protein 2 |
13798 |
0.17 |
| chr14_69015594_69016105 | 0.55 |
CTD-2325P2.4 |
|
79313 |
0.1 |
| chr9_4137653_4137928 | 0.48 |
GLIS3 |
GLIS family zinc finger 3 |
7403 |
0.28 |
| chr8_41906632_41907752 | 0.47 |
KAT6A |
K(lysine) acetyltransferase 6A |
2313 |
0.3 |
| chr18_12598337_12598791 | 0.45 |
SPIRE1 |
spire-type actin nucleation factor 1 |
54520 |
0.09 |
| chr17_46080195_46080690 | 0.45 |
RP11-6N17.10 |
|
6880 |
0.1 |
| chr3_152787726_152787975 | 0.44 |
RP11-529G21.2 |
|
91714 |
0.08 |
| chr5_9622384_9622535 | 0.43 |
TAS2R1 |
taste receptor, type 2, member 1 |
8004 |
0.22 |
| chr14_35629047_35629198 | 0.43 |
KIAA0391 |
KIAA0391 |
37157 |
0.15 |
| chr14_72179521_72179805 | 0.43 |
SIPA1L1 |
signal-induced proliferation-associated 1 like 1 |
94190 |
0.09 |
| chr11_9927954_9928253 | 0.42 |
ENSG00000252568 |
. |
62680 |
0.11 |
| chr13_77518573_77518724 | 0.41 |
IRG1 |
immunoresponsive 1 homolog (mouse) |
3984 |
0.24 |
| chr4_78502666_78502963 | 0.41 |
CXCL13 |
chemokine (C-X-C motif) ligand 13 |
69907 |
0.13 |
| chr10_75987768_75988344 | 0.40 |
ADK |
adenosine kinase |
51535 |
0.15 |
| chr13_31440489_31440640 | 0.40 |
MEDAG |
mesenteric estrogen-dependent adipogenesis |
39764 |
0.16 |
| chr15_101143578_101144308 | 0.40 |
ASB7 |
ankyrin repeat and SOCS box containing 7 |
1129 |
0.36 |
| chr6_2103859_2104525 | 0.39 |
GMDS |
GDP-mannose 4,6-dehydratase |
72033 |
0.13 |
| chr3_111820782_111820933 | 0.39 |
ENSG00000207940 |
. |
10791 |
0.15 |
| chr7_101457741_101458764 | 0.39 |
CUX1 |
cut-like homeobox 1 |
707 |
0.73 |
| chr7_46949633_46949784 | 0.38 |
AC011294.3 |
Uncharacterized protein |
212988 |
0.03 |
| chr16_18980279_18980782 | 0.38 |
TMC7 |
transmembrane channel-like 7 |
14726 |
0.14 |
| chr5_110849348_110849548 | 0.38 |
STARD4 |
StAR-related lipid transfer (START) domain containing 4 |
1160 |
0.42 |
| chr14_42683704_42683855 | 0.38 |
ENSG00000222084 |
. |
86175 |
0.1 |
| chr4_146467395_146467619 | 0.38 |
SMAD1-AS1 |
SMAD1 antisense RNA 1 |
29237 |
0.17 |
| chr5_66356025_66356176 | 0.38 |
MAST4 |
microtubule associated serine/threonine kinase family member 4 |
55617 |
0.15 |
| chr4_170582263_170582414 | 0.37 |
CLCN3 |
chloride channel, voltage-sensitive 3 |
1125 |
0.6 |
| chr22_26717807_26718076 | 0.37 |
SEZ6L |
seizure related 6 homolog (mouse)-like |
25740 |
0.18 |
| chr2_227312371_227312522 | 0.37 |
ENSG00000263363 |
. |
211063 |
0.02 |
| chr13_103360569_103360720 | 0.37 |
ENSG00000238869 |
. |
13508 |
0.13 |
| chr3_188251532_188251683 | 0.37 |
LPP-AS1 |
LPP antisense RNA 1 |
34847 |
0.2 |
| chr7_116347170_116347321 | 0.37 |
MET |
met proto-oncogene |
8106 |
0.26 |
| chr12_109195761_109195912 | 0.37 |
ENSG00000264043 |
. |
23789 |
0.13 |
| chr5_34052813_34052964 | 0.37 |
C1QTNF3 |
C1q and tumor necrosis factor related protein 3 |
8951 |
0.23 |
| chr17_60897719_60897987 | 0.37 |
MARCH10 |
membrane-associated ring finger (C3HC4) 10, E3 ubiquitin protein ligase |
12148 |
0.25 |
| chr7_42030328_42030605 | 0.36 |
GLI3 |
GLI family zinc finger 3 |
236854 |
0.02 |
| chr19_13147341_13147492 | 0.36 |
NFIX |
nuclear factor I/X (CCAAT-binding transcription factor) |
11605 |
0.1 |
| chr12_91290582_91290865 | 0.36 |
CCER1 |
coiled-coil glutamate-rich protein 1 |
58230 |
0.15 |
| chr1_234754493_234754778 | 0.35 |
IRF2BP2 |
interferon regulatory factor 2 binding protein 2 |
9364 |
0.19 |
| chr2_47006358_47006650 | 0.35 |
LINC01118 |
long intergenic non-protein coding RNA 1118 |
37318 |
0.13 |
| chr20_44409069_44409220 | 0.35 |
ENSG00000221046 |
. |
1807 |
0.19 |
| chr17_32580574_32580862 | 0.34 |
CCL2 |
chemokine (C-C motif) ligand 2 |
1586 |
0.24 |
| chr15_78594610_78594888 | 0.34 |
WDR61 |
WD repeat domain 61 |
2613 |
0.18 |
| chr6_139729934_139730085 | 0.34 |
CITED2 |
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 |
34252 |
0.22 |
| chr9_111235960_111236111 | 0.34 |
ENSG00000222512 |
. |
114826 |
0.07 |
| chr4_157860965_157861230 | 0.34 |
PDGFC |
platelet derived growth factor C |
30958 |
0.19 |
| chr10_60420713_60420913 | 0.34 |
BICC1 |
bicaudal C homolog 1 (Drosophila) |
132482 |
0.05 |
| chr1_224076219_224076370 | 0.34 |
TP53BP2 |
tumor protein p53 binding protein, 2 |
42620 |
0.17 |
| chr4_139232146_139232297 | 0.34 |
SLC7A11 |
solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11 |
68718 |
0.14 |
| chr5_53683581_53683732 | 0.33 |
HSPB3 |
heat shock 27kDa protein 3 |
67789 |
0.12 |
| chr3_114959246_114959397 | 0.33 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
93203 |
0.1 |
| chr3_12550506_12550657 | 0.33 |
ENSG00000200114 |
. |
2131 |
0.22 |
| chr10_95202028_95202179 | 0.33 |
MYOF |
myoferlin |
39848 |
0.15 |
| chr9_111207476_111207627 | 0.33 |
ENSG00000222512 |
. |
86342 |
0.11 |
| chr1_113088087_113088238 | 0.33 |
RP4-671G15.2 |
|
27099 |
0.14 |
| chr6_37850154_37850385 | 0.33 |
ENSG00000200597 |
. |
33080 |
0.16 |
| chr1_224820105_224820256 | 0.33 |
CNIH3 |
cornichon family AMPA receptor auxiliary protein 3 |
16185 |
0.19 |
| chr22_28144785_28145032 | 0.32 |
RP11-375H17.1 |
|
32440 |
0.2 |
| chr1_8106369_8106520 | 0.32 |
ERRFI1 |
ERBB receptor feedback inhibitor 1 |
20076 |
0.2 |
| chr7_102477303_102477454 | 0.32 |
ENSG00000252643 |
. |
2326 |
0.29 |
| chr2_150601107_150601258 | 0.32 |
ENSG00000207270 |
. |
133894 |
0.05 |
| chr6_72113075_72113298 | 0.32 |
ENSG00000207827 |
. |
138 |
0.97 |
| chr3_8979557_8979708 | 0.32 |
ENSG00000199815 |
. |
6453 |
0.22 |
| chr10_63734512_63734894 | 0.32 |
ENSG00000221272 |
. |
47926 |
0.17 |
| chr21_30331573_30331724 | 0.32 |
LTN1 |
listerin E3 ubiquitin protein ligase 1 |
33558 |
0.14 |
| chr2_224950724_224950875 | 0.32 |
SERPINE2 |
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 |
46763 |
0.17 |
| chr2_38918276_38918577 | 0.32 |
GALM |
galactose mutarotase (aldose 1-epimerase) |
9989 |
0.17 |
| chr11_130451238_130451517 | 0.32 |
C11orf44 |
chromosome 11 open reading frame 44 |
91474 |
0.09 |
| chr7_80512457_80512608 | 0.32 |
SEMA3C |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
35967 |
0.24 |
| chr16_54960016_54960167 | 0.32 |
IRX5 |
iroquois homeobox 5 |
4683 |
0.36 |
| chr11_46071541_46071813 | 0.31 |
PHF21A |
PHD finger protein 21A |
42079 |
0.15 |
| chrX_24166137_24167088 | 0.31 |
ZFX |
zinc finger protein, X-linked |
1134 |
0.35 |
| chr3_81474618_81474769 | 0.31 |
ENSG00000222389 |
. |
83934 |
0.11 |
| chr11_102321739_102322277 | 0.31 |
TMEM123 |
transmembrane protein 123 |
46 |
0.96 |
| chr3_16024682_16024833 | 0.31 |
ENSG00000207815 |
. |
109479 |
0.06 |
| chr7_151190654_151190805 | 0.31 |
RHEB |
Ras homolog enriched in brain |
24426 |
0.13 |
| chr20_36815816_36815967 | 0.31 |
TGM2 |
transglutaminase 2 |
20911 |
0.17 |
| chr5_72713863_72714014 | 0.31 |
FOXD1 |
forkhead box D1 |
30414 |
0.18 |
| chr2_225543203_225543354 | 0.31 |
CUL3 |
cullin 3 |
93168 |
0.09 |
| chr12_45627192_45627343 | 0.31 |
ANO6 |
anoctamin 6 |
17364 |
0.28 |
| chr15_90950843_90951750 | 0.31 |
RP11-154B12.3 |
|
10183 |
0.16 |
| chr5_73497876_73498136 | 0.31 |
ENSG00000222551 |
. |
69584 |
0.13 |
| chr2_242318504_242318655 | 0.31 |
FARP2 |
FERM, RhoGEF and pleckstrin domain protein 2 |
6315 |
0.17 |
| chr10_65024052_65024675 | 0.31 |
JMJD1C |
jumonji domain containing 1C |
4463 |
0.31 |
| chr11_121532407_121532558 | 0.30 |
SORL1 |
sortilin-related receptor, L(DLR class) A repeats containing |
71354 |
0.13 |
| chr1_172290576_172290727 | 0.30 |
ENSG00000252354 |
. |
26632 |
0.17 |
| chr9_14281034_14281328 | 0.30 |
NFIB |
nuclear factor I/B |
26831 |
0.22 |
| chr6_52289660_52289856 | 0.30 |
EFHC1 |
EF-hand domain (C-terminal) containing 1 |
1288 |
0.54 |
| chr9_81321983_81322311 | 0.30 |
PSAT1 |
phosphoserine aminotransferase 1 |
410088 |
0.01 |
| chr10_50760988_50761139 | 0.30 |
ERCC6 |
excision repair cross-complementing rodent repair deficiency, complementation group 6 |
13479 |
0.15 |
| chr13_47207440_47207736 | 0.30 |
LRCH1 |
leucine-rich repeats and calponin homology (CH) domain containing 1 |
46383 |
0.18 |
| chrX_45570754_45570905 | 0.30 |
ENSG00000207870 |
. |
34865 |
0.19 |
| chr13_45294845_45294996 | 0.30 |
ENSG00000238932 |
. |
92111 |
0.09 |
| chr10_117532445_117532596 | 0.30 |
ATRNL1 |
attractin-like 1 |
471037 |
0.01 |
| chr12_89339824_89340184 | 0.30 |
ENSG00000238302 |
. |
336058 |
0.01 |
| chr4_152277546_152277697 | 0.30 |
FAM160A1 |
family with sequence similarity 160, member A1 |
52777 |
0.13 |
| chr2_65581901_65582416 | 0.30 |
SPRED2 |
sprouty-related, EVH1 domain containing 2 |
10238 |
0.23 |
| chr9_137251018_137251277 | 0.30 |
ENSG00000263897 |
. |
20110 |
0.22 |
| chr4_174132836_174132987 | 0.30 |
RP11-10K16.1 |
|
42108 |
0.13 |
| chr11_96059054_96059205 | 0.30 |
ENSG00000266192 |
. |
15473 |
0.19 |
| chr5_153826475_153827364 | 0.30 |
SAP30L |
SAP30-like |
814 |
0.51 |
| chr3_97636799_97636950 | 0.30 |
CRYBG3 |
beta-gamma crystallin domain containing 3 |
6182 |
0.25 |
| chr12_58923583_58923734 | 0.30 |
RP11-362K2.2 |
Protein LOC100506869 |
14249 |
0.3 |
| chr4_184558434_184558585 | 0.30 |
ENSG00000252157 |
. |
16320 |
0.14 |
| chr17_39222775_39222926 | 0.30 |
KRTAP2-4 |
keratin associated protein 2-4 |
719 |
0.36 |
| chr17_53804938_53805089 | 0.30 |
TMEM100 |
transmembrane protein 100 |
4469 |
0.29 |
| chr13_107156553_107156704 | 0.30 |
EFNB2 |
ephrin-B2 |
30834 |
0.23 |
| chr2_43941438_43941589 | 0.30 |
ENSG00000252490 |
. |
57976 |
0.11 |
| chr14_100376837_100376988 | 0.30 |
EML1 |
echinoderm microtubule associated protein like 1 |
12342 |
0.2 |
| chr7_80350905_80351077 | 0.30 |
CD36 |
CD36 molecule (thrombospondin receptor) |
48299 |
0.19 |
| chr8_42409885_42410036 | 0.29 |
SMIM19 |
small integral membrane protein 19 |
8389 |
0.18 |
| chr4_14728667_14728818 | 0.29 |
ENSG00000202449 |
. |
36408 |
0.24 |
| chr7_29662924_29663075 | 0.29 |
DPY19L2P3 |
DPY19L2 pseudogene 3 |
26844 |
0.17 |
| chr6_121988152_121988303 | 0.29 |
ENSG00000222659 |
. |
86560 |
0.1 |
| chr9_74299304_74300206 | 0.29 |
TMEM2 |
transmembrane protein 2 |
20093 |
0.28 |
| chr16_24742094_24743166 | 0.29 |
TNRC6A |
trinucleotide repeat containing 6A |
1596 |
0.5 |
| chr2_68420623_68420967 | 0.29 |
RP11-474G23.2 |
|
13830 |
0.14 |
| chr4_148693217_148693552 | 0.29 |
ENSG00000264274 |
. |
10362 |
0.18 |
| chr5_136117041_136117192 | 0.29 |
ENSG00000222285 |
. |
187879 |
0.03 |
| chr12_64565372_64565523 | 0.29 |
RP11-196H14.3 |
|
23810 |
0.14 |
| chr11_13310143_13310461 | 0.29 |
ARNTL |
aryl hydrocarbon receptor nuclear translocator-like |
10870 |
0.27 |
| chr3_187985246_187985463 | 0.29 |
LPP |
LIM domain containing preferred translocation partner in lipoma |
27702 |
0.23 |
| chr17_26610027_26610178 | 0.29 |
ENSG00000207844 |
. |
6523 |
0.1 |
| chr3_156469526_156469677 | 0.29 |
TIPARP |
TCDD-inducible poly(ADP-ribose) polymerase |
70646 |
0.1 |
| chr21_18145527_18145678 | 0.29 |
ENSG00000239023 |
. |
54285 |
0.17 |
| chr8_89288937_89289088 | 0.28 |
RP11-586K2.1 |
|
50053 |
0.16 |
| chr12_52604417_52604648 | 0.28 |
KRT80 |
keratin 80 |
18748 |
0.1 |
| chr9_21590451_21590602 | 0.28 |
MIR31HG |
MIR31 host gene (non-protein coding) |
30858 |
0.16 |
| chr4_140958813_140959347 | 0.28 |
RP11-392B6.1 |
|
90089 |
0.09 |
| chr1_199825379_199825530 | 0.28 |
ENSG00000200139 |
. |
31833 |
0.23 |
| chr14_94839858_94840009 | 0.28 |
SERPINA1 |
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 |
14993 |
0.18 |
| chr12_132267707_132268092 | 0.28 |
ENSG00000200516 |
. |
18080 |
0.16 |
| chr11_123089847_123089998 | 0.28 |
CLMP |
CXADR-like membrane protein |
23933 |
0.18 |
| chr12_6332006_6332305 | 0.28 |
ENSG00000202318 |
. |
11466 |
0.17 |
| chr8_141734362_141734513 | 0.28 |
PTK2 |
protein tyrosine kinase 2 |
2550 |
0.32 |
| chr4_169330808_169330959 | 0.28 |
DDX60L |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 60-like |
8754 |
0.24 |
| chr11_108742969_108743120 | 0.28 |
ENSG00000201243 |
. |
133200 |
0.05 |
| chr10_95236478_95236629 | 0.28 |
MYOF |
myoferlin |
5398 |
0.21 |
| chr4_126245178_126245329 | 0.28 |
FAT4 |
FAT atypical cadherin 4 |
7699 |
0.29 |
| chr3_188067044_188067254 | 0.28 |
LPP |
LIM domain containing preferred translocation partner in lipoma |
109497 |
0.07 |
| chr8_25771009_25771433 | 0.28 |
EBF2 |
early B-cell factor 2 |
25789 |
0.27 |
| chr4_187597285_187597436 | 0.28 |
FAT1 |
FAT atypical cadherin 1 |
47649 |
0.17 |
| chr3_17719701_17719852 | 0.28 |
ENSG00000271841 |
. |
13321 |
0.25 |
| chr12_107256430_107256581 | 0.28 |
RP11-144F15.1 |
Uncharacterized protein |
87809 |
0.07 |
| chr7_129304567_129304757 | 0.28 |
NRF1 |
nuclear respiratory factor 1 |
7471 |
0.24 |
| chr2_48571922_48572463 | 0.28 |
ENSG00000251889 |
. |
4227 |
0.27 |
| chr14_52001291_52001686 | 0.28 |
ENSG00000252400 |
. |
11906 |
0.22 |
| chr21_39664180_39664501 | 0.28 |
KCNJ15 |
potassium inwardly-rectifying channel, subfamily J, member 15 |
4139 |
0.33 |
| chr2_147739330_147739481 | 0.28 |
ENSG00000238860 |
. |
342134 |
0.01 |
| chr12_32748094_32748245 | 0.28 |
FGD4 |
FYVE, RhoGEF and PH domain containing 4 |
3291 |
0.33 |
| chr11_114224456_114224725 | 0.28 |
RP11-64D24.2 |
|
2703 |
0.24 |
| chr6_52382258_52382409 | 0.28 |
TRAM2 |
translocation associated membrane protein 2 |
59380 |
0.12 |
| chr5_57483908_57484059 | 0.28 |
ENSG00000238899 |
. |
117968 |
0.07 |
| chr17_27574011_27574162 | 0.28 |
CRYBA1 |
crystallin, beta A1 |
205 |
0.94 |
| chr7_18809523_18809674 | 0.28 |
ENSG00000222164 |
. |
38304 |
0.21 |
| chr14_78060038_78060870 | 0.28 |
SPTLC2 |
serine palmitoyltransferase, long chain base subunit 2 |
3210 |
0.25 |
| chr7_77496191_77496692 | 0.28 |
PHTF2 |
putative homeodomain transcription factor 2 |
26944 |
0.18 |
| chr1_98793274_98793457 | 0.28 |
ENSG00000221777 |
. |
45576 |
0.21 |
| chr1_202251770_202252122 | 0.28 |
ENSG00000239046 |
. |
51154 |
0.1 |
| chr15_60699546_60699697 | 0.28 |
ANXA2 |
annexin A2 |
4539 |
0.29 |
| chr2_225955794_225955945 | 0.28 |
DOCK10 |
dedicator of cytokinesis 10 |
48710 |
0.18 |
| chr8_67472669_67472820 | 0.28 |
MYBL1 |
v-myb avian myeloblastosis viral oncogene homolog-like 1 |
52398 |
0.1 |
| chr13_76299059_76299210 | 0.28 |
LMO7 |
LIM domain 7 |
35369 |
0.19 |
| chr5_86453164_86453315 | 0.27 |
ENSG00000265919 |
. |
42468 |
0.15 |
| chr13_76867098_76867386 | 0.27 |
ENSG00000243274 |
. |
158175 |
0.04 |
| chr7_80127442_80127593 | 0.27 |
GNAT3 |
guanine nucleotide binding protein, alpha transducing 3 |
13819 |
0.27 |
| chr11_27407572_27407723 | 0.27 |
CCDC34 |
coiled-coil domain containing 34 |
22232 |
0.21 |
| chr10_89435270_89435421 | 0.27 |
PAPSS2 |
3'-phosphoadenosine 5'-phosphosulfate synthase 2 |
14637 |
0.2 |
| chr3_58034413_58034564 | 0.27 |
FLNB |
filamin B, beta |
29568 |
0.21 |
| chr11_35606785_35606936 | 0.27 |
FJX1 |
four jointed box 1 (Drosophila) |
32875 |
0.18 |
| chr6_53389704_53389855 | 0.27 |
GCLC |
glutamate-cysteine ligase, catalytic subunit |
1657 |
0.39 |
| chr9_83643160_83643311 | 0.27 |
ENSG00000221581 |
. |
293991 |
0.01 |
| chr2_55327280_55327923 | 0.27 |
ENSG00000266376 |
. |
8245 |
0.2 |
| chr9_134918134_134918364 | 0.27 |
MED27 |
mediator complex subunit 27 |
37004 |
0.2 |
| chr11_130717130_130717402 | 0.27 |
SNX19 |
sorting nexin 19 |
46503 |
0.19 |
| chr20_46815691_46815842 | 0.27 |
ENSG00000252227 |
. |
51000 |
0.18 |
| chr14_59804366_59804548 | 0.27 |
ENSG00000252869 |
. |
62284 |
0.12 |
| chr1_206164682_206164833 | 0.27 |
FAM72A |
family with sequence similarity 72, member A |
25846 |
0.15 |
| chr1_157977722_157977873 | 0.27 |
KIRREL |
kin of IRRE like (Drosophila) |
14362 |
0.2 |
| chr9_38026523_38026674 | 0.27 |
SHB |
Src homology 2 domain containing adaptor protein B |
42610 |
0.17 |
| chr6_163777027_163777374 | 0.27 |
QKI |
QKI, KH domain containing, RNA binding |
58475 |
0.16 |
| chr14_74724538_74725207 | 0.27 |
VSX2 |
visual system homeobox 2 |
18697 |
0.14 |
| chr5_39072103_39073220 | 0.27 |
RICTOR |
RPTOR independent companion of MTOR, complex 2 |
1830 |
0.43 |
| chr20_11910800_11911092 | 0.27 |
BTBD3 |
BTB (POZ) domain containing 3 |
11447 |
0.24 |
| chr10_4808545_4809063 | 0.27 |
AKR1E2 |
aldo-keto reductase family 1, member E2 |
20017 |
0.25 |
| chr10_60462562_60462713 | 0.27 |
BICC1 |
bicaudal C homolog 1 (Drosophila) |
90658 |
0.1 |
| chr13_46074470_46074652 | 0.26 |
COG3 |
component of oligomeric golgi complex 3 |
35501 |
0.15 |
| chr13_43843628_43843779 | 0.26 |
ENOX1 |
ecto-NOX disulfide-thiol exchanger 1 |
91510 |
0.1 |
| chr5_14806397_14806548 | 0.26 |
ENSG00000264792 |
. |
19649 |
0.23 |
| chr1_51127497_51127778 | 0.26 |
FAF1 |
Fas (TNFRSF6) associated factor 1 |
49467 |
0.15 |
| chr1_94269708_94269859 | 0.26 |
RP11-488P3.1 |
|
28783 |
0.16 |
| chr12_120620090_120620241 | 0.26 |
ENSG00000263913 |
. |
10235 |
0.11 |
| chr9_16031215_16031463 | 0.26 |
C9orf92 |
chromosome 9 open reading frame 92 |
184558 |
0.03 |
| chr1_87174228_87174695 | 0.26 |
SH3GLB1 |
SH3-domain GRB2-like endophilin B1 |
3883 |
0.2 |
| chr17_40554278_40554429 | 0.26 |
ENSG00000221020 |
. |
3326 |
0.15 |
| chr4_159080194_159080345 | 0.26 |
FAM198B |
family with sequence similarity 198, member B |
564 |
0.76 |
| chr3_99750954_99751105 | 0.26 |
ENSG00000264897 |
. |
67787 |
0.11 |
| chr3_11550931_11551082 | 0.26 |
VGLL4 |
vestigial like 4 (Drosophila) |
59392 |
0.14 |
| chr2_33589041_33589192 | 0.26 |
ENSG00000252246 |
. |
31151 |
0.19 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0071436 | sodium ion export(GO:0071436) |
| 0.1 | 0.3 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.1 | 0.3 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.1 | 0.2 | GO:0060426 | lung vasculature development(GO:0060426) |
| 0.1 | 0.2 | GO:0051834 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.1 | 0.3 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.1 | 0.2 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.1 | 0.2 | GO:0060492 | foregut regionalization(GO:0060423) lung field specification(GO:0060424) lung induction(GO:0060492) |
| 0.1 | 0.3 | GO:0097061 | dendritic spine morphogenesis(GO:0060997) regulation of dendritic spine morphogenesis(GO:0061001) dendritic spine organization(GO:0097061) |
| 0.1 | 0.3 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.1 | 0.2 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) negative regulation of neural precursor cell proliferation(GO:2000178) |
| 0.1 | 0.2 | GO:0019934 | cGMP-mediated signaling(GO:0019934) |
| 0.1 | 0.2 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.1 | 0.2 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.1 | 0.2 | GO:0051409 | response to nitrosative stress(GO:0051409) |
| 0.1 | 0.1 | GO:0051176 | positive regulation of sulfur metabolic process(GO:0051176) |
| 0.0 | 0.2 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.1 | GO:1900115 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.3 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.2 | GO:0010837 | regulation of keratinocyte proliferation(GO:0010837) |
| 0.0 | 0.1 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.1 | GO:0002604 | dendritic cell antigen processing and presentation(GO:0002468) regulation of dendritic cell antigen processing and presentation(GO:0002604) positive regulation of dendritic cell antigen processing and presentation(GO:0002606) |
| 0.0 | 0.2 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.2 | GO:0045091 | regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045091) |
| 0.0 | 0.1 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.1 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) |
| 0.0 | 0.1 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.0 | 0.2 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.1 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 0.0 | GO:0060435 | bronchiole development(GO:0060435) |
| 0.0 | 0.1 | GO:0035385 | Roundabout signaling pathway(GO:0035385) |
| 0.0 | 0.1 | GO:0048245 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.0 | 0.1 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.1 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.1 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.0 | 0.1 | GO:0090081 | regulation of heart induction by regulation of canonical Wnt signaling pathway(GO:0090081) |
| 0.0 | 0.0 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
| 0.0 | 0.1 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.0 | 0.1 | GO:0031659 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G1/S transition of mitotic cell cycle(GO:0031657) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G1/S transition of mitotic cell cycle(GO:0031659) |
| 0.0 | 0.2 | GO:0008356 | asymmetric cell division(GO:0008356) |
| 0.0 | 0.1 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.0 | 0.1 | GO:0060839 | endothelial cell fate commitment(GO:0060839) |
| 0.0 | 0.1 | GO:0060841 | venous blood vessel development(GO:0060841) |
| 0.0 | 0.1 | GO:0060638 | mesenchymal-epithelial cell signaling(GO:0060638) |
| 0.0 | 0.1 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.1 | GO:0043578 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.1 | GO:0048669 | collateral sprouting in absence of injury(GO:0048669) |
| 0.0 | 0.1 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.0 | 0.2 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 0.1 | GO:0060594 | mammary gland specification(GO:0060594) |
| 0.0 | 0.0 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.0 | 0.2 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.1 | GO:0042501 | serine phosphorylation of STAT protein(GO:0042501) |
| 0.0 | 0.1 | GO:0003283 | atrial septum development(GO:0003283) atrial septum morphogenesis(GO:0060413) |
| 0.0 | 0.1 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.1 | GO:0045077 | negative regulation of interferon-gamma biosynthetic process(GO:0045077) |
| 0.0 | 0.1 | GO:0008300 | isoprenoid catabolic process(GO:0008300) |
| 0.0 | 0.1 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.1 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.0 | 0.1 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.0 | 0.1 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.0 | 0.1 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.0 | 0.1 | GO:0009414 | response to water deprivation(GO:0009414) |
| 0.0 | 0.1 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.0 | 0.1 | GO:0060081 | membrane hyperpolarization(GO:0060081) |
| 0.0 | 0.1 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.1 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) ribonucleoside diphosphate catabolic process(GO:0009191) |
| 0.0 | 0.0 | GO:0034625 | fatty acid elongation, monounsaturated fatty acid(GO:0034625) |
| 0.0 | 0.2 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.0 | GO:0021631 | optic nerve morphogenesis(GO:0021631) |
| 0.0 | 0.0 | GO:0072216 | positive regulation of metanephros development(GO:0072216) |
| 0.0 | 0.0 | GO:0034067 | protein localization to Golgi apparatus(GO:0034067) |
| 0.0 | 0.1 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 0.1 | GO:0071731 | response to nitric oxide(GO:0071731) |
| 0.0 | 0.1 | GO:0003032 | detection of oxygen(GO:0003032) |
| 0.0 | 0.1 | GO:2000095 | regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.0 | 0.7 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.2 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.1 | GO:0042984 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.0 | 0.1 | GO:0046512 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.0 | 0.1 | GO:0060457 | negative regulation of digestive system process(GO:0060457) |
| 0.0 | 0.1 | GO:0009128 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.0 | 0.0 | GO:0071072 | negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 0.0 | 0.1 | GO:2000757 | negative regulation of histone acetylation(GO:0035067) negative regulation of protein acetylation(GO:1901984) negative regulation of peptidyl-lysine acetylation(GO:2000757) |
| 0.0 | 0.1 | GO:0010715 | regulation of extracellular matrix disassembly(GO:0010715) negative regulation of extracellular matrix disassembly(GO:0010716) regulation of extracellular matrix organization(GO:1903053) negative regulation of extracellular matrix organization(GO:1903054) |
| 0.0 | 0.1 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.1 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 0.0 | GO:0060129 | thyroid-stimulating hormone-secreting cell differentiation(GO:0060129) |
| 0.0 | 0.0 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.1 | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.0 | 0.0 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.0 | 0.0 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 0.0 | 0.0 | GO:0071071 | regulation of phospholipid biosynthetic process(GO:0071071) |
| 0.0 | 0.2 | GO:0031000 | response to caffeine(GO:0031000) |
| 0.0 | 0.1 | GO:0009649 | entrainment of circadian clock(GO:0009649) |
| 0.0 | 0.1 | GO:0006929 | substrate-dependent cell migration(GO:0006929) |
| 0.0 | 0.0 | GO:0060592 | mammary gland formation(GO:0060592) mammary placode formation(GO:0060596) |
| 0.0 | 0.1 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.0 | GO:0048670 | regulation of collateral sprouting(GO:0048670) |
| 0.0 | 0.0 | GO:0060406 | positive regulation of penile erection(GO:0060406) |
| 0.0 | 0.0 | GO:0010659 | striated muscle cell apoptotic process(GO:0010658) cardiac muscle cell apoptotic process(GO:0010659) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) regulation of cardiac muscle cell apoptotic process(GO:0010665) negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.0 | 0.1 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.0 | 0.0 | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.0 | 0.1 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) |
| 0.0 | 0.0 | GO:0060174 | limb bud formation(GO:0060174) |
| 0.0 | 0.1 | GO:0033603 | positive regulation of dopamine secretion(GO:0033603) |
| 0.0 | 0.1 | GO:0032462 | regulation of protein homooligomerization(GO:0032462) |
| 0.0 | 0.0 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.0 | 0.1 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.0 | 0.0 | GO:0050942 | positive regulation of melanocyte differentiation(GO:0045636) positive regulation of pigment cell differentiation(GO:0050942) |
| 0.0 | 0.3 | GO:0048008 | platelet-derived growth factor receptor signaling pathway(GO:0048008) |
| 0.0 | 0.0 | GO:0036445 | forebrain neuroblast division(GO:0021873) neuronal stem cell division(GO:0036445) neuroblast division(GO:0055057) |
| 0.0 | 0.0 | GO:2000649 | regulation of sodium:proton antiporter activity(GO:0032415) regulation of sodium ion transmembrane transport(GO:1902305) regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.0 | 0.1 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.0 | GO:0090200 | positive regulation of mitochondrion organization(GO:0010822) positive regulation of release of cytochrome c from mitochondria(GO:0090200) positive regulation of apoptotic signaling pathway(GO:2001235) |
| 0.0 | 0.1 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 0.2 | GO:0046459 | short-chain fatty acid metabolic process(GO:0046459) |
| 0.0 | 0.0 | GO:0009111 | vitamin catabolic process(GO:0009111) fat-soluble vitamin catabolic process(GO:0042363) |
| 0.0 | 0.1 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 0.0 | 0.0 | GO:0003079 | obsolete positive regulation of natriuresis(GO:0003079) |
| 0.0 | 0.0 | GO:0007132 | meiotic metaphase I(GO:0007132) |
| 0.0 | 0.0 | GO:0060083 | smooth muscle contraction involved in micturition(GO:0060083) |
| 0.0 | 0.0 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.0 | GO:0036037 | CD8-positive, alpha-beta T cell activation(GO:0036037) CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.0 | GO:0060179 | male courtship behavior(GO:0008049) male mating behavior(GO:0060179) |
| 0.0 | 0.1 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 0.1 | GO:0060438 | trachea development(GO:0060438) |
| 0.0 | 0.0 | GO:0070091 | glucagon secretion(GO:0070091) regulation of glucagon secretion(GO:0070092) |
| 0.0 | 0.2 | GO:0019321 | pentose metabolic process(GO:0019321) |
| 0.0 | 0.0 | GO:0009448 | gamma-aminobutyric acid metabolic process(GO:0009448) |
| 0.0 | 0.0 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.0 | 0.1 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.0 | GO:0046882 | negative regulation of follicle-stimulating hormone secretion(GO:0046882) |
| 0.0 | 0.0 | GO:1901374 | acetylcholine transport(GO:0015870) acetate ester transport(GO:1901374) |
| 0.0 | 0.0 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.0 | 0.1 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.0 | 0.0 | GO:0032236 | obsolete positive regulation of calcium ion transport via store-operated calcium channel activity(GO:0032236) |
| 0.0 | 0.0 | GO:0010891 | negative regulation of sequestering of triglyceride(GO:0010891) |
| 0.0 | 0.1 | GO:0061384 | heart trabecula formation(GO:0060347) heart trabecula morphogenesis(GO:0061384) |
| 0.0 | 0.0 | GO:2000008 | regulation of protein localization to cell surface(GO:2000008) |
| 0.0 | 0.2 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.0 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 | 0.0 | GO:0061525 | hindgut morphogenesis(GO:0007442) hindgut development(GO:0061525) |
| 0.0 | 0.0 | GO:0010661 | positive regulation of muscle cell apoptotic process(GO:0010661) positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.0 | 0.1 | GO:0009200 | deoxyribonucleoside triphosphate metabolic process(GO:0009200) |
| 0.0 | 0.0 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) |
| 0.0 | 0.1 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.0 | 0.0 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.0 | GO:0002385 | innate immune response in mucosa(GO:0002227) mucosal immune response(GO:0002385) |
| 0.0 | 0.0 | GO:0001921 | positive regulation of receptor recycling(GO:0001921) |
| 0.0 | 0.1 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.0 | 0.1 | GO:0090504 | wound healing, spreading of epidermal cells(GO:0035313) wound healing, spreading of cells(GO:0044319) epiboly(GO:0090504) epiboly involved in wound healing(GO:0090505) |
| 0.0 | 0.1 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.1 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 | 0.1 | GO:0007635 | chemosensory behavior(GO:0007635) |
| 0.0 | 0.0 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.0 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.0 | 0.0 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.1 | GO:0060013 | righting reflex(GO:0060013) |
| 0.0 | 0.1 | GO:0009886 | post-embryonic morphogenesis(GO:0009886) |
| 0.0 | 0.0 | GO:1902803 | regulation of synaptic vesicle transport(GO:1902803) regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.0 | 0.0 | GO:0014848 | urinary tract smooth muscle contraction(GO:0014848) |
| 0.0 | 0.2 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 | 0.0 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.0 | GO:0061004 | pattern specification involved in kidney development(GO:0061004) renal system pattern specification(GO:0072048) pattern specification involved in metanephros development(GO:0072268) |
| 0.0 | 0.1 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 0.0 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) |
| 0.0 | 0.0 | GO:0060449 | bud elongation involved in lung branching(GO:0060449) |
| 0.0 | 0.1 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.0 | 0.0 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.0 | GO:0006210 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.0 | 0.0 | GO:0051280 | negative regulation of release of sequestered calcium ion into cytosol(GO:0051280) positive regulation of sequestering of calcium ion(GO:0051284) negative regulation of calcium ion transmembrane transport(GO:1903170) |
| 0.0 | 0.2 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.0 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.0 | 0.0 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.0 | 0.0 | GO:0070254 | mucus secretion(GO:0070254) |
| 0.0 | 0.0 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.0 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.0 | GO:0030033 | microvillus assembly(GO:0030033) microvillus organization(GO:0032528) |
| 0.0 | 0.1 | GO:0008611 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.0 | GO:0055094 | response to lipoprotein particle(GO:0055094) |
| 0.0 | 0.0 | GO:0022605 | oogenesis stage(GO:0022605) |
| 0.0 | 0.1 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.0 | GO:0048643 | positive regulation of skeletal muscle tissue development(GO:0048643) |
| 0.0 | 0.1 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.0 | GO:0009151 | purine deoxyribonucleotide metabolic process(GO:0009151) |
| 0.0 | 0.0 | GO:0048087 | positive regulation of developmental pigmentation(GO:0048087) |
| 0.0 | 0.1 | GO:0000768 | syncytium formation by plasma membrane fusion(GO:0000768) myoblast fusion(GO:0007520) |
| 0.0 | 0.0 | GO:0010766 | negative regulation of sodium ion transport(GO:0010766) |
| 0.0 | 0.0 | GO:0097435 | extracellular fibril organization(GO:0043206) fibril organization(GO:0097435) |
| 0.0 | 0.0 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.0 | 0.0 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.0 | GO:0001302 | replicative cell aging(GO:0001302) |
| 0.0 | 0.0 | GO:0006388 | RNA splicing, via endonucleolytic cleavage and ligation(GO:0000394) tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.0 | GO:0097094 | cranial suture morphogenesis(GO:0060363) craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.1 | GO:0016246 | RNA interference(GO:0016246) |
| 0.0 | 0.0 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.1 | GO:0043277 | apoptotic cell clearance(GO:0043277) |
| 0.0 | 0.0 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.0 | GO:0070296 | sarcoplasmic reticulum calcium ion transport(GO:0070296) |
| 0.0 | 0.0 | GO:0010896 | regulation of triglyceride catabolic process(GO:0010896) positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.0 | 0.0 | GO:0007500 | mesodermal cell fate determination(GO:0007500) |
| 0.0 | 0.1 | GO:0021955 | central nervous system neuron axonogenesis(GO:0021955) |
| 0.0 | 0.1 | GO:0001945 | lymph vessel development(GO:0001945) |
| 0.0 | 0.0 | GO:0006498 | N-terminal protein lipidation(GO:0006498) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.1 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.0 | 0.1 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.2 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.8 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.1 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.2 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.1 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.0 | GO:0018995 | host(GO:0018995) host cell part(GO:0033643) host intracellular part(GO:0033646) intracellular region of host(GO:0043656) host cell(GO:0043657) other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) |
| 0.0 | 0.1 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.2 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.1 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.1 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.1 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.5 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.1 | GO:0060200 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.0 | 0.1 | GO:0098642 | network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.0 | 0.1 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.1 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.1 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.0 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 0.0 | 0.1 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.0 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.0 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.1 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.1 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.1 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.1 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 0.0 | 0.5 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.1 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.1 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.0 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.0 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.1 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.1 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.8 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.0 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.1 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.1 | GO:0000145 | exocyst(GO:0000145) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 0.3 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 0.3 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.1 | 0.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 0.3 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.1 | 0.3 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.1 | 0.2 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 0.2 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.1 | 0.2 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 0.2 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.2 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.2 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.1 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.1 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.2 | GO:0004470 | malic enzyme activity(GO:0004470) |
| 0.0 | 0.1 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.1 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.0 | 0.1 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.3 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.1 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.2 | GO:0004661 | protein geranylgeranyltransferase activity(GO:0004661) |
| 0.0 | 0.1 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.2 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.0 | 0.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.1 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) xenobiotic transporter activity(GO:0042910) |
| 0.0 | 0.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.2 | GO:0016595 | glutamate binding(GO:0016595) |
| 0.0 | 0.1 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.1 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.0 | 0.0 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.1 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.1 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.0 | 0.1 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.1 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.0 | 0.1 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.1 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.0 | 0.1 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.1 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.0 | 0.1 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.0 | 0.1 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.1 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.5 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.1 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.1 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.0 | 0.1 | GO:0004904 | interferon receptor activity(GO:0004904) |
| 0.0 | 0.1 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.1 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.2 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.0 | 0.1 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.1 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.0 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.0 | 0.1 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.1 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.1 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.0 | 0.1 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.1 | GO:0004645 | phosphorylase activity(GO:0004645) |
| 0.0 | 0.1 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.2 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.1 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.0 | 0.1 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.1 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.1 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.0 | 0.2 | GO:0004312 | fatty acid synthase activity(GO:0004312) |
| 0.0 | 0.2 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.1 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.0 | 0.0 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.2 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.1 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.1 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 0.0 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.0 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.0 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.0 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.0 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.0 | 0.1 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.0 | 0.1 | GO:0042153 | obsolete RPTP-like protein binding(GO:0042153) |
| 0.0 | 0.0 | GO:0052659 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) inositol-1,3,4,5-tetrakisphosphate 5-phosphatase activity(GO:0052659) |
| 0.0 | 0.2 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.0 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.0 | 0.2 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.1 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.8 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 0.2 | GO:0043498 | obsolete cell surface binding(GO:0043498) |
| 0.0 | 0.1 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.1 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.0 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.1 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.0 | 0.1 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.1 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.1 | GO:0048019 | receptor inhibitor activity(GO:0030547) receptor antagonist activity(GO:0048019) |
| 0.0 | 0.7 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.0 | GO:0050543 | icosatetraenoic acid binding(GO:0050543) |
| 0.0 | 0.1 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.1 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.1 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.1 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.1 | GO:0015385 | sodium:proton antiporter activity(GO:0015385) |
| 0.0 | 0.0 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 0.1 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.1 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.0 | 0.2 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.0 | GO:0032450 | maltose alpha-glucosidase activity(GO:0032450) |
| 0.0 | 0.0 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.2 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.1 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.1 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.1 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.2 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.0 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.1 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.0 | 0.0 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.0 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.0 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.0 | 0.1 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.0 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.2 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.1 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.1 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.0 | 0.0 | GO:0016149 | translation release factor activity, codon specific(GO:0016149) |
| 0.0 | 0.1 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.0 | GO:0008252 | nucleotidase activity(GO:0008252) |
| 0.0 | 0.1 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.0 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.1 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 1.0 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.2 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.0 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.4 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.0 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 0.0 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.0 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 0.3 | PID NECTIN PATHWAY | Nectin adhesion pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.0 | REACTOME REGULATION OF APOPTOSIS | Genes involved in Regulation of Apoptosis |
| 0.0 | 0.0 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.0 | 0.3 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.3 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.2 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.0 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.0 | 0.2 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.0 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.4 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.3 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.1 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.2 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.0 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.2 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.1 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.0 | REACTOME PKB MEDIATED EVENTS | Genes involved in PKB-mediated events |
| 0.0 | 0.1 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.0 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.0 | 0.3 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.1 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.0 | 0.1 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.0 | REACTOME ADP SIGNALLING THROUGH P2RY1 | Genes involved in ADP signalling through P2Y purinoceptor 1 |
| 0.0 | 0.1 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.0 | 0.1 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.1 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.0 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |