Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
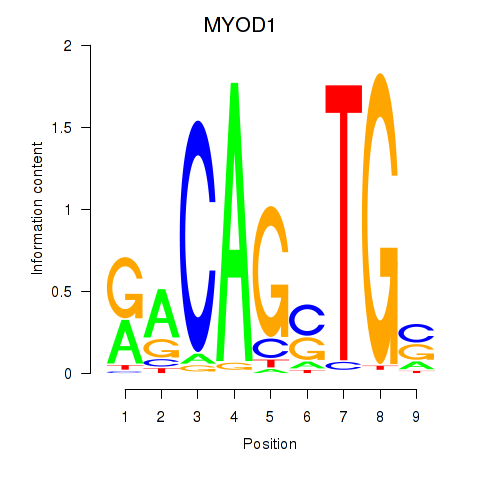
Results for MYOD1
Z-value: 0.34
Transcription factors associated with MYOD1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MYOD1
|
ENSG00000129152.3 | myogenic differentiation 1 |
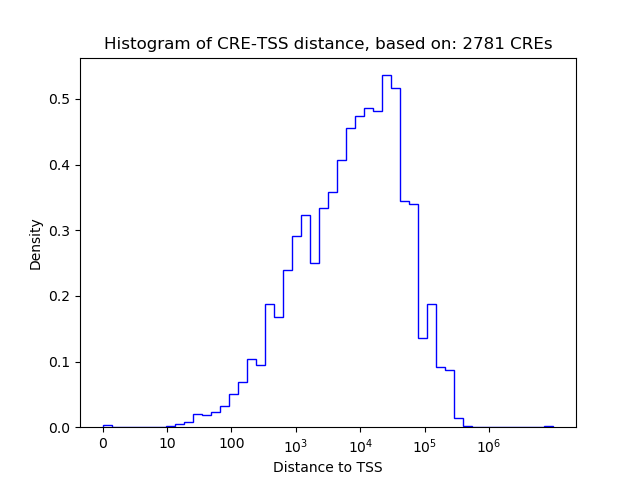
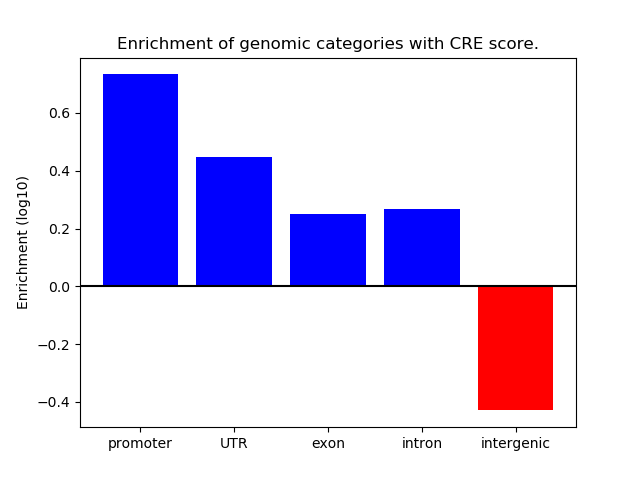
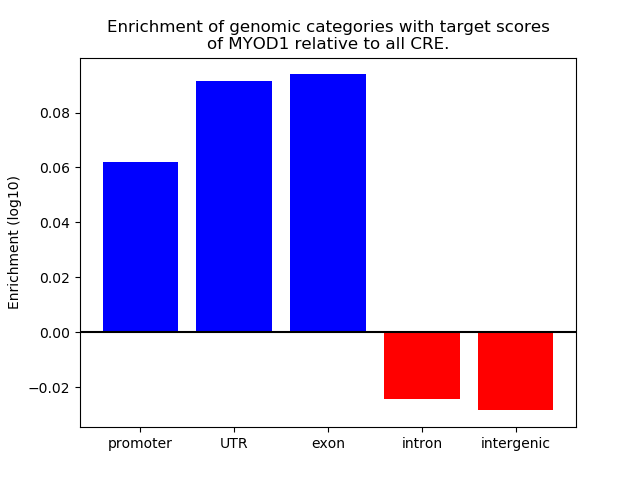
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr11_17742683_17742834 | MYOD1 | 1643 | 0.375160 | 0.68 | 4.2e-02 | Click! |
| chr11_17741919_17742070 | MYOD1 | 879 | 0.600829 | 0.38 | 3.2e-01 | Click! |
| chr11_17742082_17742233 | MYOD1 | 1042 | 0.533606 | -0.19 | 6.2e-01 | Click! |
| chr11_17742914_17743065 | MYOD1 | 1874 | 0.339728 | -0.13 | 7.3e-01 | Click! |
| chr11_17741707_17741858 | MYOD1 | 667 | 0.705995 | -0.02 | 9.6e-01 | Click! |
Activity of the MYOD1 motif across conditions
Conditions sorted by the z-value of the MYOD1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
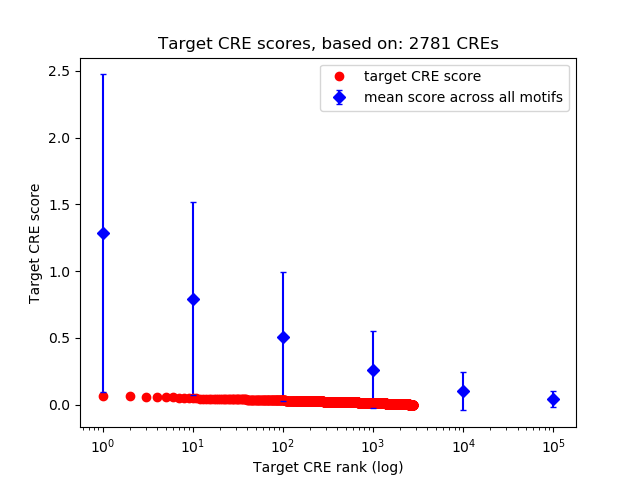
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr6_112302873_112303024 | 0.06 |
WISP3 |
WNT1 inducible signaling pathway protein 3 |
72327 |
0.1 |
| chr8_36999886_37000356 | 0.06 |
KCNU1 |
potassium channel, subfamily U, member 1 |
213698 |
0.02 |
| chr16_84210365_84210716 | 0.06 |
DNAAF1 |
dynein, axonemal, assembly factor 1 |
793 |
0.49 |
| chr19_18794913_18795747 | 0.06 |
CRTC1 |
CREB regulated transcription coactivator 1 |
842 |
0.53 |
| chr1_7531258_7531628 | 0.06 |
RP4-549F15.1 |
|
29829 |
0.21 |
| chr8_78032492_78032643 | 0.06 |
PEX2 |
peroxisomal biogenesis factor 2 |
119287 |
0.06 |
| chr17_19912897_19913158 | 0.05 |
SPECC1 |
sperm antigen with calponin homology and coiled-coil domains 1 |
149 |
0.82 |
| chr1_201430876_201431027 | 0.05 |
PHLDA3 |
pleckstrin homology-like domain, family A, member 3 |
7361 |
0.16 |
| chr15_75954147_75954338 | 0.05 |
SNX33 |
sorting nexin 33 |
12145 |
0.09 |
| chr16_70795018_70795169 | 0.05 |
VAC14-AS1 |
VAC14 antisense RNA 1 |
6057 |
0.16 |
| chr4_111008729_111008880 | 0.05 |
ENSG00000263940 |
. |
30088 |
0.21 |
| chr7_127804538_127804689 | 0.05 |
ENSG00000207705 |
. |
43311 |
0.14 |
| chr1_202558845_202559092 | 0.05 |
RP11-569A11.1 |
|
14428 |
0.2 |
| chr3_139255001_139255152 | 0.05 |
RBP1 |
retinol binding protein 1, cellular |
3485 |
0.18 |
| chr4_1003833_1004600 | 0.04 |
FGFRL1 |
fibroblast growth factor receptor-like 1 |
449 |
0.75 |
| chr1_22196645_22196796 | 0.04 |
HSPG2 |
heparan sulfate proteoglycan 2 |
4479 |
0.22 |
| chr12_113797562_113797776 | 0.04 |
SLC8B1 |
solute carrier family 8 (sodium/lithium/calcium exchanger), member B1 |
371 |
0.78 |
| chr12_52585492_52585885 | 0.04 |
KRT80 |
keratin 80 |
96 |
0.95 |
| chr21_46737404_46737555 | 0.04 |
ENSG00000215447 |
. |
29512 |
0.14 |
| chr5_121499966_121500239 | 0.04 |
CTC-441N14.1 |
|
8392 |
0.19 |
| chr7_44224030_44224181 | 0.04 |
GCK |
glucokinase (hexokinase 4) |
4481 |
0.15 |
| chr20_47299343_47299823 | 0.04 |
ENSG00000251876 |
. |
56402 |
0.16 |
| chr13_114563717_114564199 | 0.04 |
GAS6 |
growth arrest-specific 6 |
3082 |
0.32 |
| chr1_214666364_214666654 | 0.04 |
PTPN14 |
protein tyrosine phosphatase, non-receptor type 14 |
28363 |
0.24 |
| chrX_70842306_70842457 | 0.04 |
ENSG00000264855 |
. |
3324 |
0.23 |
| chr1_224752371_224752739 | 0.04 |
RP11-100E13.1 |
|
51367 |
0.13 |
| chr14_68698275_68698426 | 0.04 |
ENSG00000243546 |
. |
4906 |
0.3 |
| chr8_49229512_49229861 | 0.04 |
ENSG00000252710 |
. |
9096 |
0.32 |
| chr5_148942473_148942978 | 0.04 |
ARHGEF37 |
Rho guanine nucleotide exchange factor (GEF) 37 |
11215 |
0.15 |
| chr15_35985538_35985689 | 0.04 |
DPH6 |
diphthamine biosynthesis 6 |
147220 |
0.05 |
| chr9_35695879_35696173 | 0.04 |
TPM2 |
tropomyosin 2 (beta) |
5009 |
0.08 |
| chr5_167831136_167831287 | 0.04 |
WWC1 |
WW and C2 domain containing 1 |
2073 |
0.41 |
| chr5_166722386_166722537 | 0.04 |
CTB-180C19.1 |
|
1183 |
0.45 |
| chr18_8800055_8800206 | 0.04 |
RP11-674N23.4 |
|
1524 |
0.42 |
| chr1_183260943_183261094 | 0.04 |
NMNAT2 |
nicotinamide nucleotide adenylyltransferase 2 |
12991 |
0.26 |
| chr17_76787623_76787888 | 0.04 |
USP36 |
ubiquitin specific peptidase 36 |
7311 |
0.18 |
| chr16_67441027_67441178 | 0.04 |
ZDHHC1 |
zinc finger, DHHC-type containing 1 |
6208 |
0.11 |
| chr19_9957964_9958210 | 0.04 |
ENSG00000266247 |
. |
4017 |
0.11 |
| chr1_115865777_115865928 | 0.04 |
NGF |
nerve growth factor (beta polypeptide) |
15005 |
0.24 |
| chr6_154720634_154720785 | 0.04 |
CNKSR3 |
CNKSR family member 3 |
30920 |
0.21 |
| chr17_78933014_78933165 | 0.04 |
CTD-2561B21.3 |
|
2424 |
0.18 |
| chr11_20017061_20017212 | 0.04 |
NAV2-AS3 |
NAV2 antisense RNA 3 |
14253 |
0.21 |
| chr1_77415551_77415702 | 0.04 |
ENSG00000221720 |
. |
57803 |
0.14 |
| chr12_122235900_122236051 | 0.04 |
RHOF |
ras homolog family member F (in filopodia) |
2608 |
0.23 |
| chr22_46942701_46942994 | 0.04 |
RP3-439F8.1 |
|
5455 |
0.21 |
| chr1_177895954_177896105 | 0.04 |
SEC16B |
SEC16 homolog B (S. cerevisiae) |
32009 |
0.25 |
| chr6_4734376_4734527 | 0.04 |
CDYL |
chromodomain protein, Y-like |
28058 |
0.2 |
| chr6_11609999_11610289 | 0.04 |
TMEM170B |
transmembrane protein 170B |
71633 |
0.11 |
| chr15_40631527_40631678 | 0.04 |
C15orf52 |
chromosome 15 open reading frame 52 |
641 |
0.48 |
| chr5_33613140_33613291 | 0.04 |
TARS |
threonyl-tRNA synthetase |
171728 |
0.03 |
| chr11_130761204_130761355 | 0.04 |
SNX19 |
sorting nexin 19 |
2490 |
0.42 |
| chr11_12028563_12028714 | 0.04 |
DKK3 |
dickkopf WNT signaling pathway inhibitor 3 |
1492 |
0.54 |
| chr22_46404101_46404436 | 0.04 |
WNT7B |
wingless-type MMTV integration site family, member 7B |
31259 |
0.1 |
| chr10_123900032_123900183 | 0.04 |
TACC2 |
transforming, acidic coiled-coil containing protein 2 |
13878 |
0.27 |
| chr9_117050436_117050587 | 0.04 |
ORM1 |
orosomucoid 1 |
34825 |
0.15 |
| chr3_157503107_157503258 | 0.04 |
ENSG00000251751 |
. |
145332 |
0.05 |
| chr4_99443439_99443625 | 0.04 |
RP11-724M22.1 |
|
26017 |
0.24 |
| chr1_115844836_115844987 | 0.04 |
RP4-663N10.1 |
|
19256 |
0.23 |
| chr3_149955178_149955329 | 0.04 |
RP11-167H9.4 |
|
139434 |
0.04 |
| chr2_101482521_101482672 | 0.04 |
AC092168.2 |
|
43473 |
0.15 |
| chr14_93503097_93503248 | 0.04 |
ITPK1 |
inositol-tetrakisphosphate 1-kinase |
29531 |
0.17 |
| chr17_73811280_73811431 | 0.04 |
UNK |
unkempt family zinc finger |
4465 |
0.1 |
| chr7_30844366_30844720 | 0.04 |
AC004691.5 |
|
1517 |
0.41 |
| chr12_96866193_96866344 | 0.04 |
C12orf55 |
chromosome 12 open reading frame 55 |
17085 |
0.19 |
| chr17_25711228_25711379 | 0.04 |
TBC1D3P5 |
TBC1 domain family, member 3 pseudogene 5 |
35261 |
0.17 |
| chr11_44622766_44622917 | 0.04 |
RP11-58K22.5 |
|
4046 |
0.22 |
| chr11_20057064_20057215 | 0.04 |
NAV2 |
neuron navigator 2 |
12503 |
0.2 |
| chr19_33737197_33737348 | 0.04 |
SLC7A10 |
solute carrier family 7 (neutral amino acid transporter light chain, asc system), member 10 |
20516 |
0.12 |
| chr4_26023741_26023892 | 0.03 |
SMIM20 |
small integral membrane protein 20 |
107885 |
0.07 |
| chr1_8149763_8149914 | 0.03 |
ERRFI1 |
ERBB receptor feedback inhibitor 1 |
63470 |
0.11 |
| chr21_44144933_44145084 | 0.03 |
AP001627.1 |
|
16860 |
0.21 |
| chr9_95509831_95509982 | 0.03 |
BICD2 |
bicaudal D homolog 2 (Drosophila) |
17188 |
0.19 |
| chr19_1168901_1169806 | 0.03 |
SBNO2 |
strawberry notch homolog 2 (Drosophila) |
411 |
0.76 |
| chr10_92404426_92404577 | 0.03 |
HTR7 |
5-hydroxytryptamine (serotonin) receptor 7, adenylate cyclase-coupled |
212954 |
0.02 |
| chr14_73074403_73074554 | 0.03 |
RGS6 |
regulator of G-protein signaling 6 |
148101 |
0.04 |
| chr11_131851218_131851369 | 0.03 |
RP11-697E14.2 |
|
3262 |
0.35 |
| chr2_129063259_129063630 | 0.03 |
HS6ST1 |
heparan sulfate 6-O-sulfotransferase 1 |
12707 |
0.24 |
| chr7_129214409_129214560 | 0.03 |
NRF1 |
nuclear respiratory factor 1 |
37071 |
0.16 |
| chr7_46851027_46851178 | 0.03 |
AC011294.3 |
Uncharacterized protein |
114382 |
0.07 |
| chr3_137762283_137762711 | 0.03 |
DZIP1L |
DAZ interacting zinc finger protein 1-like |
23985 |
0.17 |
| chr15_101227024_101227175 | 0.03 |
ENSG00000212306 |
. |
48531 |
0.13 |
| chr16_70721238_70721389 | 0.03 |
MTSS1L |
metastasis suppressor 1-like |
1344 |
0.35 |
| chr10_22887513_22887753 | 0.03 |
PIP4K2A |
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha |
6991 |
0.31 |
| chr15_89425540_89425746 | 0.03 |
ACAN |
aggrecan |
9443 |
0.19 |
| chr1_7462341_7462492 | 0.03 |
RP3-453P22.2 |
|
12602 |
0.21 |
| chr1_68638974_68639247 | 0.03 |
ENSG00000221203 |
. |
10183 |
0.22 |
| chrX_153713483_153713883 | 0.03 |
UBL4A |
ubiquitin-like 4A |
1271 |
0.2 |
| chr2_178065_178216 | 0.03 |
AC079779.7 |
|
19429 |
0.21 |
| chr20_60943487_60943729 | 0.03 |
LAMA5 |
laminin, alpha 5 |
1240 |
0.33 |
| chr11_72500407_72500904 | 0.03 |
STARD10 |
StAR-related lipid transfer (START) domain containing 10 |
3643 |
0.14 |
| chr7_73120336_73120487 | 0.03 |
ENSG00000265724 |
. |
5236 |
0.11 |
| chr3_151644390_151644541 | 0.03 |
RP11-454C18.2 |
|
1491 |
0.5 |
| chr17_17734899_17735050 | 0.03 |
SREBF1 |
sterol regulatory element binding transcription factor 1 |
5157 |
0.15 |
| chr2_239836120_239836271 | 0.03 |
ENSG00000211566 |
. |
58511 |
0.11 |
| chr22_44317614_44317765 | 0.03 |
PNPLA3 |
patatin-like phospholipase domain containing 3 |
1930 |
0.28 |
| chr14_96519676_96519827 | 0.03 |
C14orf132 |
chromosome 14 open reading frame 132 |
14090 |
0.23 |
| chr19_4377503_4377654 | 0.03 |
SH3GL1 |
SH3-domain GRB2-like 1 |
2711 |
0.11 |
| chr20_43322075_43322226 | 0.03 |
RP11-445H22.4 |
|
2575 |
0.19 |
| chr10_115426849_115427000 | 0.03 |
NRAP |
nebulin-related anchoring protein |
3038 |
0.26 |
| chr11_113544621_113544772 | 0.03 |
TMPRSS5 |
transmembrane protease, serine 5 |
32328 |
0.18 |
| chr11_82744912_82745063 | 0.03 |
RAB30 |
RAB30, member RAS oncogene family |
256 |
0.91 |
| chr19_15363852_15364003 | 0.03 |
EPHX3 |
epoxide hydrolase 3 |
19681 |
0.15 |
| chr1_181064835_181064986 | 0.03 |
IER5 |
immediate early response 5 |
7272 |
0.24 |
| chr3_149322526_149322677 | 0.03 |
WWTR1 |
WW domain containing transcription regulator 1 |
28562 |
0.16 |
| chrX_45665420_45666160 | 0.03 |
ENSG00000207725 |
. |
59260 |
0.13 |
| chr20_43968652_43968803 | 0.03 |
SDC4 |
syndecan 4 |
8337 |
0.12 |
| chr1_22463906_22464057 | 0.03 |
WNT4 |
wingless-type MMTV integration site family, member 4 |
5478 |
0.23 |
| chr4_152882169_152882320 | 0.03 |
ENSG00000253077 |
. |
10521 |
0.3 |
| chr20_1398658_1398906 | 0.03 |
FKBP1A |
FK506 binding protein 1A, 12kDa |
24976 |
0.13 |
| chr1_148930426_148930577 | 0.03 |
ENSG00000222854 |
. |
17119 |
0.25 |
| chr10_11799148_11799299 | 0.03 |
ECHDC3 |
enoyl CoA hydratase domain containing 3 |
9949 |
0.25 |
| chr10_116615267_116615418 | 0.03 |
FAM160B1 |
family with sequence similarity 160, member B1 |
5208 |
0.3 |
| chr1_46654010_46654161 | 0.03 |
POMGNT1 |
protein O-linked mannose N-acetylglucosaminyltransferase 1 (beta 1,2-) |
9062 |
0.12 |
| chr11_19702330_19702481 | 0.03 |
NAV2 |
neuron navigator 2 |
32476 |
0.19 |
| chr9_110803338_110803489 | 0.03 |
ENSG00000222459 |
. |
122154 |
0.06 |
| chr4_141860055_141860206 | 0.03 |
RNF150 |
ring finger protein 150 |
58060 |
0.15 |
| chr8_13016830_13016981 | 0.03 |
ENSG00000200630 |
. |
782 |
0.71 |
| chr16_28394741_28394892 | 0.03 |
NPIPB6 |
nuclear pore complex interacting protein family, member B6 |
19987 |
0.15 |
| chr7_156838975_156839126 | 0.03 |
MNX1-AS1 |
MNX1 antisense RNA 1 (head to head) |
35551 |
0.13 |
| chr17_78252953_78253155 | 0.03 |
RNF213 |
ring finger protein 213 |
18385 |
0.14 |
| chr12_66197112_66197263 | 0.03 |
HMGA2 |
high mobility group AT-hook 2 |
20724 |
0.21 |
| chr17_36621931_36622082 | 0.03 |
ARHGAP23 |
Rho GTPase activating protein 23 |
6174 |
0.18 |
| chr15_74716383_74716612 | 0.03 |
SEMA7A |
semaphorin 7A, GPI membrane anchor (John Milton Hagen blood group) |
9504 |
0.14 |
| chr6_24832416_24832567 | 0.03 |
ENSG00000263391 |
. |
7802 |
0.17 |
| chr4_169787971_169788122 | 0.03 |
ENSG00000201176 |
. |
18460 |
0.19 |
| chr5_154072169_154072320 | 0.03 |
ENSG00000221552 |
. |
6908 |
0.16 |
| chr16_28743058_28743209 | 0.03 |
EIF3C |
eukaryotic translation initiation factor 3, subunit C |
18553 |
0.11 |
| chr8_128962754_128962905 | 0.03 |
TMEM75 |
transmembrane protein 75 |
2238 |
0.3 |
| chr10_128038987_128039138 | 0.03 |
ADAM12 |
ADAM metallopeptidase domain 12 |
37962 |
0.21 |
| chr3_50138493_50138644 | 0.03 |
RBM5 |
RNA binding motif protein 5 |
10765 |
0.14 |
| chr11_86194975_86195126 | 0.03 |
RP11-317J19.1 |
|
52418 |
0.14 |
| chr17_74525285_74525534 | 0.03 |
CYGB |
cytoglobin |
2738 |
0.12 |
| chr13_111029669_111029820 | 0.03 |
ENSG00000238629 |
. |
36808 |
0.16 |
| chr5_133787860_133788011 | 0.03 |
CDKN2AIPNL |
CDKN2A interacting protein N-terminal like |
40346 |
0.11 |
| chr5_131594695_131594846 | 0.03 |
PDLIM4 |
PDZ and LIM domain 4 |
1370 |
0.38 |
| chr1_209792070_209792327 | 0.03 |
LAMB3 |
laminin, beta 3 |
55 |
0.97 |
| chr3_66413563_66413714 | 0.03 |
ENSG00000241587 |
. |
39359 |
0.19 |
| chr10_3797419_3797677 | 0.03 |
RP11-184A2.3 |
|
4289 |
0.25 |
| chr15_64813692_64814396 | 0.03 |
ZNF609 |
zinc finger protein 609 |
22553 |
0.15 |
| chr9_136556701_136556852 | 0.03 |
SARDH |
sarcosine dehydrogenase |
12047 |
0.18 |
| chr9_131682870_131683022 | 0.03 |
PHYHD1 |
phytanoyl-CoA dioxygenase domain containing 1 |
228 |
0.88 |
| chr5_83220337_83220488 | 0.03 |
ENSG00000216125 |
. |
49293 |
0.18 |
| chr7_41926654_41927141 | 0.03 |
AC005027.3 |
|
181964 |
0.03 |
| chr11_78949120_78949271 | 0.03 |
TENM4 |
teneurin transmembrane protein 4 |
22241 |
0.27 |
| chr1_210053632_210053783 | 0.03 |
DIEXF |
digestive organ expansion factor homolog (zebrafish) |
41557 |
0.14 |
| chr6_37423751_37423902 | 0.03 |
CMTR1 |
cap methyltransferase 1 |
19269 |
0.19 |
| chr5_98397400_98397551 | 0.03 |
ENSG00000200351 |
. |
125024 |
0.06 |
| chr3_48777654_48777856 | 0.03 |
PRKAR2A |
protein kinase, cAMP-dependent, regulatory, type II, alpha |
11996 |
0.13 |
| chr17_29899687_29899838 | 0.03 |
AC003101.1 |
Uncharacterized protein |
1601 |
0.22 |
| chr5_157567459_157567610 | 0.03 |
ENSG00000222626 |
. |
163570 |
0.04 |
| chr8_99158779_99158930 | 0.03 |
POP1 |
processing of precursor 1, ribonuclease P/MRP subunit (S. cerevisiae) |
28786 |
0.14 |
| chr17_39643176_39643459 | 0.03 |
KRT36 |
keratin 36 |
2799 |
0.11 |
| chr16_11172637_11172993 | 0.03 |
RP11-66H6.3 |
|
7856 |
0.21 |
| chr13_99189047_99189198 | 0.03 |
STK24 |
serine/threonine kinase 24 |
14870 |
0.21 |
| chr3_29739467_29739618 | 0.03 |
RBMS3-AS2 |
RBMS3 antisense RNA 2 |
55242 |
0.17 |
| chr11_12039741_12039892 | 0.03 |
DKK3 |
dickkopf WNT signaling pathway inhibitor 3 |
8500 |
0.28 |
| chr10_3088518_3088669 | 0.03 |
PFKP |
phosphofructokinase, platelet |
19932 |
0.26 |
| chr6_44664207_44664358 | 0.03 |
SUPT3H |
suppressor of Ty 3 homolog (S. cerevisiae) |
258965 |
0.02 |
| chr10_126105708_126105859 | 0.03 |
OAT |
ornithine aminotransferase |
1722 |
0.37 |
| chr6_7641658_7641809 | 0.03 |
SNRNP48 |
small nuclear ribonucleoprotein 48kDa (U11/U12) |
51301 |
0.14 |
| chr12_52890097_52890248 | 0.03 |
KRT6A |
keratin 6A |
3131 |
0.14 |
| chr7_157102085_157102478 | 0.03 |
ENSG00000266453 |
. |
3794 |
0.28 |
| chr3_112365560_112365711 | 0.03 |
CCDC80 |
coiled-coil domain containing 80 |
5488 |
0.27 |
| chr22_31479008_31479415 | 0.03 |
RP3-412A9.16 |
|
340 |
0.68 |
| chr1_21868245_21868396 | 0.03 |
ALPL |
alkaline phosphatase, liver/bone/kidney |
9452 |
0.2 |
| chr2_235195728_235195879 | 0.03 |
ARL4C |
ADP-ribosylation factor-like 4C |
209441 |
0.02 |
| chr2_128334300_128334515 | 0.03 |
MYO7B |
myosin VIIB |
17141 |
0.16 |
| chr13_22257583_22257734 | 0.03 |
FGF9 |
fibroblast growth factor 9 |
12136 |
0.25 |
| chr6_44029671_44029954 | 0.03 |
RP5-1120P11.1 |
|
12577 |
0.17 |
| chr8_23780024_23780175 | 0.03 |
STC1 |
stanniocalcin 1 |
67779 |
0.11 |
| chr3_122817242_122817393 | 0.03 |
PDIA5 |
protein disulfide isomerase family A, member 5 |
31122 |
0.2 |
| chr1_7539238_7539389 | 0.03 |
RP4-549F15.1 |
|
37699 |
0.19 |
| chr20_48766064_48766215 | 0.03 |
TMEM189-UBE2V1 |
TMEM189-UBE2V1 readthrough |
4035 |
0.2 |
| chrX_122933914_122934065 | 0.03 |
ENSG00000252178 |
. |
28000 |
0.15 |
| chr17_79037545_79037696 | 0.03 |
BAIAP2 |
BAI1-associated protein 2 |
6086 |
0.14 |
| chr19_19928396_19928679 | 0.03 |
ZNF506 |
zinc finger protein 506 |
3964 |
0.17 |
| chr3_134086085_134086236 | 0.03 |
AMOTL2 |
angiomotin like 2 |
4594 |
0.25 |
| chr2_28055526_28056020 | 0.03 |
RBKS |
ribokinase |
10255 |
0.18 |
| chr12_57999143_57999314 | 0.03 |
DTX3 |
deltex homolog 3 (Drosophila) |
438 |
0.64 |
| chr17_70382891_70383042 | 0.03 |
SOX9 |
SRY (sex determining region Y)-box 9 |
265805 |
0.02 |
| chr20_35473773_35473929 | 0.03 |
SOGA1 |
suppressor of glucose, autophagy associated 1 |
14425 |
0.16 |
| chr2_216973110_216973261 | 0.03 |
XRCC5 |
X-ray repair complementing defective repair in Chinese hamster cells 5 (double-strand-break rejoining) |
835 |
0.62 |
| chr18_53525572_53525723 | 0.03 |
TCF4 |
transcription factor 4 |
193629 |
0.03 |
| chr9_37938333_37938484 | 0.03 |
ENSG00000251745 |
. |
1643 |
0.43 |
| chr18_8812965_8813116 | 0.03 |
RP11-674N23.4 |
|
11386 |
0.21 |
| chrX_106958207_106958942 | 0.03 |
TSC22D3 |
TSC22 domain family, member 3 |
1057 |
0.6 |
| chr1_115843065_115843263 | 0.03 |
RP4-663N10.1 |
|
17509 |
0.23 |
| chr8_37379902_37380053 | 0.03 |
RP11-150O12.6 |
|
5438 |
0.29 |
| chr11_704821_704972 | 0.03 |
EPS8L2 |
EPS8-like 2 |
321 |
0.76 |
| chr15_33300880_33301031 | 0.03 |
ENSG00000212415 |
. |
38027 |
0.18 |
| chr16_65046532_65046683 | 0.03 |
CDH11 |
cadherin 11, type 2, OB-cadherin (osteoblast) |
46974 |
0.21 |
| chr2_99533263_99533414 | 0.03 |
KIAA1211L |
KIAA1211-like |
8354 |
0.26 |
| chr1_1502439_1502726 | 0.03 |
SSU72 |
SSU72 RNA polymerase II CTD phosphatase homolog (S. cerevisiae) |
7382 |
0.1 |
| chr2_220360347_220360718 | 0.03 |
RP11-316O14.1 |
|
2436 |
0.13 |
| chr3_159572255_159572681 | 0.03 |
SCHIP1 |
schwannomin interacting protein 1 |
1740 |
0.37 |
| chr16_67960353_67960504 | 0.03 |
CTRL |
chymotrypsin-like |
5337 |
0.09 |
| chr1_38604043_38604194 | 0.03 |
ENSG00000265596 |
. |
49215 |
0.14 |
| chr1_26629293_26629444 | 0.03 |
UBXN11 |
UBX domain protein 11 |
139 |
0.93 |
| chr20_10617346_10617497 | 0.03 |
JAG1 |
jagged 1 |
25733 |
0.22 |
| chr18_33887538_33887689 | 0.03 |
FHOD3 |
formin homology 2 domain containing 3 |
9814 |
0.28 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0010566 | regulation of ketone biosynthetic process(GO:0010566) regulation of aldosterone metabolic process(GO:0032344) regulation of aldosterone biosynthetic process(GO:0032347) |