Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
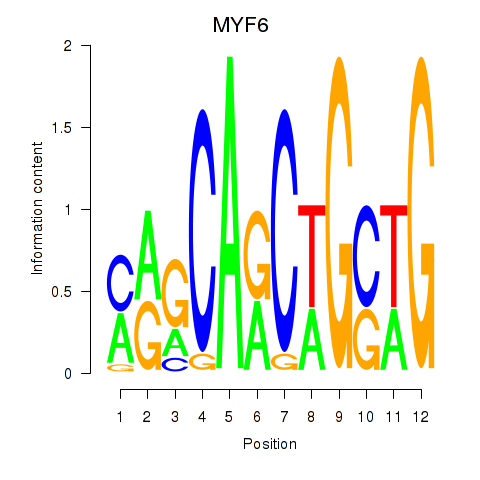
Results for MYF6
Z-value: 0.80
Transcription factors associated with MYF6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MYF6
|
ENSG00000111046.3 | myogenic factor 6 |
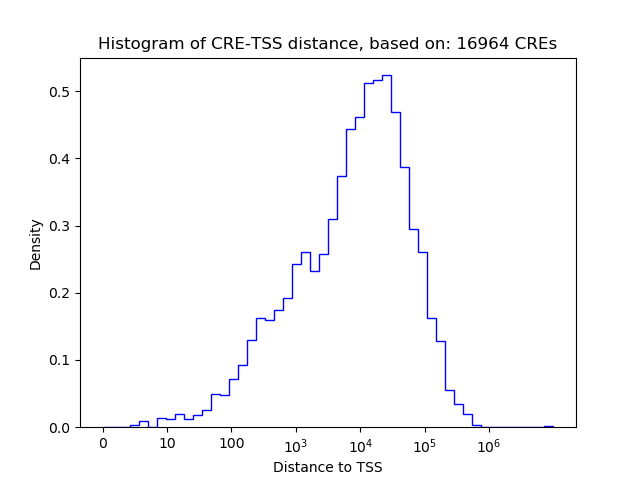
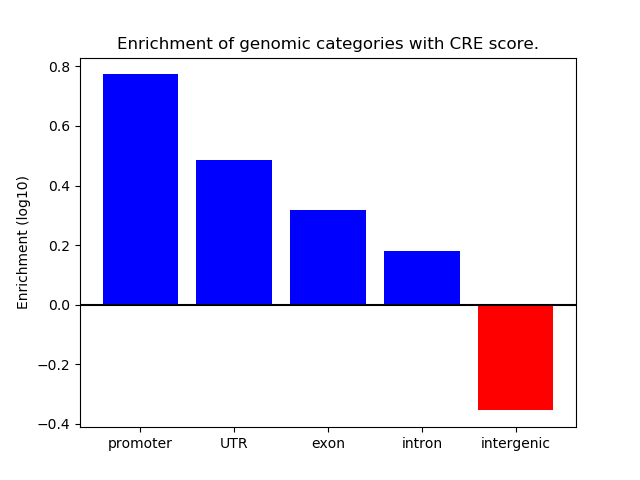
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr12_81101667_81101818 | MYF6 | 465 | 0.835408 | 0.60 | 8.5e-02 | Click! |
| chr12_81102661_81102850 | MYF6 | 1478 | 0.436473 | 0.41 | 2.8e-01 | Click! |
| chr12_81101957_81102111 | MYF6 | 757 | 0.686123 | 0.26 | 5.0e-01 | Click! |
| chr12_81102321_81102512 | MYF6 | 1139 | 0.529472 | 0.15 | 7.0e-01 | Click! |
| chr12_81097094_81097245 | MYF6 | 4108 | 0.246428 | 0.07 | 8.5e-01 | Click! |
Activity of the MYF6 motif across conditions
Conditions sorted by the z-value of the MYF6 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
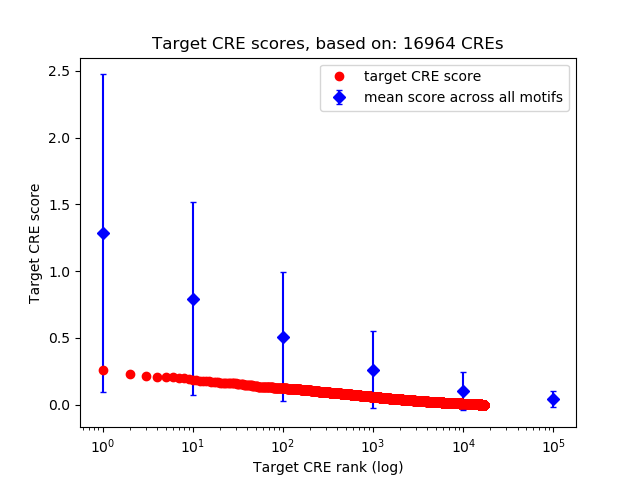
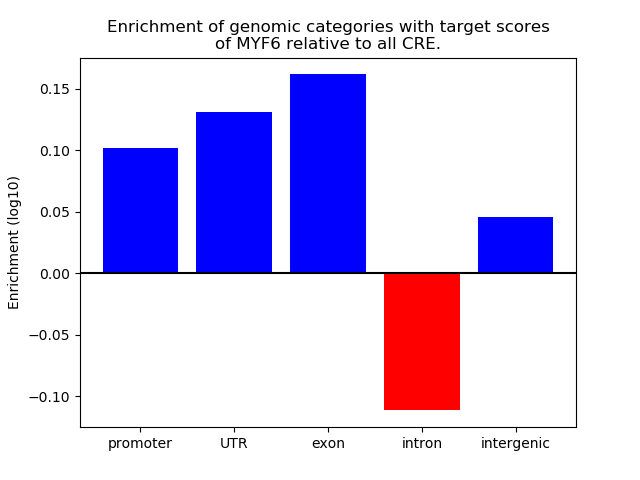
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr20_42186030_42186402 | 0.26 |
SGK2 |
serum/glucocorticoid regulated kinase 2 |
1392 |
0.36 |
| chr12_120427135_120427403 | 0.23 |
CCDC64 |
coiled-coil domain containing 64 |
404 |
0.87 |
| chr2_27305313_27305464 | 0.22 |
EMILIN1 |
elastin microfibril interfacer 1 |
1075 |
0.26 |
| chr6_6636840_6637139 | 0.21 |
LY86-AS1 |
LY86 antisense RNA 1 |
13985 |
0.26 |
| chr3_171547801_171548005 | 0.20 |
TMEM212 |
transmembrane protein 212 |
13236 |
0.18 |
| chr15_90583479_90583717 | 0.20 |
ZNF710 |
zinc finger protein 710 |
27648 |
0.12 |
| chr3_107148782_107148933 | 0.20 |
CCDC54 |
coiled-coil domain containing 54 |
52669 |
0.15 |
| chr5_95108981_95109222 | 0.20 |
RHOBTB3 |
Rho-related BTB domain containing 3 |
5257 |
0.19 |
| chr18_374955_375106 | 0.19 |
RP11-720L2.2 |
|
49386 |
0.15 |
| chr2_74783730_74784343 | 0.18 |
LOXL3 |
lysyl oxidase-like 3 |
1219 |
0.25 |
| chr17_43278136_43278287 | 0.18 |
CTD-2020K17.1 |
|
19488 |
0.09 |
| chr2_71291273_71291652 | 0.18 |
AC007040.8 |
|
411 |
0.73 |
| chr5_158655488_158655639 | 0.18 |
ENSG00000252458 |
. |
1639 |
0.34 |
| chr10_7124084_7124395 | 0.18 |
SFMBT2 |
Scm-like with four mbt domains 2 |
326468 |
0.01 |
| chr1_212844814_212845399 | 0.18 |
ENSG00000207491 |
. |
20500 |
0.14 |
| chr4_100780978_100781132 | 0.17 |
LAMTOR3 |
late endosomal/lysosomal adaptor, MAPK and MTOR activator 3 |
34478 |
0.16 |
| chr6_51167860_51168049 | 0.17 |
ENSG00000212532 |
. |
161534 |
0.04 |
| chr9_37993909_37994194 | 0.17 |
ENSG00000251745 |
. |
57286 |
0.12 |
| chr16_48653717_48653876 | 0.17 |
N4BP1 |
NEDD4 binding protein 1 |
9676 |
0.19 |
| chrX_19915221_19915598 | 0.17 |
SH3KBP1 |
SH3-domain kinase binding protein 1 |
9690 |
0.25 |
| chr12_654579_654730 | 0.17 |
B4GALNT3 |
beta-1,4-N-acetyl-galactosaminyl transferase 3 |
2355 |
0.27 |
| chr16_2556960_2557241 | 0.17 |
ATP6V0C |
ATPase, H+ transporting, lysosomal 16kDa, V0 subunit c |
6771 |
0.06 |
| chr17_40670908_40671198 | 0.17 |
ENSG00000265611 |
. |
4847 |
0.09 |
| chr6_146755943_146756197 | 0.16 |
ENSG00000222971 |
. |
94514 |
0.08 |
| chr7_73240850_73241001 | 0.16 |
CLDN4 |
claudin 4 |
1149 |
0.42 |
| chr7_26590451_26590692 | 0.16 |
KIAA0087 |
KIAA0087 |
12164 |
0.27 |
| chr1_233217531_233217682 | 0.16 |
PCNXL2 |
pecanex-like 2 (Drosophila) |
23342 |
0.26 |
| chr9_21119427_21119782 | 0.16 |
IFNW1 |
interferon, omega 1 |
22540 |
0.12 |
| chr1_25439261_25439439 | 0.16 |
ENSG00000264371 |
. |
89356 |
0.07 |
| chr20_4930731_4930882 | 0.16 |
SLC23A2 |
solute carrier family 23 (ascorbic acid transporter), member 2 |
50513 |
0.12 |
| chr22_46427472_46427760 | 0.16 |
RP6-109B7.5 |
|
21357 |
0.1 |
| chr13_111175044_111175195 | 0.16 |
COL4A2-AS1 |
COL4A2 antisense RNA 1 |
14593 |
0.2 |
| chr1_110306049_110306352 | 0.16 |
EPS8L3 |
EPS8-like 3 |
326 |
0.84 |
| chr10_35776302_35776593 | 0.16 |
GJD4 |
gap junction protein, delta 4, 40.1kDa |
117891 |
0.05 |
| chr8_91657659_91657810 | 0.15 |
TMEM64 |
transmembrane protein 64 |
6 |
0.96 |
| chr11_113847463_113847737 | 0.15 |
HTR3A |
5-hydroxytryptamine (serotonin) receptor 3A, ionotropic |
622 |
0.76 |
| chr15_69652696_69652882 | 0.15 |
ENSG00000252972 |
. |
7135 |
0.13 |
| chr10_73620300_73620451 | 0.15 |
PSAP |
prosaposin |
9249 |
0.2 |
| chr3_197878509_197878660 | 0.15 |
AC073135.3 |
|
41507 |
0.14 |
| chr20_48889044_48889195 | 0.15 |
CEBPB |
CCAAT/enhancer binding protein (C/EBP), beta |
81743 |
0.08 |
| chr5_149867249_149867416 | 0.15 |
NDST1 |
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
1475 |
0.4 |
| chr2_145440654_145440805 | 0.15 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
162108 |
0.04 |
| chr9_35042246_35042534 | 0.15 |
C9orf131 |
chromosome 9 open reading frame 131 |
180 |
0.73 |
| chr11_77213533_77213871 | 0.14 |
PAK1 |
p21 protein (Cdc42/Rac)-activated kinase 1 |
28022 |
0.15 |
| chr7_6474034_6474265 | 0.14 |
DAGLB |
diacylglycerol lipase, beta |
13414 |
0.16 |
| chr13_95254337_95254566 | 0.14 |
GPR180 |
G protein-coupled receptor 180 |
294 |
0.91 |
| chr1_182113646_182113797 | 0.14 |
ZNF648 |
zinc finger protein 648 |
82874 |
0.1 |
| chr1_21399249_21399400 | 0.14 |
EIF4G3 |
eukaryotic translation initiation factor 4 gamma, 3 |
16377 |
0.24 |
| chrX_70390205_70390391 | 0.14 |
NLGN3 |
neuroligin 3 |
25586 |
0.1 |
| chr22_18210286_18210437 | 0.14 |
ENSG00000264757 |
. |
36664 |
0.14 |
| chr3_4520289_4520508 | 0.14 |
SUMF1 |
sulfatase modifying factor 1 |
11433 |
0.2 |
| chr4_144435798_144435959 | 0.14 |
SMARCA5-AS1 |
SMARCA5 antisense RNA 1 |
90 |
0.96 |
| chr9_136038241_136038401 | 0.14 |
GBGT1 |
globoside alpha-1,3-N-acetylgalactosaminyltransferase 1 |
961 |
0.35 |
| chr11_69457785_69457945 | 0.14 |
CCND1 |
cyclin D1 |
1891 |
0.38 |
| chr20_30666848_30667078 | 0.14 |
HCK |
hemopoietic cell kinase |
26899 |
0.1 |
| chr8_122140841_122140992 | 0.14 |
ENSG00000221644 |
. |
58216 |
0.14 |
| chr7_27615419_27615570 | 0.13 |
HIBADH |
3-hydroxyisobutyrate dehydrogenase |
71881 |
0.11 |
| chr15_40628103_40628665 | 0.13 |
C15orf52 |
chromosome 15 open reading frame 52 |
1857 |
0.16 |
| chr4_139335299_139335467 | 0.13 |
SLC7A11 |
solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11 |
171880 |
0.04 |
| chr15_101709991_101710273 | 0.13 |
CHSY1 |
chondroitin sulfate synthase 1 |
82005 |
0.08 |
| chr5_139727515_139728049 | 0.13 |
HBEGF |
heparin-binding EGF-like growth factor |
1566 |
0.26 |
| chr6_106957352_106957633 | 0.13 |
AIM1 |
absent in melanoma 1 |
2238 |
0.32 |
| chr7_47641377_47641730 | 0.13 |
TNS3 |
tensin 3 |
19397 |
0.24 |
| chr20_32551295_32551527 | 0.13 |
RALY |
RALY heterogeneous nuclear ribonucleoprotein |
30041 |
0.18 |
| chr14_58765318_58765585 | 0.13 |
ARID4A |
AT rich interactive domain 4A (RBP1-like) |
145 |
0.84 |
| chr10_89875342_89875493 | 0.13 |
ENSG00000200891 |
. |
120965 |
0.06 |
| chr10_82355785_82355994 | 0.13 |
SH2D4B |
SH2 domain containing 4B |
55314 |
0.14 |
| chr1_172792534_172792808 | 0.13 |
FASLG |
Fas ligand (TNF superfamily, member 6) |
164513 |
0.04 |
| chr7_26344110_26344302 | 0.13 |
SNX10 |
sorting nexin 10 |
11532 |
0.23 |
| chr8_37761152_37761429 | 0.13 |
RAB11FIP1 |
RAB11 family interacting protein 1 (class I) |
4305 |
0.16 |
| chr20_30641676_30641832 | 0.13 |
HCK |
hemopoietic cell kinase |
1690 |
0.25 |
| chr5_77793836_77794265 | 0.13 |
LHFPL2 |
lipoma HMGIC fusion partner-like 2 |
50924 |
0.17 |
| chr7_95237066_95237395 | 0.13 |
AC002451.3 |
|
1878 |
0.37 |
| chr17_40990067_40990218 | 0.13 |
PSME3 |
proteasome (prosome, macropain) activator subunit 3 (PA28 gamma; Ki) |
4629 |
0.09 |
| chr20_1356162_1356422 | 0.13 |
RP11-314N13.3 |
|
14026 |
0.14 |
| chr4_74864141_74864430 | 0.13 |
CXCL5 |
chemokine (C-X-C motif) ligand 5 |
211 |
0.91 |
| chr19_16263728_16264013 | 0.13 |
HSH2D |
hematopoietic SH2 domain containing |
9324 |
0.12 |
| chr8_19743814_19744105 | 0.13 |
LPL |
lipoprotein lipase |
15269 |
0.24 |
| chr17_18933418_18933712 | 0.13 |
GRAP |
GRB2-related adaptor protein |
12233 |
0.11 |
| chr6_41744687_41744838 | 0.13 |
FRS3 |
fibroblast growth factor receptor substrate 3 |
1157 |
0.31 |
| chr17_76908326_76908701 | 0.13 |
CTD-2373H9.5 |
|
9168 |
0.14 |
| chr20_1359967_1360118 | 0.13 |
FKBP1A |
FK506 binding protein 1A, 12kDa |
13564 |
0.14 |
| chr2_27110278_27110603 | 0.13 |
DPYSL5 |
dihydropyrimidinase-like 5 |
39064 |
0.12 |
| chr4_83943782_83943933 | 0.13 |
LIN54 |
lin-54 homolog (C. elegans) |
9778 |
0.16 |
| chr22_26800846_26801092 | 0.13 |
ENSG00000252267 |
. |
14227 |
0.17 |
| chr21_40455880_40456270 | 0.13 |
PSMG1 |
proteasome (prosome, macropain) assembly chaperone 1 |
99339 |
0.07 |
| chr17_42849030_42849346 | 0.13 |
ADAM11 |
ADAM metallopeptidase domain 11 |
12495 |
0.13 |
| chr17_1959980_1960184 | 0.13 |
HIC1 |
hypermethylated in cancer 1 |
478 |
0.62 |
| chr1_66777257_66777408 | 0.13 |
PDE4B |
phosphodiesterase 4B, cAMP-specific |
19079 |
0.28 |
| chr12_112212431_112212814 | 0.13 |
ALDH2 |
aldehyde dehydrogenase 2 family (mitochondrial) |
7896 |
0.19 |
| chr6_2825832_2826072 | 0.12 |
SERPINB1 |
serpin peptidase inhibitor, clade B (ovalbumin), member 1 |
16142 |
0.17 |
| chr8_37757773_37757924 | 0.12 |
RAB11FIP1 |
RAB11 family interacting protein 1 (class I) |
863 |
0.51 |
| chr6_6887247_6887398 | 0.12 |
ENSG00000240936 |
. |
51808 |
0.17 |
| chr9_124273187_124273484 | 0.12 |
ENSG00000240299 |
. |
16538 |
0.18 |
| chr11_34354091_34354256 | 0.12 |
ABTB2 |
ankyrin repeat and BTB (POZ) domain containing 2 |
24629 |
0.23 |
| chr5_137801463_137801649 | 0.12 |
EGR1 |
early growth response 1 |
377 |
0.84 |
| chr12_112628379_112628530 | 0.12 |
HECTD4 |
HECT domain containing E3 ubiquitin protein ligase 4 |
13948 |
0.17 |
| chr15_94408923_94409174 | 0.12 |
ENSG00000222409 |
. |
31208 |
0.26 |
| chr6_137211819_137211970 | 0.12 |
SLC35D3 |
solute carrier family 35, member D3 |
31508 |
0.14 |
| chr17_15845936_15846239 | 0.12 |
ADORA2B |
adenosine A2b receptor |
2144 |
0.31 |
| chr20_36824784_36825086 | 0.12 |
TGM2 |
transglutaminase 2 |
29955 |
0.14 |
| chr21_45123059_45123298 | 0.12 |
PDXK |
pyridoxal (pyridoxine, vitamin B6) kinase |
15797 |
0.17 |
| chr19_33770830_33771059 | 0.12 |
CTD-2540B15.11 |
|
19896 |
0.11 |
| chr2_191506139_191506290 | 0.12 |
NAB1 |
NGFI-A binding protein 1 (EGR1 binding protein 1) |
5258 |
0.25 |
| chr12_27702924_27703232 | 0.12 |
PPFIBP1 |
PTPRF interacting protein, binding protein 1 (liprin beta 1) |
12022 |
0.23 |
| chr2_71687770_71687921 | 0.12 |
DYSF |
dysferlin |
5987 |
0.27 |
| chr1_109681194_109681359 | 0.12 |
KIAA1324 |
KIAA1324 |
24499 |
0.11 |
| chr5_154229551_154229702 | 0.12 |
FAXDC2 |
fatty acid hydroxylase domain containing 2 |
537 |
0.74 |
| chr11_45944455_45944606 | 0.12 |
GYLTL1B |
glycosyltransferase-like 1B |
201 |
0.9 |
| chr21_44817450_44817601 | 0.12 |
SIK1 |
salt-inducible kinase 1 |
29483 |
0.23 |
| chr14_51544517_51544668 | 0.12 |
RP11-1140I5.1 |
|
11083 |
0.21 |
| chr12_123921421_123921707 | 0.12 |
RILPL2 |
Rab interacting lysosomal protein-like 2 |
300 |
0.87 |
| chr15_64283322_64283528 | 0.12 |
DAPK2 |
death-associated protein kinase 2 |
49041 |
0.12 |
| chr6_6798807_6798985 | 0.12 |
ENSG00000240936 |
. |
140234 |
0.05 |
| chr11_65075775_65076171 | 0.12 |
CDC42EP2 |
CDC42 effector protein (Rho GTPase binding) 2 |
6316 |
0.13 |
| chr14_59931229_59931380 | 0.12 |
GPR135 |
G protein-coupled receptor 135 |
756 |
0.65 |
| chr10_93870631_93870837 | 0.12 |
ENSG00000252993 |
. |
34735 |
0.16 |
| chr10_30092663_30093124 | 0.12 |
SVIL |
supervillin |
68160 |
0.12 |
| chr19_6059466_6059704 | 0.12 |
RFX2 |
regulatory factor X, 2 (influences HLA class II expression) |
2291 |
0.23 |
| chr6_150184862_150185013 | 0.12 |
LRP11 |
low density lipoprotein receptor-related protein 11 |
225 |
0.57 |
| chr1_160254501_160254652 | 0.12 |
PEX19 |
peroxisomal biogenesis factor 19 |
340 |
0.46 |
| chr10_82012381_82012532 | 0.12 |
AL359195.1 |
Uncharacterized protein; cDNA FLJ46261 fis, clone TESTI4025062 |
2990 |
0.26 |
| chr15_29561544_29561695 | 0.12 |
NDNL2 |
necdin-like 2 |
414 |
0.88 |
| chr9_99693594_99693748 | 0.12 |
NUTM2G |
NUT family member 2G |
2385 |
0.36 |
| chr8_22953881_22954118 | 0.12 |
TNFRSF10C |
tumor necrosis factor receptor superfamily, member 10c, decoy without an intracellular domain |
6104 |
0.14 |
| chr10_104957863_104958014 | 0.12 |
NT5C2 |
5'-nucleotidase, cytosolic II |
4882 |
0.23 |
| chr2_112400633_112400819 | 0.12 |
ENSG00000266063 |
. |
127985 |
0.06 |
| chr2_174889480_174890119 | 0.12 |
SP3 |
Sp3 transcription factor |
59369 |
0.15 |
| chr20_23008934_23009272 | 0.12 |
RP4-753D10.5 |
|
2319 |
0.23 |
| chr9_113683953_113684408 | 0.12 |
ENSG00000207401 |
. |
12506 |
0.22 |
| chr5_142980288_142980439 | 0.12 |
CTB-57H20.1 |
|
4830 |
0.29 |
| chr1_244505150_244505410 | 0.12 |
C1orf100 |
chromosome 1 open reading frame 100 |
10657 |
0.25 |
| chr11_43810486_43810637 | 0.12 |
RP11-613D13.5 |
|
44906 |
0.15 |
| chr9_97088395_97088740 | 0.12 |
NUTM2F |
NUT family member 2F |
2359 |
0.3 |
| chr2_97870529_97870680 | 0.12 |
ANKRD36 |
ankyrin repeat domain 36 |
40976 |
0.13 |
| chr8_72088568_72088719 | 0.12 |
RP11-326E22.1 |
|
20886 |
0.28 |
| chr18_60646350_60646591 | 0.12 |
PHLPP1 |
PH domain and leucine rich repeat protein phosphatase 1 |
76087 |
0.1 |
| chr19_7720613_7720936 | 0.12 |
STXBP2 |
syntaxin binding protein 2 |
9929 |
0.08 |
| chr8_9938557_9938708 | 0.12 |
ENSG00000221320 |
. |
6799 |
0.21 |
| chr12_48168957_48169108 | 0.12 |
SLC48A1 |
solute carrier family 48 (heme transporter), member 1 |
2041 |
0.23 |
| chr6_43775268_43775445 | 0.12 |
VEGFA |
vascular endothelial growth factor A |
33266 |
0.15 |
| chr8_1873489_1873640 | 0.12 |
ARHGEF10 |
Rho guanine nucleotide exchange factor (GEF) 10 |
42742 |
0.16 |
| chr14_75754946_75755194 | 0.12 |
FOS |
FBJ murine osteosarcoma viral oncogene homolog |
8174 |
0.17 |
| chr1_27973206_27973357 | 0.12 |
FGR |
feline Gardner-Rasheed sarcoma viral oncogene homolog |
11493 |
0.12 |
| chr4_172263778_172264007 | 0.12 |
ENSG00000251961 |
. |
451274 |
0.01 |
| chr9_97567295_97567893 | 0.12 |
ENSG00000252153 |
. |
4650 |
0.2 |
| chr20_19403915_19404216 | 0.11 |
ENSG00000221748 |
. |
82125 |
0.1 |
| chr4_86954647_86954929 | 0.11 |
RP13-514E23.1 |
|
19761 |
0.19 |
| chr11_67859868_67860019 | 0.11 |
CHKA |
choline kinase alpha |
28352 |
0.1 |
| chr16_85112017_85112168 | 0.11 |
KIAA0513 |
KIAA0513 |
67 |
0.98 |
| chr16_85855178_85855329 | 0.11 |
COX4I1 |
cytochrome c oxidase subunit IV isoform 1 |
20445 |
0.12 |
| chr12_96564682_96564833 | 0.11 |
ELK3 |
ELK3, ETS-domain protein (SRF accessory protein 2) |
23403 |
0.18 |
| chr10_3612660_3612862 | 0.11 |
RP11-184A2.3 |
|
180498 |
0.03 |
| chr1_204940620_204940796 | 0.11 |
NFASC |
neurofascin |
10276 |
0.21 |
| chr15_41225289_41225440 | 0.11 |
DLL4 |
delta-like 4 (Drosophila) |
3773 |
0.15 |
| chr1_17609322_17609523 | 0.11 |
ENSG00000266634 |
. |
5038 |
0.18 |
| chr2_55347968_55348289 | 0.11 |
RTN4 |
reticulon 4 |
8371 |
0.19 |
| chr15_81033301_81033659 | 0.11 |
KIAA1199 |
KIAA1199 |
38204 |
0.15 |
| chr9_80564462_80564644 | 0.11 |
GNAQ |
guanine nucleotide binding protein (G protein), q polypeptide |
80967 |
0.11 |
| chr20_19300587_19300738 | 0.11 |
ENSG00000221748 |
. |
21278 |
0.21 |
| chr17_14651949_14652297 | 0.11 |
HS3ST3B1 |
heparan sulfate (glucosamine) 3-O-sulfotransferase 3B1 |
447723 |
0.01 |
| chr19_10341033_10341420 | 0.11 |
ENSG00000264266 |
. |
137 |
0.8 |
| chr2_86642212_86642427 | 0.11 |
KDM3A |
lysine (K)-specific demethylase 3A |
25451 |
0.16 |
| chr5_95003135_95003286 | 0.11 |
RFESD |
Rieske (Fe-S) domain containing |
13445 |
0.16 |
| chr17_54040448_54040734 | 0.11 |
ENSG00000264277 |
. |
91557 |
0.09 |
| chr2_208497932_208498495 | 0.11 |
METTL21A |
methyltransferase like 21A |
7562 |
0.19 |
| chr13_29170887_29171038 | 0.11 |
POMP |
proteasome maturation protein |
62279 |
0.13 |
| chr12_109117281_109117726 | 0.11 |
CORO1C |
coronin, actin binding protein, 1C |
6824 |
0.16 |
| chr19_35948719_35948870 | 0.11 |
FFAR2 |
free fatty acid receptor 2 |
8177 |
0.11 |
| chr21_43723122_43723420 | 0.11 |
ENSG00000216197 |
. |
2816 |
0.21 |
| chr9_97338725_97338876 | 0.11 |
FBP2 |
fructose-1,6-bisphosphatase 2 |
17275 |
0.22 |
| chr2_95941719_95941977 | 0.11 |
PROM2 |
prominin 2 |
1603 |
0.42 |
| chr10_73628074_73628344 | 0.11 |
PSAP |
prosaposin |
17083 |
0.19 |
| chr21_39640704_39640894 | 0.11 |
KCNJ15 |
potassium inwardly-rectifying channel, subfamily J, member 15 |
3373 |
0.34 |
| chr20_22904829_22904980 | 0.11 |
RP4-753D10.5 |
|
106518 |
0.06 |
| chr2_242843726_242843877 | 0.11 |
AC131097.4 |
Protein LOC285095 |
901 |
0.5 |
| chr12_11992163_11992414 | 0.11 |
ETV6 |
ets variant 6 |
46583 |
0.19 |
| chr21_38895077_38895228 | 0.11 |
AP001421.1 |
Uncharacterized protein |
6412 |
0.26 |
| chr3_53740550_53740701 | 0.11 |
CACNA1D |
calcium channel, voltage-dependent, L type, alpha 1D subunit |
40197 |
0.16 |
| chr6_13480251_13480456 | 0.11 |
AL583828.1 |
|
6062 |
0.17 |
| chr9_130910949_130911100 | 0.11 |
LCN2 |
lipocalin 2 |
326 |
0.76 |
| chr12_94566333_94566520 | 0.11 |
RP11-74K11.2 |
|
3754 |
0.26 |
| chr9_83733305_83733456 | 0.11 |
ENSG00000221581 |
. |
384136 |
0.01 |
| chr3_46172431_46172582 | 0.11 |
CCR3 |
chemokine (C-C motif) receptor 3 |
32590 |
0.18 |
| chr16_66262660_66262838 | 0.11 |
ENSG00000201999 |
. |
72963 |
0.11 |
| chr1_94018769_94019015 | 0.11 |
FNBP1L |
formin binding protein 1-like |
23625 |
0.16 |
| chr10_119460300_119460451 | 0.11 |
EMX2OS |
EMX2 opposite strand/antisense RNA |
155796 |
0.04 |
| chr6_144080973_144081159 | 0.11 |
PHACTR2 |
phosphatase and actin regulator 2 |
81861 |
0.09 |
| chr4_6855596_6855818 | 0.11 |
TBC1D14 |
TBC1 domain family, member 14 |
55262 |
0.11 |
| chr3_184335510_184335694 | 0.11 |
EPHB3 |
EPH receptor B3 |
56030 |
0.11 |
| chr1_11004143_11004582 | 0.11 |
C1orf127 |
chromosome 1 open reading frame 127 |
3565 |
0.22 |
| chr20_56045926_56046077 | 0.11 |
CTCFL |
CCCTC-binding factor (zinc finger protein)-like |
53312 |
0.11 |
| chr19_12893778_12893929 | 0.11 |
ENSG00000263800 |
. |
4089 |
0.08 |
| chr22_39239357_39239520 | 0.11 |
NPTXR |
neuronal pentraxin receptor |
549 |
0.69 |
| chr5_70883327_70883478 | 0.11 |
MCCC2 |
methylcrotonoyl-CoA carboxylase 2 (beta) |
207 |
0.96 |
| chr2_102733095_102733246 | 0.11 |
IL1R1 |
interleukin 1 receptor, type I |
12141 |
0.23 |
| chr14_69509818_69509969 | 0.11 |
ACTN1-AS1 |
ACTN1 antisense RNA 1 |
63135 |
0.11 |
| chr20_55061855_55062036 | 0.11 |
ENSG00000238294 |
. |
11246 |
0.12 |
| chr1_161058959_161059110 | 0.11 |
PVRL4 |
poliovirus receptor-related 4 |
355 |
0.7 |
| chr5_169908402_169908553 | 0.11 |
CTB-147C13.1 |
|
4934 |
0.24 |
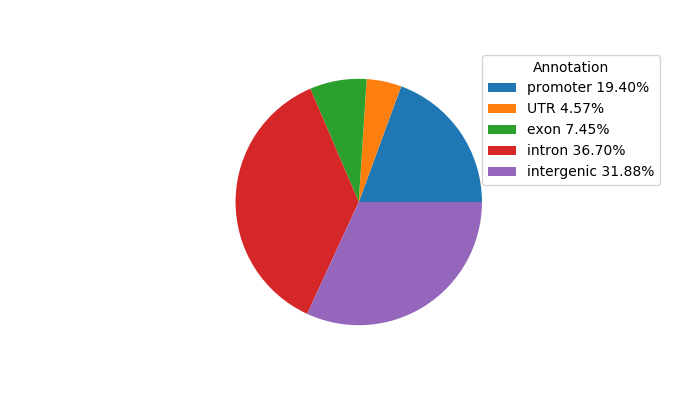
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0032455 | nerve growth factor processing(GO:0032455) |
| 0.0 | 0.2 | GO:0071731 | response to nitric oxide(GO:0071731) |
| 0.0 | 0.1 | GO:0032959 | inositol trisphosphate biosynthetic process(GO:0032959) |
| 0.0 | 0.1 | GO:0071504 | cellular response to heparin(GO:0071504) |
| 0.0 | 0.1 | GO:0052308 | pathogen-associated molecular pattern dependent induction by symbiont of host innate immune response(GO:0052033) positive regulation by symbiont of host innate immune response(GO:0052166) modulation by symbiont of host innate immune response(GO:0052167) pathogen-associated molecular pattern dependent modulation by symbiont of host innate immune response(GO:0052169) pathogen-associated molecular pattern dependent induction by organism of innate immune response of other organism involved in symbiotic interaction(GO:0052257) positive regulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052305) modulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052306) pathogen-associated molecular pattern dependent modulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052308) |
| 0.0 | 0.1 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.0 | 0.1 | GO:0021827 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) substrate-dependent cerebral cortex tangential migration(GO:0021825) postnatal olfactory bulb interneuron migration(GO:0021827) |
| 0.0 | 0.1 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.1 | GO:0051547 | regulation of keratinocyte migration(GO:0051547) positive regulation of keratinocyte migration(GO:0051549) |
| 0.0 | 0.1 | GO:0001878 | response to yeast(GO:0001878) |
| 0.0 | 0.1 | GO:0045425 | positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) |
| 0.0 | 0.1 | GO:0002238 | response to molecule of fungal origin(GO:0002238) |
| 0.0 | 0.1 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.0 | 0.1 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.0 | 0.1 | GO:0060087 | relaxation of smooth muscle(GO:0044557) relaxation of vascular smooth muscle(GO:0060087) |
| 0.0 | 0.1 | GO:0018101 | protein citrullination(GO:0018101) |
| 0.0 | 0.1 | GO:0021801 | cerebral cortex radial glia guided migration(GO:0021801) telencephalon glial cell migration(GO:0022030) |
| 0.0 | 0.1 | GO:0033197 | response to vitamin E(GO:0033197) |
| 0.0 | 0.1 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.1 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.0 | 0.1 | GO:0042663 | endodermal cell fate specification(GO:0001714) regulation of endodermal cell fate specification(GO:0042663) regulation of endodermal cell differentiation(GO:1903224) |
| 0.0 | 0.1 | GO:0002538 | arachidonic acid metabolite production involved in inflammatory response(GO:0002538) leukotriene production involved in inflammatory response(GO:0002540) |
| 0.0 | 0.2 | GO:0015936 | coenzyme A metabolic process(GO:0015936) |
| 0.0 | 0.1 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.0 | 0.1 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.0 | GO:0002282 | microglial cell activation involved in immune response(GO:0002282) |
| 0.0 | 0.1 | GO:0060633 | negative regulation of transcription initiation from RNA polymerase II promoter(GO:0060633) negative regulation of DNA-templated transcription, initiation(GO:2000143) |
| 0.0 | 0.1 | GO:0051570 | regulation of histone H3-K9 methylation(GO:0051570) |
| 0.0 | 0.1 | GO:0002002 | regulation of angiotensin levels in blood(GO:0002002) |
| 0.0 | 0.1 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.0 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 | 0.0 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.0 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.0 | 0.0 | GO:0015864 | pyrimidine nucleoside transport(GO:0015864) |
| 0.0 | 0.1 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.1 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.0 | 0.0 | GO:0016048 | detection of temperature stimulus(GO:0016048) |
| 0.0 | 0.1 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 | 0.0 | GO:0031558 | obsolete induction of apoptosis in response to chemical stimulus(GO:0031558) |
| 0.0 | 0.0 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.0 | 0.1 | GO:0043551 | regulation of phosphatidylinositol 3-kinase activity(GO:0043551) positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.0 | 0.1 | GO:0042832 | defense response to protozoan(GO:0042832) |
| 0.0 | 0.1 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.1 | GO:1901678 | iron coordination entity transport(GO:1901678) |
| 0.0 | 0.1 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) telomere maintenance in response to DNA damage(GO:0043247) |
| 0.0 | 0.1 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.0 | 0.1 | GO:0010591 | regulation of lamellipodium assembly(GO:0010591) |
| 0.0 | 0.0 | GO:0002323 | natural killer cell activation involved in immune response(GO:0002323) natural killer cell degranulation(GO:0043320) |
| 0.0 | 0.0 | GO:0045341 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) |
| 0.0 | 0.1 | GO:0050849 | negative regulation of calcium-mediated signaling(GO:0050849) |
| 0.0 | 0.1 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) positive regulation of neutrophil migration(GO:1902624) |
| 0.0 | 0.0 | GO:0051541 | elastin metabolic process(GO:0051541) |
| 0.0 | 0.0 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.0 | 0.0 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.0 | 0.0 | GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0043619) |
| 0.0 | 0.0 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.0 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) |
| 0.0 | 0.1 | GO:0050872 | white fat cell differentiation(GO:0050872) |
| 0.0 | 0.1 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.1 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.0 | GO:0060536 | trachea cartilage morphogenesis(GO:0060535) cartilage morphogenesis(GO:0060536) |
| 0.0 | 0.0 | GO:0001821 | histamine secretion(GO:0001821) |
| 0.0 | 0.1 | GO:0046504 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.0 | GO:0021800 | cerebral cortex tangential migration(GO:0021800) |
| 0.0 | 0.1 | GO:0006000 | fructose metabolic process(GO:0006000) |
| 0.0 | 0.1 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.0 | GO:0015803 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.0 | 0.0 | GO:0021529 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) oligodendrocyte cell fate specification(GO:0021778) glial cell fate specification(GO:0021780) |
| 0.0 | 0.0 | GO:0042268 | regulation of cytolysis(GO:0042268) |
| 0.0 | 0.1 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.0 | 0.0 | GO:0032495 | response to muramyl dipeptide(GO:0032495) |
| 0.0 | 0.0 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.0 | 0.1 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.0 | GO:0070296 | sarcoplasmic reticulum calcium ion transport(GO:0070296) |
| 0.0 | 0.0 | GO:0071503 | response to heparin(GO:0071503) |
| 0.0 | 0.0 | GO:0051770 | positive regulation of nitric-oxide synthase biosynthetic process(GO:0051770) |
| 0.0 | 0.0 | GO:0034371 | chylomicron remodeling(GO:0034371) |
| 0.0 | 0.1 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.0 | 0.0 | GO:0021932 | hindbrain radial glia guided cell migration(GO:0021932) |
| 0.0 | 0.1 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.1 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.0 | 0.1 | GO:0014049 | positive regulation of glutamate secretion(GO:0014049) |
| 0.0 | 0.0 | GO:0043276 | anoikis(GO:0043276) |
| 0.0 | 0.0 | GO:0002085 | inhibition of neuroepithelial cell differentiation(GO:0002085) |
| 0.0 | 0.0 | GO:0019255 | glucose 1-phosphate metabolic process(GO:0019255) |
| 0.0 | 0.0 | GO:0048867 | stem cell fate commitment(GO:0048865) stem cell fate determination(GO:0048867) |
| 0.0 | 0.0 | GO:0071397 | cellular response to sterol(GO:0036315) cellular response to cholesterol(GO:0071397) |
| 0.0 | 0.0 | GO:0001845 | phagolysosome assembly(GO:0001845) phagosome maturation(GO:0090382) |
| 0.0 | 0.0 | GO:0019348 | polyprenol metabolic process(GO:0016093) dolichol metabolic process(GO:0019348) |
| 0.0 | 0.0 | GO:0034114 | heterotypic cell-cell adhesion(GO:0034113) regulation of heterotypic cell-cell adhesion(GO:0034114) |
| 0.0 | 0.0 | GO:0035608 | protein deglutamylation(GO:0035608) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0002141 | stereocilia coupling link(GO:0002139) stereocilia ankle link(GO:0002141) stereocilia ankle link complex(GO:0002142) |
| 0.0 | 0.2 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 0.1 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.2 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.2 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.0 | GO:0031313 | extrinsic component of vacuolar membrane(GO:0000306) extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.1 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.0 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.1 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.1 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.1 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.1 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.1 | GO:0042827 | platelet dense granule(GO:0042827) |
| 0.0 | 0.0 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.1 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.1 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.2 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.0 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.1 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.1 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.1 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.1 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.0 | 0.1 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.0 | 0.1 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.0 | 0.1 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.1 | GO:0022821 | potassium ion antiporter activity(GO:0022821) |
| 0.0 | 0.1 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 0.1 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.2 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.0 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.1 | GO:0004331 | fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.0 | 0.0 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.0 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.0 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.1 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.0 | GO:1901567 | icosanoid binding(GO:0050542) fatty acid derivative binding(GO:1901567) |
| 0.0 | 0.0 | GO:0008898 | S-adenosylmethionine-homocysteine S-methyltransferase activity(GO:0008898) |
| 0.0 | 0.1 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.1 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.1 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.0 | 0.0 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.0 | 0.0 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) inositol-1,3,4,5-tetrakisphosphate 5-phosphatase activity(GO:0052659) |
| 0.0 | 0.1 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.0 | 0.1 | GO:0043176 | amine binding(GO:0043176) |
| 0.0 | 0.0 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.1 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.1 | GO:0004340 | glucokinase activity(GO:0004340) |
| 0.0 | 0.0 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.0 | 0.1 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.0 | GO:0033765 | steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) |
| 0.0 | 0.0 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.0 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.0 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.0 | 0.1 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.1 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.0 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.1 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 0.0 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.0 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 0.0 | 0.0 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.1 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.0 | GO:0043175 | RNA polymerase core enzyme binding(GO:0043175) |
| 0.0 | 0.0 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.0 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.1 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.1 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.2 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.0 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.1 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.0 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.0 | 0.0 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.0 | 0.2 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.1 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.0 | 0.0 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.1 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.1 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.0 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.1 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.0 | 0.0 | REACTOME SIGNALING BY THE B CELL RECEPTOR BCR | Genes involved in Signaling by the B Cell Receptor (BCR) |
| 0.0 | 0.0 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.0 | REACTOME SIGNALING BY FGFR IN DISEASE | Genes involved in Signaling by FGFR in disease |
| 0.0 | 0.0 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 0.1 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |