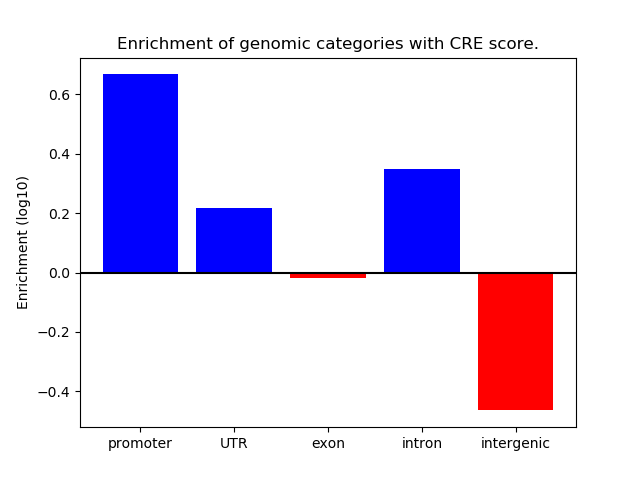
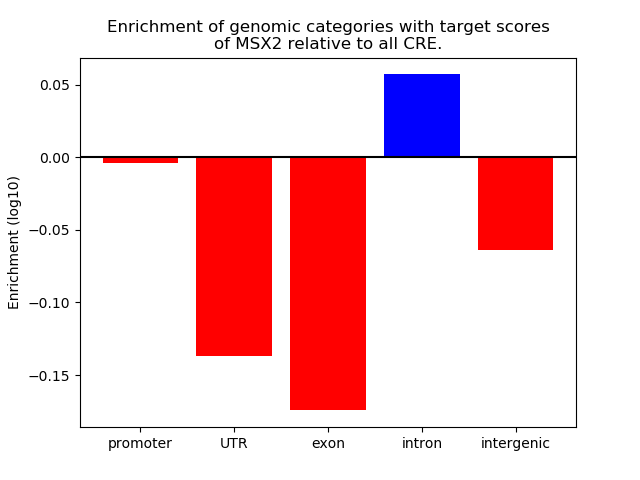
Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
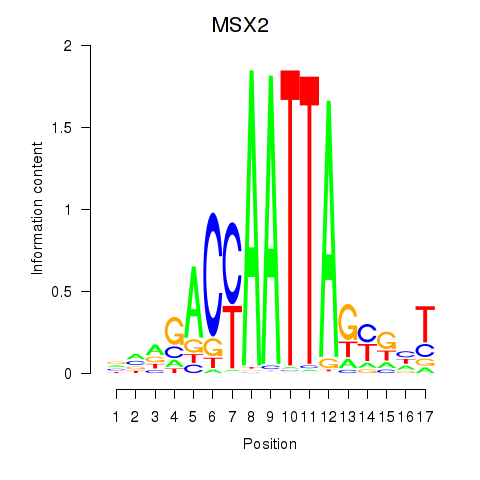
Results for MSX2
Z-value: 1.03
Transcription factors associated with MSX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MSX2
|
ENSG00000120149.7 | msh homeobox 2 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr5_174151057_174151248 | MSX2 | 384 | 0.906182 | 0.81 | 8.5e-03 | Click! |
| chr5_173916383_173916534 | MSX2 | 235078 | 0.019896 | -0.69 | 3.8e-02 | Click! |
| chr5_173918124_173918275 | MSX2 | 233337 | 0.020178 | -0.69 | 4.1e-02 | Click! |
| chr5_173888101_173888252 | MSX2 | 263360 | 0.016023 | 0.67 | 4.6e-02 | Click! |
| chr5_173882231_173882382 | MSX2 | 269230 | 0.015351 | 0.67 | 4.6e-02 | Click! |
Activity of the MSX2 motif across conditions
Conditions sorted by the z-value of the MSX2 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr17_64419641_64419918 | 0.70 |
RP11-4F22.2 |
|
6807 |
0.26 |
| chr18_59619334_59619485 | 0.65 |
RNF152 |
ring finger protein 152 |
57945 |
0.16 |
| chr3_17274716_17274867 | 0.60 |
ENSG00000252976 |
. |
97577 |
0.08 |
| chr2_235231609_235231760 | 0.56 |
ARL4C |
ADP-ribosylation factor-like 4C |
173560 |
0.03 |
| chr7_150635354_150635936 | 0.53 |
KCNH2 |
potassium voltage-gated channel, subfamily H (eag-related), member 2 |
17349 |
0.15 |
| chr14_99720519_99720759 | 0.47 |
AL109767.1 |
|
8646 |
0.22 |
| chr10_98301601_98301752 | 0.47 |
TLL2 |
tolloid-like 2 |
28008 |
0.18 |
| chr22_19463070_19463221 | 0.44 |
UFD1L |
ubiquitin fusion degradation 1 like (yeast) |
3309 |
0.13 |
| chr12_112779348_112779499 | 0.44 |
HECTD4 |
HECT domain containing E3 ubiquitin protein ligase 4 |
40473 |
0.14 |
| chr11_20135030_20135181 | 0.42 |
NAV2-AS1 |
NAV2 antisense RNA 1 |
7073 |
0.24 |
| chrX_107068310_107068461 | 0.42 |
MID2 |
midline 2 |
600 |
0.75 |
| chr1_180549413_180549564 | 0.41 |
ENSG00000266683 |
. |
34520 |
0.16 |
| chr3_9445704_9445855 | 0.41 |
SETD5-AS1 |
SETD5 antisense RNA 1 |
5516 |
0.16 |
| chr15_81248608_81248759 | 0.41 |
KIAA1199 |
KIAA1199 |
22916 |
0.14 |
| chr5_142562188_142562394 | 0.40 |
ARHGAP26-IT1 |
ARHGAP26 intronic transcript 1 (non-protein coding) |
9774 |
0.25 |
| chr6_3159984_3160135 | 0.39 |
TUBB2A |
tubulin, beta 2A class IIa |
2299 |
0.23 |
| chr1_92019254_92019429 | 0.39 |
CDC7 |
cell division cycle 7 |
52646 |
0.17 |
| chr5_96000301_96000453 | 0.39 |
CAST |
calpastatin |
1620 |
0.38 |
| chr15_50648645_50649024 | 0.39 |
ENSG00000244879 |
. |
166 |
0.9 |
| chr16_50219117_50219379 | 0.38 |
PAPD5 |
PAP associated domain containing 5 |
31469 |
0.14 |
| chr12_95196881_95197032 | 0.37 |
ENSG00000208038 |
. |
31218 |
0.23 |
| chr2_23603358_23603509 | 0.37 |
KLHL29 |
kelch-like family member 29 |
4655 |
0.35 |
| chr19_31557220_31557371 | 0.37 |
AC020952.1 |
Uncharacterized protein |
83067 |
0.11 |
| chr14_75672848_75673038 | 0.36 |
ENSG00000252013 |
. |
4071 |
0.19 |
| chr10_96161826_96161977 | 0.36 |
TBC1D12 |
TBC1 domain family, member 12 |
360 |
0.9 |
| chr1_181121299_181121450 | 0.36 |
IER5 |
immediate early response 5 |
63736 |
0.13 |
| chr20_62042443_62042594 | 0.36 |
RP11-358D14.2 |
|
36886 |
0.09 |
| chr5_140998829_140999245 | 0.36 |
DIAPH1 |
diaphanous-related formin 1 |
415 |
0.66 |
| chr3_160940381_160941002 | 0.35 |
NMD3 |
NMD3 ribosome export adaptor |
1344 |
0.59 |
| chr1_172644665_172644877 | 0.35 |
FASLG |
Fas ligand (TNF superfamily, member 6) |
16613 |
0.25 |
| chr16_67618812_67618963 | 0.35 |
CTCF |
CCCTC-binding factor (zinc finger protein) |
22577 |
0.08 |
| chr11_117917434_117917585 | 0.35 |
ENSG00000272075 |
. |
20443 |
0.14 |
| chr22_20226344_20226543 | 0.35 |
RTN4R |
reticulon 4 receptor |
4764 |
0.15 |
| chrX_106928490_106928775 | 0.35 |
TSC22D3 |
TSC22 domain family, member 3 |
30999 |
0.18 |
| chr5_39231622_39231809 | 0.34 |
FYB |
FYN binding protein |
12007 |
0.28 |
| chr20_57231559_57231809 | 0.34 |
STX16 |
syntaxin 16 |
4621 |
0.19 |
| chr10_130706677_130706967 | 0.34 |
MGMT |
O-6-methylguanine-DNA methyltransferase |
558626 |
0.0 |
| chr15_60871759_60871981 | 0.33 |
RORA |
RAR-related orphan receptor A |
12870 |
0.21 |
| chr2_99954705_99955291 | 0.33 |
EIF5B |
eukaryotic translation initiation factor 5B |
1182 |
0.4 |
| chr2_119188712_119188863 | 0.33 |
INSIG2 |
insulin induced gene 2 |
342737 |
0.01 |
| chr15_48251538_48251689 | 0.33 |
SLC24A5 |
solute carrier family 24 (sodium/potassium/calcium exchanger), member 5 |
161556 |
0.03 |
| chr6_112306754_112307040 | 0.33 |
WISP3 |
WNT1 inducible signaling pathway protein 3 |
68378 |
0.11 |
| chr6_119227091_119227242 | 0.32 |
ASF1A |
anti-silencing function 1A histone chaperone |
11782 |
0.19 |
| chr18_60458887_60459075 | 0.32 |
AC015989.1 |
Uncharacterized protein |
32442 |
0.17 |
| chr3_122048704_122048855 | 0.32 |
CSTA |
cystatin A (stefin A) |
4670 |
0.19 |
| chr8_71124474_71124716 | 0.32 |
NCOA2 |
nuclear receptor coactivator 2 |
33015 |
0.21 |
| chr4_99563113_99563379 | 0.32 |
TSPAN5 |
tetraspanin 5 |
15540 |
0.22 |
| chr4_110483700_110484182 | 0.32 |
CCDC109B |
coiled-coil domain containing 109B |
1795 |
0.42 |
| chr17_47440332_47440604 | 0.32 |
ZNF652 |
zinc finger protein 652 |
633 |
0.61 |
| chr9_20320271_20320422 | 0.31 |
ENSG00000221744 |
. |
25344 |
0.21 |
| chr10_33425697_33425907 | 0.31 |
ENSG00000263576 |
. |
38238 |
0.16 |
| chr14_52795656_52796058 | 0.31 |
PTGER2 |
prostaglandin E receptor 2 (subtype EP2), 53kDa |
14744 |
0.25 |
| chr12_74884186_74884337 | 0.31 |
ATXN7L3B |
ataxin 7-like 3B |
47290 |
0.17 |
| chr14_53158829_53159146 | 0.31 |
ERO1L |
ERO1-like (S. cerevisiae) |
3260 |
0.21 |
| chr2_62806762_62807072 | 0.30 |
TMEM17 |
transmembrane protein 17 |
73441 |
0.1 |
| chr1_12291292_12291653 | 0.30 |
VPS13D |
vacuolar protein sorting 13 homolog D (S. cerevisiae) |
1348 |
0.41 |
| chr6_139492633_139492784 | 0.30 |
HECA |
headcase homolog (Drosophila) |
36459 |
0.17 |
| chr2_202297300_202297558 | 0.30 |
TRAK2 |
trafficking protein, kinesin binding 2 |
848 |
0.59 |
| chr9_20490846_20490997 | 0.30 |
ENSG00000264941 |
. |
11419 |
0.23 |
| chr8_144097895_144098282 | 0.30 |
LY6E |
lymphocyte antigen 6 complex, locus E |
1311 |
0.42 |
| chr10_29772721_29772872 | 0.29 |
SVIL |
supervillin |
12651 |
0.23 |
| chr17_62180290_62180441 | 0.29 |
ERN1 |
endoplasmic reticulum to nucleus signaling 1 |
27120 |
0.14 |
| chr7_73265347_73265568 | 0.29 |
WBSCR27 |
Williams Beuren syndrome chromosome region 27 |
8592 |
0.15 |
| chr5_36949315_36949466 | 0.29 |
NIPBL |
Nipped-B homolog (Drosophila) |
72498 |
0.13 |
| chr4_10107887_10108195 | 0.29 |
ENSG00000223086 |
. |
9467 |
0.16 |
| chr14_65380506_65380744 | 0.29 |
CHURC1 |
churchill domain containing 1 |
454 |
0.38 |
| chr17_63031973_63032289 | 0.29 |
RP11-583F2.5 |
|
15896 |
0.16 |
| chr7_156809109_156809289 | 0.29 |
MNX1-AS1 |
MNX1 antisense RNA 1 (head to head) |
5700 |
0.18 |
| chr20_37165542_37165693 | 0.29 |
ENSG00000263473 |
. |
20411 |
0.13 |
| chr8_25033335_25033486 | 0.29 |
ENSG00000241811 |
. |
5825 |
0.25 |
| chr16_4674083_4674234 | 0.29 |
MGRN1 |
mahogunin ring finger 1, E3 ubiquitin protein ligase |
633 |
0.59 |
| chr3_183544116_183544267 | 0.28 |
MAP6D1 |
MAP6 domain containing 1 |
809 |
0.53 |
| chr5_140934914_140935065 | 0.28 |
CTD-2024I7.13 |
|
2889 |
0.17 |
| chr15_83733229_83733380 | 0.28 |
BTBD1 |
BTB (POZ) domain containing 1 |
2599 |
0.17 |
| chr20_56969877_56970028 | 0.28 |
VAPB |
VAMP (vesicle-associated membrane protein)-associated protein B and C |
5583 |
0.26 |
| chr1_112491159_112491310 | 0.28 |
KCND3 |
potassium voltage-gated channel, Shal-related subfamily, member 3 |
34184 |
0.17 |
| chr9_33431365_33431516 | 0.28 |
AQP3 |
aquaporin 3 (Gill blood group) |
16169 |
0.14 |
| chr12_105956829_105956980 | 0.28 |
C12orf75 |
chromosome 12 open reading frame 75 |
232258 |
0.02 |
| chr1_193428730_193429035 | 0.28 |
ENSG00000252241 |
. |
272192 |
0.01 |
| chr2_228735932_228736236 | 0.28 |
DAW1 |
dynein assembly factor with WDR repeat domains 1 |
239 |
0.94 |
| chr6_139482795_139483056 | 0.28 |
HECA |
headcase homolog (Drosophila) |
26676 |
0.21 |
| chr12_64798932_64799366 | 0.28 |
XPOT |
exportin, tRNA |
323 |
0.88 |
| chr13_53027549_53027700 | 0.28 |
CKAP2 |
cytoskeleton associated protein 2 |
1940 |
0.3 |
| chr8_120219756_120219907 | 0.28 |
MAL2 |
mal, T-cell differentiation protein 2 (gene/pseudogene) |
779 |
0.76 |
| chr1_172149548_172149911 | 0.28 |
DNM3OS |
DNM3 opposite strand/antisense RNA |
35795 |
0.15 |
| chr3_119015914_119016065 | 0.28 |
ARHGAP31 |
Rho GTPase activating protein 31 |
2769 |
0.24 |
| chr6_35149420_35149571 | 0.27 |
SCUBE3 |
signal peptide, CUB domain, EGF-like 3 |
32695 |
0.14 |
| chr4_88929574_88929774 | 0.27 |
PKD2 |
polycystic kidney disease 2 (autosomal dominant) |
854 |
0.61 |
| chr5_170763650_170763947 | 0.27 |
ENSG00000265740 |
. |
22323 |
0.14 |
| chr5_95294789_95295005 | 0.27 |
ELL2 |
elongation factor, RNA polymerase II, 2 |
2655 |
0.26 |
| chr3_156478754_156479058 | 0.27 |
LEKR1 |
leucine, glutamate and lysine rich 1 |
65183 |
0.12 |
| chr14_66404533_66404789 | 0.27 |
CTD-2014B16.3 |
Uncharacterized protein |
66580 |
0.13 |
| chr18_46918320_46918471 | 0.27 |
DYM |
dymeclin |
13376 |
0.18 |
| chr13_26475136_26475287 | 0.27 |
AL138815.2 |
Uncharacterized protein |
22532 |
0.17 |
| chr22_36232750_36232901 | 0.27 |
RBFOX2 |
RNA binding protein, fox-1 homolog (C. elegans) 2 |
3440 |
0.35 |
| chr5_40511051_40511202 | 0.26 |
ENSG00000199552 |
. |
143939 |
0.04 |
| chr15_42352123_42352335 | 0.26 |
PLA2G4E |
phospholipase A2, group IVE |
8841 |
0.16 |
| chr22_32046621_32046810 | 0.26 |
PISD |
phosphatidylserine decarboxylase |
11451 |
0.2 |
| chr2_148804260_148804811 | 0.26 |
ORC4 |
origin recognition complex, subunit 4 |
25388 |
0.19 |
| chr22_40654247_40654479 | 0.26 |
TNRC6B |
trinucleotide repeat containing 6B |
6644 |
0.26 |
| chr16_70097897_70098048 | 0.26 |
PDXDC2P |
|
1707 |
0.31 |
| chr1_100889654_100889805 | 0.26 |
ENSG00000216067 |
. |
45398 |
0.13 |
| chr6_73135306_73135457 | 0.26 |
RIMS1 |
regulating synaptic membrane exocytosis 1 |
58949 |
0.16 |
| chr21_19188629_19188846 | 0.26 |
C21orf91 |
chromosome 21 open reading frame 91 |
2599 |
0.3 |
| chr1_116185862_116186059 | 0.26 |
VANGL1 |
VANGL planar cell polarity protein 1 |
710 |
0.71 |
| chr4_39364717_39364868 | 0.26 |
RFC1 |
replication factor C (activator 1) 1, 145kDa |
3177 |
0.21 |
| chr8_87524414_87524565 | 0.26 |
RMDN1 |
regulator of microtubule dynamics 1 |
2078 |
0.28 |
| chrX_16661091_16661259 | 0.26 |
S100G |
S100 calcium binding protein G |
7106 |
0.18 |
| chr8_91783068_91783219 | 0.26 |
NECAB1 |
N-terminal EF-hand calcium binding protein 1 |
20635 |
0.19 |
| chr9_5531438_5531729 | 0.26 |
PDCD1LG2 |
programmed cell death 1 ligand 2 |
21013 |
0.22 |
| chr11_2966895_2967126 | 0.25 |
PHLDA2 |
pleckstrin homology-like domain, family A, member 2 |
16325 |
0.11 |
| chr15_34873680_34873831 | 0.25 |
GOLGA8B |
golgin A8 family, member B |
2016 |
0.29 |
| chr14_69238852_69239003 | 0.25 |
ZFP36L1 |
ZFP36 ring finger protein-like 1 |
19033 |
0.19 |
| chr9_118131626_118131777 | 0.25 |
DEC1 |
deleted in esophageal cancer 1 |
227604 |
0.02 |
| chr1_10569828_10570301 | 0.25 |
PEX14 |
peroxisomal biogenesis factor 14 |
35025 |
0.11 |
| chr14_76605047_76605198 | 0.25 |
GPATCH2L |
G patch domain containing 2-like |
13140 |
0.26 |
| chr5_150644691_150644842 | 0.25 |
GM2A |
GM2 ganglioside activator |
5406 |
0.2 |
| chr2_31440406_31440557 | 0.25 |
CAPN14 |
calpain 14 |
70 |
0.97 |
| chr18_33647690_33647852 | 0.25 |
RPRD1A |
regulation of nuclear pre-mRNA domain containing 1A |
232 |
0.94 |
| chr6_4804997_4805148 | 0.25 |
CDYL |
chromodomain protein, Y-like |
28403 |
0.2 |
| chr17_39874084_39874250 | 0.25 |
ENSG00000201920 |
. |
239 |
0.85 |
| chr2_143918611_143918880 | 0.25 |
RP11-190J23.1 |
|
10996 |
0.25 |
| chr11_82617435_82617696 | 0.25 |
C11orf82 |
chromosome 11 open reading frame 82 |
2735 |
0.27 |
| chr13_99867467_99867663 | 0.25 |
ENSG00000201793 |
. |
9603 |
0.19 |
| chr3_142296047_142296384 | 0.25 |
ATR |
ataxia telangiectasia and Rad3 related |
1453 |
0.37 |
| chr3_132137362_132137693 | 0.24 |
DNAJC13 |
DnaJ (Hsp40) homolog, subfamily C, member 13 |
1157 |
0.6 |
| chr11_14378772_14379652 | 0.24 |
RRAS2 |
related RAS viral (r-ras) oncogene homolog 2 |
819 |
0.73 |
| chr8_61918765_61918944 | 0.24 |
CLVS1 |
clavesin 1 |
50863 |
0.18 |
| chr17_41668721_41668956 | 0.24 |
ETV4 |
ets variant 4 |
11850 |
0.16 |
| chr8_110987376_110987527 | 0.24 |
KCNV1 |
potassium channel, subfamily V, member 1 |
492 |
0.75 |
| chr9_100744268_100744520 | 0.24 |
ANP32B |
acidic (leucine-rich) nuclear phosphoprotein 32 family, member B |
1249 |
0.39 |
| chr1_223084470_223084621 | 0.24 |
ENSG00000239054 |
. |
744 |
0.72 |
| chr6_48037013_48037164 | 0.24 |
PTCHD4 |
patched domain containing 4 |
663 |
0.83 |
| chr16_85369002_85369321 | 0.24 |
RP11-680G10.1 |
Uncharacterized protein |
21908 |
0.2 |
| chr6_37665777_37665940 | 0.24 |
MDGA1 |
MAM domain containing glycosylphosphatidylinositol anchor 1 |
92 |
0.98 |
| chr12_104329327_104329478 | 0.24 |
ENSG00000265072 |
. |
5199 |
0.15 |
| chr5_54605298_54605894 | 0.24 |
SKIV2L2 |
superkiller viralicidic activity 2-like 2 (S. cerevisiae) |
1767 |
0.29 |
| chr14_63940495_63940646 | 0.24 |
ENSG00000252463 |
. |
3922 |
0.18 |
| chr1_171702173_171702324 | 0.24 |
VAMP4 |
vesicle-associated membrane protein 4 |
8954 |
0.19 |
| chr12_50580729_50580880 | 0.24 |
RP3-405J10.3 |
|
4342 |
0.14 |
| chr17_48618789_48619185 | 0.24 |
ENSG00000250286 |
. |
2872 |
0.13 |
| chr3_15687980_15688217 | 0.24 |
BTD |
biotinidase |
44622 |
0.11 |
| chr17_64644644_64644795 | 0.24 |
ENSG00000251698 |
. |
9332 |
0.19 |
| chr4_38092282_38092433 | 0.24 |
TBC1D1 |
TBC1 (tre-2/USP6, BUB2, cdc16) domain family, member 1 |
44865 |
0.19 |
| chr5_55911226_55911525 | 0.24 |
AC022431.2 |
Homo sapiens uncharacterized LOC101928448 (LOC101928448), mRNA. |
9316 |
0.26 |
| chr12_51015541_51015966 | 0.24 |
ENSG00000207136 |
. |
11303 |
0.21 |
| chr5_141503692_141503877 | 0.23 |
NDFIP1 |
Nedd4 family interacting protein 1 |
15714 |
0.23 |
| chr7_23036448_23036599 | 0.23 |
FAM126A |
family with sequence similarity 126, member A |
17170 |
0.24 |
| chr2_228275751_228276175 | 0.23 |
TM4SF20 |
transmembrane 4 L six family member 20 |
29252 |
0.16 |
| chr3_45209403_45209586 | 0.23 |
CDCP1 |
CUB domain containing protein 1 |
21580 |
0.2 |
| chr18_2588153_2588403 | 0.23 |
NDC80 |
NDC80 kinetochore complex component |
10430 |
0.14 |
| chr3_30664719_30665032 | 0.23 |
TGFBR2 |
transforming growth factor, beta receptor II (70/80kDa) |
16782 |
0.27 |
| chr7_99005041_99005202 | 0.23 |
BUD31 |
BUD31 homolog (S. cerevisiae) |
1143 |
0.26 |
| chr1_10555616_10555775 | 0.23 |
PEX14 |
peroxisomal biogenesis factor 14 |
20656 |
0.12 |
| chr2_238787700_238787851 | 0.23 |
ENSG00000263723 |
. |
9228 |
0.2 |
| chr2_101171486_101171889 | 0.23 |
ENSG00000266005 |
. |
825 |
0.59 |
| chr15_89170331_89170558 | 0.23 |
AEN |
apoptosis enhancing nuclease |
5843 |
0.16 |
| chr8_128412696_128412847 | 0.23 |
POU5F1B |
POU class 5 homeobox 1B |
13764 |
0.24 |
| chr4_146102106_146102613 | 0.23 |
OTUD4 |
OTU domain containing 4 |
1046 |
0.59 |
| chr10_105525632_105525787 | 0.23 |
SH3PXD2A |
SH3 and PX domains 2A |
35346 |
0.17 |
| chr9_32524416_32524567 | 0.23 |
DDX58 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 58 |
1702 |
0.32 |
| chr11_14375636_14375787 | 0.23 |
RRAS2 |
related RAS viral (r-ras) oncogene homolog 2 |
335 |
0.93 |
| chr21_46496959_46497110 | 0.23 |
ADARB1 |
adenosine deaminase, RNA-specific, B1 |
2519 |
0.22 |
| chr14_103270514_103270708 | 0.23 |
ENSG00000238853 |
. |
14683 |
0.17 |
| chr2_10025449_10025600 | 0.23 |
TAF1B |
TATA box binding protein (TBP)-associated factor, RNA polymerase I, B, 63kDa |
9397 |
0.23 |
| chr3_152888395_152888546 | 0.23 |
RAP2B |
RAP2B, member of RAS oncogene family |
8441 |
0.21 |
| chr4_36396689_36396840 | 0.23 |
RP11-431M7.2 |
|
2732 |
0.4 |
| chr1_175139081_175139232 | 0.23 |
KIAA0040 |
KIAA0040 |
22734 |
0.22 |
| chr1_235406790_235406941 | 0.23 |
ARID4B |
AT rich interactive domain 4B (RBP1-like) |
29634 |
0.16 |
| chr7_29605198_29605734 | 0.23 |
PRR15 |
proline rich 15 |
400 |
0.84 |
| chr14_92973076_92973375 | 0.23 |
RIN3 |
Ras and Rab interactor 3 |
6893 |
0.29 |
| chr6_106590141_106590398 | 0.23 |
RP1-134E15.3 |
|
42254 |
0.16 |
| chr2_99095585_99095903 | 0.22 |
INPP4A |
inositol polyphosphate-4-phosphatase, type I, 107kDa |
34331 |
0.18 |
| chr7_45962099_45962436 | 0.22 |
IGFBP3 |
insulin-like growth factor binding protein 3 |
794 |
0.65 |
| chr1_3619930_3620081 | 0.22 |
TP73 |
tumor protein p73 |
5383 |
0.13 |
| chr5_64919668_64919819 | 0.22 |
TRIM23 |
tripartite motif containing 23 |
380 |
0.59 |
| chr1_24971768_24971919 | 0.22 |
SRRM1 |
serine/arginine repetitive matrix 1 |
1793 |
0.32 |
| chr3_16522193_16522470 | 0.22 |
RFTN1 |
raftlin, lipid raft linker 1 |
2041 |
0.43 |
| chr7_4819835_4819986 | 0.22 |
AP5Z1 |
adaptor-related protein complex 5, zeta 1 subunit |
4646 |
0.17 |
| chr2_145081239_145081390 | 0.22 |
GTDC1 |
glycosyltransferase-like domain containing 1 |
3877 |
0.36 |
| chr10_71981169_71981370 | 0.22 |
PPA1 |
pyrophosphatase (inorganic) 1 |
11891 |
0.2 |
| chr7_100477718_100477869 | 0.22 |
SRRT |
serrate RNA effector molecule homolog (Arabidopsis) |
4782 |
0.1 |
| chr10_35050362_35050513 | 0.22 |
PARD3 |
par-3 family cell polarity regulator |
53812 |
0.14 |
| chr19_39686992_39687200 | 0.22 |
NCCRP1 |
non-specific cytotoxic cell receptor protein 1 homolog (zebrafish) |
505 |
0.69 |
| chr4_39638786_39639138 | 0.22 |
SMIM14 |
small integral membrane protein 14 |
1551 |
0.38 |
| chr8_58807157_58807308 | 0.22 |
FAM110B |
family with sequence similarity 110, member B |
99881 |
0.09 |
| chr11_34948593_34948744 | 0.22 |
PDHX |
pyruvate dehydrogenase complex, component X |
10480 |
0.19 |
| chr16_27185640_27185898 | 0.22 |
KDM8 |
lysine (K)-specific demethylase 8 |
29038 |
0.17 |
| chr13_74644571_74644841 | 0.22 |
KLF12 |
Kruppel-like factor 12 |
63688 |
0.15 |
| chr11_96059054_96059205 | 0.22 |
ENSG00000266192 |
. |
15473 |
0.19 |
| chr2_235401803_235401954 | 0.22 |
ARL4C |
ADP-ribosylation factor-like 4C |
3366 |
0.4 |
| chr16_3865090_3865241 | 0.22 |
CREBBP |
CREB binding protein |
34297 |
0.17 |
| chr17_17640963_17641296 | 0.22 |
RAI1-AS1 |
RAI1 antisense RNA 1 |
33006 |
0.11 |
| chr5_114504885_114505107 | 0.21 |
TRIM36 |
tripartite motif containing 36 |
610 |
0.73 |
| chr3_128601669_128601820 | 0.21 |
ACAD9 |
acyl-CoA dehydrogenase family, member 9 |
3272 |
0.23 |
| chr5_133389529_133390012 | 0.21 |
VDAC1 |
voltage-dependent anion channel 1 |
48946 |
0.13 |
| chr10_131583616_131583767 | 0.21 |
EBF3 |
early B-cell factor 3 |
56744 |
0.14 |
| chr10_63603062_63603213 | 0.21 |
ARID5B |
AT rich interactive domain 5B (MRF1-like) |
57922 |
0.14 |
| chr2_197084220_197084574 | 0.21 |
ENSG00000239161 |
. |
4351 |
0.23 |
| chr3_71686752_71686903 | 0.21 |
FOXP1 |
forkhead box P1 |
53687 |
0.13 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0051138 | NK T cell differentiation(GO:0001865) regulation of NK T cell differentiation(GO:0051136) positive regulation of NK T cell differentiation(GO:0051138) |
| 0.1 | 0.2 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.1 | 0.1 | GO:0051284 | negative regulation of release of sequestered calcium ion into cytosol(GO:0051280) positive regulation of sequestering of calcium ion(GO:0051284) negative regulation of calcium ion transmembrane transport(GO:1903170) |
| 0.1 | 0.2 | GO:0070266 | necroptotic process(GO:0070266) programmed necrotic cell death(GO:0097300) |
| 0.1 | 0.2 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.1 | 0.2 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.0 | 0.1 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.0 | 0.2 | GO:0043374 | CD8-positive, alpha-beta T cell activation(GO:0036037) CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.3 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.0 | 0.2 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.1 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.0 | 0.2 | GO:0003323 | type B pancreatic cell development(GO:0003323) |
| 0.0 | 0.1 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.0 | 0.1 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.0 | 0.1 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.0 | 0.2 | GO:0016246 | RNA interference(GO:0016246) |
| 0.0 | 0.1 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.0 | GO:0033689 | negative regulation of osteoblast proliferation(GO:0033689) |
| 0.0 | 0.1 | GO:0006498 | N-terminal protein lipidation(GO:0006498) |
| 0.0 | 0.2 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.1 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.2 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.3 | GO:0021527 | spinal cord association neuron differentiation(GO:0021527) |
| 0.0 | 0.1 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.2 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.1 | GO:0009296 | obsolete flagellum assembly(GO:0009296) |
| 0.0 | 0.1 | GO:2000171 | negative regulation of dendrite development(GO:2000171) |
| 0.0 | 0.1 | GO:0061029 | eyelid development in camera-type eye(GO:0061029) |
| 0.0 | 0.1 | GO:0006065 | UDP-glucuronate biosynthetic process(GO:0006065) |
| 0.0 | 0.1 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.1 | GO:0071436 | sodium ion export(GO:0071436) |
| 0.0 | 0.1 | GO:0072131 | kidney mesenchyme morphogenesis(GO:0072131) metanephric mesenchyme morphogenesis(GO:0072133) |
| 0.0 | 0.1 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 0.2 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.1 | GO:0019605 | butyrate metabolic process(GO:0019605) |
| 0.0 | 0.1 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.2 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.1 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 0.1 | GO:0060836 | lymphatic endothelial cell differentiation(GO:0060836) |
| 0.0 | 0.1 | GO:0034139 | regulation of toll-like receptor 3 signaling pathway(GO:0034139) |
| 0.0 | 0.0 | GO:0070602 | centromeric sister chromatid cohesion(GO:0070601) regulation of centromeric sister chromatid cohesion(GO:0070602) |
| 0.0 | 0.2 | GO:0048242 | epinephrine transport(GO:0048241) epinephrine secretion(GO:0048242) |
| 0.0 | 0.1 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.1 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.1 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.0 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) histone H3 deacetylation(GO:0070932) histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.0 | GO:0021798 | forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.0 | 0.1 | GO:1901798 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) positive regulation of signal transduction by p53 class mediator(GO:1901798) |
| 0.0 | 0.1 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.1 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.1 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.0 | 0.1 | GO:0000089 | mitotic metaphase(GO:0000089) |
| 0.0 | 0.1 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.0 | 0.0 | GO:0046101 | hypoxanthine biosynthetic process(GO:0046101) |
| 0.0 | 0.1 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 0.2 | GO:0045070 | positive regulation of viral genome replication(GO:0045070) |
| 0.0 | 0.0 | GO:0034392 | negative regulation of smooth muscle cell apoptotic process(GO:0034392) |
| 0.0 | 0.3 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.0 | 0.2 | GO:0043576 | regulation of respiratory gaseous exchange(GO:0043576) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.0 | GO:0030011 | maintenance of cell polarity(GO:0030011) |
| 0.0 | 0.1 | GO:0032075 | positive regulation of nuclease activity(GO:0032075) |
| 0.0 | 0.1 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 0.1 | GO:0071731 | response to nitric oxide(GO:0071731) |
| 0.0 | 0.0 | GO:0072033 | renal vesicle formation(GO:0072033) |
| 0.0 | 0.0 | GO:0010748 | negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) negative regulation of anion transmembrane transport(GO:1903960) negative regulation of fatty acid transport(GO:2000192) |
| 0.0 | 0.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.1 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.0 | 0.1 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 0.0 | GO:0060972 | left/right pattern formation(GO:0060972) |
| 0.0 | 0.1 | GO:0002713 | negative regulation of B cell mediated immunity(GO:0002713) negative regulation of immunoglobulin mediated immune response(GO:0002890) |
| 0.0 | 0.1 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.0 | GO:0002068 | glandular epithelial cell development(GO:0002068) |
| 0.0 | 0.1 | GO:0010956 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.0 | 0.0 | GO:0045591 | positive regulation of regulatory T cell differentiation(GO:0045591) |
| 0.0 | 0.0 | GO:0044415 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.0 | 0.1 | GO:0032986 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.0 | 0.0 | GO:0045988 | negative regulation of striated muscle contraction(GO:0045988) |
| 0.0 | 0.1 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.2 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.0 | 0.1 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.0 | 0.0 | GO:0032070 | regulation of deoxyribonuclease activity(GO:0032070) regulation of endodeoxyribonuclease activity(GO:0032071) |
| 0.0 | 0.1 | GO:0010002 | cardioblast differentiation(GO:0010002) |
| 0.0 | 0.0 | GO:0032353 | negative regulation of hormone metabolic process(GO:0032351) negative regulation of hormone biosynthetic process(GO:0032353) |
| 0.0 | 0.1 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.1 | GO:0016999 | antibiotic metabolic process(GO:0016999) antibiotic biosynthetic process(GO:0017000) |
| 0.0 | 0.0 | GO:0009299 | mRNA transcription(GO:0009299) |
| 0.0 | 0.0 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.1 | GO:0046049 | UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.0 | 0.2 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.0 | 0.1 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.0 | GO:1902101 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.0 | 0.1 | GO:0046655 | folic acid metabolic process(GO:0046655) |
| 0.0 | 0.1 | GO:0042339 | keratan sulfate metabolic process(GO:0042339) |
| 0.0 | 0.0 | GO:0046607 | positive regulation of centrosome cycle(GO:0046607) |
| 0.0 | 0.0 | GO:0021801 | cerebral cortex radial glia guided migration(GO:0021801) telencephalon glial cell migration(GO:0022030) |
| 0.0 | 0.0 | GO:0000491 | small nucleolar ribonucleoprotein complex assembly(GO:0000491) box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.0 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.0 | 0.1 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.0 | 0.0 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.0 | 0.0 | GO:2000095 | regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.0 | 0.0 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.0 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.0 | 0.0 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 0.0 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 0.1 | GO:1904031 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.0 | 0.1 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 0.1 | GO:0010819 | T cell chemotaxis(GO:0010818) regulation of T cell chemotaxis(GO:0010819) positive regulation of T cell chemotaxis(GO:0010820) regulation of lymphocyte chemotaxis(GO:1901623) regulation of lymphocyte migration(GO:2000401) positive regulation of lymphocyte migration(GO:2000403) regulation of T cell migration(GO:2000404) positive regulation of T cell migration(GO:2000406) |
| 0.0 | 0.0 | GO:0010715 | regulation of extracellular matrix disassembly(GO:0010715) negative regulation of extracellular matrix disassembly(GO:0010716) regulation of extracellular matrix organization(GO:1903053) negative regulation of extracellular matrix organization(GO:1903054) |
| 0.0 | 0.0 | GO:0046520 | sphingoid biosynthetic process(GO:0046520) |
| 0.0 | 0.1 | GO:1901797 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) negative regulation of signal transduction by p53 class mediator(GO:1901797) |
| 0.0 | 0.1 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 0.0 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.0 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.1 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.0 | GO:0060287 | epithelial cilium movement(GO:0003351) epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.1 | GO:0014721 | voluntary skeletal muscle contraction(GO:0003010) twitch skeletal muscle contraction(GO:0014721) |
| 0.0 | 0.0 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 | 0.0 | GO:0060686 | negative regulation of prostatic bud formation(GO:0060686) |
| 0.0 | 0.0 | GO:0060057 | apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) regulation of apoptotic process involved in morphogenesis(GO:1902337) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) regulation of apoptotic process involved in development(GO:1904748) |
| 0.0 | 0.0 | GO:0045836 | positive regulation of meiotic nuclear division(GO:0045836) positive regulation of meiotic cell cycle(GO:0051446) |
| 0.0 | 0.1 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.0 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.0 | 0.1 | GO:0000132 | establishment of mitotic spindle orientation(GO:0000132) establishment of spindle orientation(GO:0051294) |
| 0.0 | 0.0 | GO:0072112 | renal filtration cell differentiation(GO:0061318) glomerular epithelium development(GO:0072010) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.0 | 0.0 | GO:0002385 | innate immune response in mucosa(GO:0002227) mucosal immune response(GO:0002385) |
| 0.0 | 0.1 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 0.1 | GO:0071265 | amino acid salvage(GO:0043102) L-methionine biosynthetic process(GO:0071265) L-methionine salvage(GO:0071267) |
| 0.0 | 0.1 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.0 | 0.1 | GO:0014010 | Schwann cell proliferation(GO:0014010) |
| 0.0 | 0.3 | GO:0000018 | regulation of DNA recombination(GO:0000018) |
| 0.0 | 0.1 | GO:0045197 | establishment or maintenance of epithelial cell apical/basal polarity(GO:0045197) |
| 0.0 | 0.0 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.0 | 0.3 | GO:0097306 | cellular response to alcohol(GO:0097306) |
| 0.0 | 0.0 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 0.1 | GO:0006855 | drug transmembrane transport(GO:0006855) |
| 0.0 | 0.1 | GO:0048808 | male genitalia morphogenesis(GO:0048808) male anatomical structure morphogenesis(GO:0090598) |
| 0.0 | 0.1 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.0 | 0.1 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.0 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.1 | GO:0042640 | anagen(GO:0042640) |
| 0.0 | 0.0 | GO:0071379 | cellular response to prostaglandin stimulus(GO:0071379) |
| 0.0 | 0.0 | GO:0046599 | regulation of centriole replication(GO:0046599) |
| 0.0 | 0.0 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.0 | 0.1 | GO:0006002 | fructose 6-phosphate metabolic process(GO:0006002) |
| 0.0 | 0.0 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 0.0 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.0 | GO:0009128 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.0 | 0.1 | GO:0009649 | entrainment of circadian clock(GO:0009649) |
| 0.0 | 0.0 | GO:0021542 | dentate gyrus development(GO:0021542) |
| 0.0 | 0.1 | GO:0010510 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.0 | 0.0 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.0 | 0.0 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.0 | 0.1 | GO:0032057 | negative regulation of translational initiation in response to stress(GO:0032057) |
| 0.0 | 0.0 | GO:0051532 | NFAT protein import into nucleus(GO:0051531) regulation of NFAT protein import into nucleus(GO:0051532) |
| 0.0 | 0.1 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.1 | GO:0001881 | receptor recycling(GO:0001881) |
| 0.0 | 0.0 | GO:0051176 | positive regulation of sulfur metabolic process(GO:0051176) |
| 0.0 | 0.2 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.0 | 0.0 | GO:0045046 | protein import into peroxisome membrane(GO:0045046) |
| 0.0 | 0.0 | GO:0060426 | lung vasculature development(GO:0060426) |
| 0.0 | 0.1 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.2 | GO:0007062 | sister chromatid cohesion(GO:0007062) |
| 0.0 | 0.1 | GO:0032527 | protein exit from endoplasmic reticulum(GO:0032527) |
| 0.0 | 0.0 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.0 | 0.0 | GO:0032455 | nerve growth factor processing(GO:0032455) |
| 0.0 | 0.0 | GO:0045627 | positive regulation of T-helper 1 cell differentiation(GO:0045627) |
| 0.0 | 0.0 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.0 | GO:0045601 | regulation of endothelial cell differentiation(GO:0045601) |
| 0.0 | 0.1 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.0 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.0 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.0 | 0.0 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.0 | 0.0 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.0 | GO:0042726 | flavin-containing compound metabolic process(GO:0042726) |
| 0.0 | 0.0 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.2 | GO:0042130 | negative regulation of T cell proliferation(GO:0042130) |
| 0.0 | 0.1 | GO:0042797 | 5S class rRNA transcription from RNA polymerase III type 1 promoter(GO:0042791) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
| 0.0 | 0.0 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.2 | GO:0030520 | intracellular estrogen receptor signaling pathway(GO:0030520) |
| 0.0 | 0.1 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.0 | GO:0051552 | flavone metabolic process(GO:0051552) |
| 0.0 | 0.0 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.0 | 0.0 | GO:0060061 | Spemann organizer formation(GO:0060061) |
| 0.0 | 0.0 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.0 | 0.0 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.0 | 0.1 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.0 | GO:0001705 | ectoderm formation(GO:0001705) |
| 0.0 | 0.3 | GO:0032480 | negative regulation of type I interferon production(GO:0032480) |
| 0.0 | 0.0 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.1 | GO:0043267 | negative regulation of potassium ion transport(GO:0043267) |
| 0.0 | 0.0 | GO:0010832 | negative regulation of myotube differentiation(GO:0010832) negative regulation of striated muscle cell differentiation(GO:0051154) |
| 0.0 | 0.1 | GO:0010833 | telomere maintenance via telomere lengthening(GO:0010833) |
| 0.0 | 0.0 | GO:0032205 | negative regulation of telomere maintenance(GO:0032205) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.1 | 0.1 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.1 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.2 | GO:0036454 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.0 | 0.2 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.1 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.1 | GO:0044453 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 0.1 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.2 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.0 | 0.1 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.3 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.1 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.1 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.1 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.1 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 0.1 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.1 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.2 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.1 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.0 | 0.2 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.1 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.1 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.0 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.0 | GO:0070188 | obsolete Stn1-Ten1 complex(GO:0070188) |
| 0.0 | 0.1 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.0 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.1 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.2 | GO:0000780 | condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.2 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.1 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.1 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.1 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.3 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.1 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.0 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.1 | GO:0032156 | septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.0 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.1 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.0 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 0.0 | 0.1 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.0 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.1 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.1 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.3 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.0 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.1 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.4 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.1 | GO:0030128 | AP-2 adaptor complex(GO:0030122) clathrin coat of endocytic vesicle(GO:0030128) |
| 0.0 | 0.3 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.1 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 0.0 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.1 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.2 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.1 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.0 | 0.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0000987 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.0 | 0.2 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.1 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.1 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.0 | 0.2 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.1 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.3 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.2 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.2 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.1 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.1 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.2 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.1 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.1 | GO:0043047 | single-stranded telomeric DNA binding(GO:0043047) sequence-specific single stranded DNA binding(GO:0098847) |
| 0.0 | 0.1 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.0 | 0.2 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.2 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.1 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.0 | 0.3 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.1 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.0 | 0.0 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.2 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.1 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.1 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.1 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.1 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.1 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.3 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.1 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.0 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.2 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.1 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.2 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.1 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.1 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.1 | GO:0001537 | N-acetylgalactosamine 4-O-sulfotransferase activity(GO:0001537) |
| 0.0 | 0.1 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.0 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.1 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.4 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.1 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) |
| 0.0 | 0.3 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 0.0 | 0.1 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.1 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.1 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.0 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.2 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 0.0 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 0.0 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.1 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.1 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.0 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 0.0 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.0 | 0.1 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.0 | 0.4 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.0 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.1 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.0 | GO:0032554 | purine deoxyribonucleotide binding(GO:0032554) adenyl deoxyribonucleotide binding(GO:0032558) |
| 0.0 | 0.2 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 0.1 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.1 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.1 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.0 | 0.3 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.0 | GO:0008934 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.1 | GO:0016417 | S-acyltransferase activity(GO:0016417) |
| 0.0 | 0.1 | GO:0034593 | phosphatidylinositol bisphosphate phosphatase activity(GO:0034593) |
| 0.0 | 0.0 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.0 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 0.0 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.0 | 0.1 | GO:0016944 | obsolete RNA polymerase II transcription elongation factor activity(GO:0016944) |
| 0.0 | 0.0 | GO:0004476 | mannose-6-phosphate isomerase activity(GO:0004476) |
| 0.0 | 0.0 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.0 | GO:0045502 | dynein binding(GO:0045502) |
| 0.0 | 0.0 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.0 | 0.0 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.0 | GO:0032453 | histone demethylase activity (H3-K4 specific)(GO:0032453) histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.0 | 0.1 | GO:0032405 | MutLalpha complex binding(GO:0032405) |
| 0.0 | 0.0 | GO:0042808 | obsolete neuronal Cdc2-like kinase binding(GO:0042808) |
| 0.0 | 0.1 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.1 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.1 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 0.2 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 0.1 | GO:0016409 | palmitoyltransferase activity(GO:0016409) |
| 0.0 | 0.1 | GO:0016273 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.0 | 0.1 | GO:0004673 | protein histidine kinase activity(GO:0004673) |
| 0.0 | 0.1 | GO:0000832 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) |
| 0.0 | 0.1 | GO:0050656 | 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.0 | 0.1 | GO:0008318 | protein prenyltransferase activity(GO:0008318) |
| 0.0 | 0.0 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.0 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.1 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.5 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.0 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.6 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.2 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.6 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.2 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.1 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.1 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.6 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.0 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 0.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.2 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.7 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.0 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.2 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.0 | 0.2 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.4 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.1 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.1 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.0 | 0.1 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.1 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 0.1 | REACTOME G ALPHA1213 SIGNALLING EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.0 | 0.0 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.4 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.4 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.1 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.0 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 0.2 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.2 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.4 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.1 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 0.2 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.3 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.3 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.0 | 0.4 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.2 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.0 | 0.1 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.0 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.0 | REACTOME AQUAPORIN MEDIATED TRANSPORT | Genes involved in Aquaporin-mediated transport |
| 0.0 | 0.0 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 0.0 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.1 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.2 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.3 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.3 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.2 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 0.1 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.1 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.1 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.2 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.0 | REACTOME P75NTR SIGNALS VIA NFKB | Genes involved in p75NTR signals via NF-kB |
| 0.0 | 0.3 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |