Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
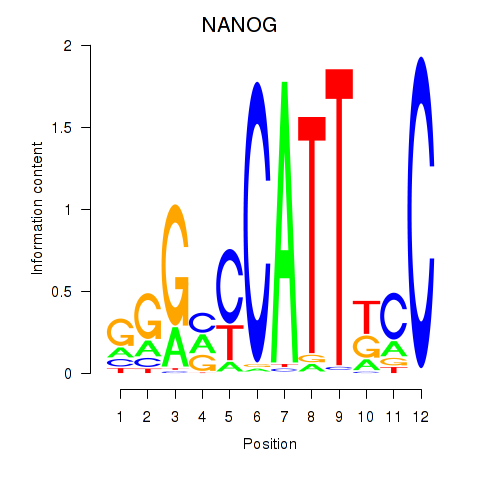
Results for NANOG
Z-value: 1.03
Transcription factors associated with NANOG
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NANOG
|
ENSG00000111704.6 | Nanog homeobox |
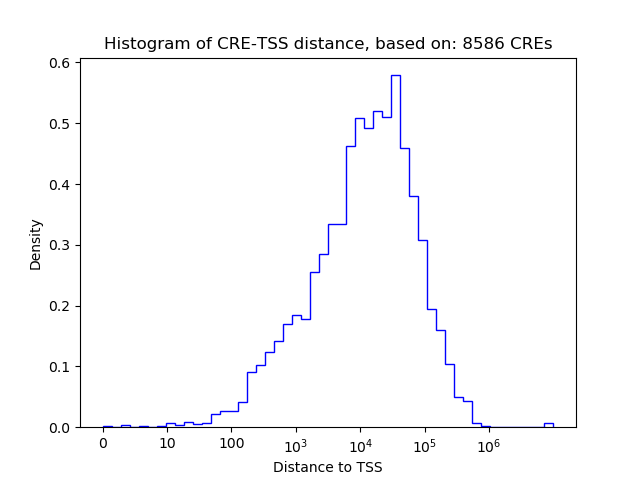
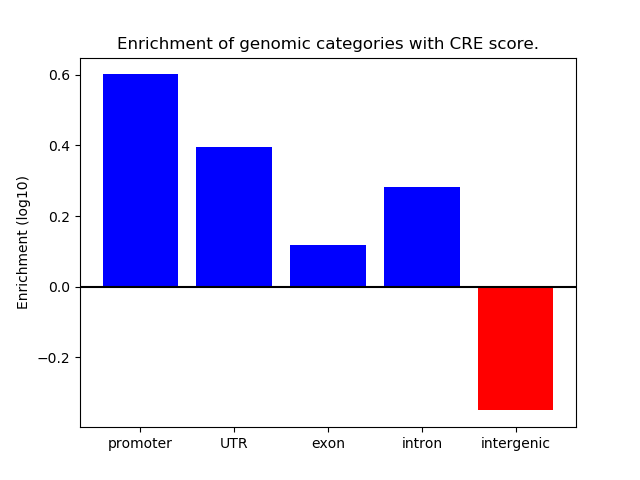
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr12_7945278_7945429 | NANOG | 3142 | 0.146781 | 0.13 | 7.3e-01 | Click! |
| chr12_7945753_7945904 | NANOG | 3617 | 0.136931 | 0.01 | 9.9e-01 | Click! |
| chr12_7946157_7946308 | NANOG | 4021 | 0.130954 | -0.01 | 9.9e-01 | Click! |
Activity of the NANOG motif across conditions
Conditions sorted by the z-value of the NANOG motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
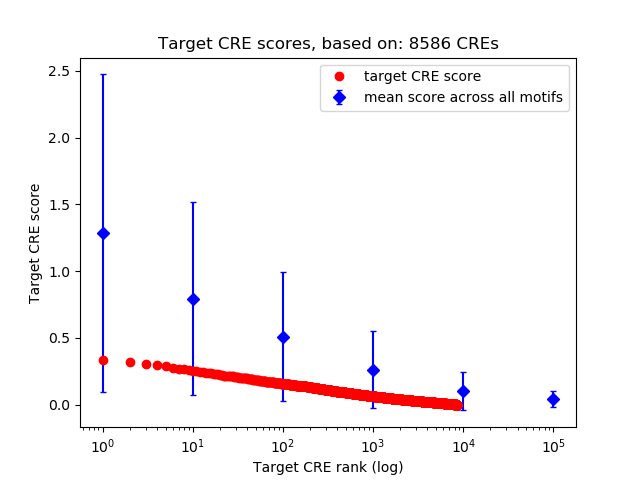
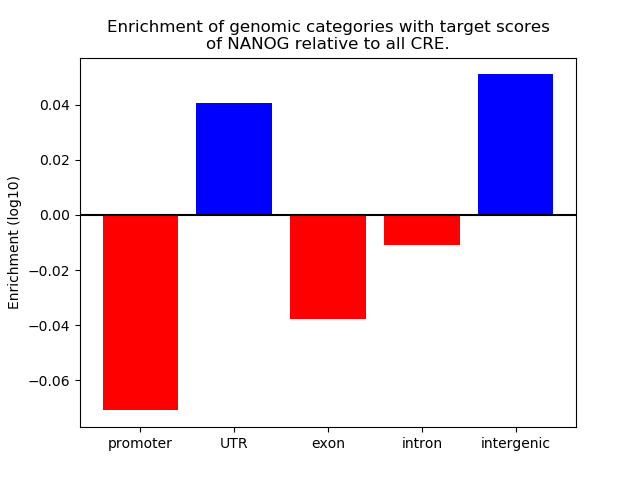
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr12_20557654_20557988 | 0.34 |
RP11-284H19.1 |
|
34625 |
0.2 |
| chr1_205670529_205670680 | 0.32 |
AC119673.1 |
|
11893 |
0.14 |
| chr3_49748675_49748961 | 0.30 |
RNF123 |
ring finger protein 123 |
1445 |
0.27 |
| chr3_149160811_149161262 | 0.30 |
TM4SF4 |
transmembrane 4 L six family member 4 |
30725 |
0.16 |
| chr16_49565236_49565387 | 0.29 |
ZNF423 |
zinc finger protein 423 |
109452 |
0.07 |
| chr11_32447864_32448015 | 0.28 |
WT1 |
Wilms tumor 1 |
2636 |
0.33 |
| chr3_121774222_121774373 | 0.27 |
CD86 |
CD86 molecule |
76 |
0.97 |
| chr15_33299614_33299765 | 0.26 |
ENSG00000212415 |
. |
36761 |
0.19 |
| chr1_25335272_25335426 | 0.26 |
ENSG00000264371 |
. |
14645 |
0.2 |
| chr4_57909352_57909761 | 0.25 |
ENSG00000251703 |
. |
40698 |
0.11 |
| chr8_96705142_96705479 | 0.25 |
ENSG00000223297 |
. |
14167 |
0.29 |
| chr11_72985767_72986011 | 0.25 |
P2RY6 |
pyrimidinergic receptor P2Y, G-protein coupled, 6 |
2424 |
0.23 |
| chr2_62356561_62356823 | 0.24 |
COMMD1 |
copper metabolism (Murr1) domain containing 1 |
6288 |
0.21 |
| chr12_113308184_113308335 | 0.24 |
OAS1 |
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
36323 |
0.15 |
| chr1_26354858_26355009 | 0.24 |
EXTL1 |
exostosin-like glycosyltransferase 1 |
6662 |
0.1 |
| chr2_202083500_202083651 | 0.24 |
CASP8 |
caspase 8, apoptosis-related cysteine peptidase |
14591 |
0.16 |
| chr2_206648577_206648850 | 0.23 |
AC007362.3 |
|
19983 |
0.24 |
| chr8_104165322_104165473 | 0.23 |
ENSG00000265657 |
. |
1445 |
0.3 |
| chr2_105594341_105594492 | 0.23 |
ENSG00000263685 |
. |
15022 |
0.19 |
| chr17_32694389_32694669 | 0.22 |
CCL1 |
chemokine (C-C motif) ligand 1 |
4279 |
0.2 |
| chr17_78723949_78724201 | 0.22 |
RP11-28G8.1 |
|
55357 |
0.14 |
| chr3_37499337_37499488 | 0.22 |
ITGA9 |
integrin, alpha 9 |
5802 |
0.25 |
| chr8_24206460_24207135 | 0.22 |
ADAM28 |
ADAM metallopeptidase domain 28 |
7632 |
0.22 |
| chr8_6149074_6149420 | 0.22 |
RP11-115C21.2 |
|
114816 |
0.06 |
| chr4_173708241_173708420 | 0.22 |
ENSG00000241652 |
. |
354464 |
0.01 |
| chr5_71768852_71769003 | 0.22 |
RP11-389C8.2 |
|
30713 |
0.19 |
| chr8_125434775_125435085 | 0.22 |
TRMT12 |
tRNA methyltransferase 12 homolog (S. cerevisiae) |
28118 |
0.16 |
| chr12_6482203_6482355 | 0.21 |
SCNN1A |
sodium channel, non-voltage-gated 1 alpha subunit |
1690 |
0.23 |
| chr14_35837591_35837748 | 0.21 |
NFKBIA |
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha |
35677 |
0.17 |
| chr6_134537452_134537663 | 0.21 |
ENSG00000238631 |
. |
9905 |
0.2 |
| chr2_231750819_231750970 | 0.21 |
AC012507.3 |
|
321 |
0.87 |
| chr10_80937857_80938019 | 0.20 |
ZMIZ1 |
zinc finger, MIZ-type containing 1 |
109146 |
0.07 |
| chr7_26685921_26686196 | 0.20 |
C7orf71 |
chromosome 7 open reading frame 71 |
8568 |
0.29 |
| chr15_81400350_81400501 | 0.20 |
C15orf26 |
chromosome 15 open reading frame 26 |
8676 |
0.24 |
| chr20_50010365_50010516 | 0.20 |
ENSG00000263645 |
. |
16582 |
0.25 |
| chr2_42304204_42304355 | 0.20 |
PKDCC |
protein kinase domain containing, cytoplasmic |
29119 |
0.18 |
| chr9_129151715_129152192 | 0.20 |
MVB12B |
multivesicular body subunit 12B |
13231 |
0.2 |
| chr17_72599092_72599393 | 0.20 |
CD300LD |
CD300 molecule-like family member d |
10820 |
0.12 |
| chr16_23907560_23907711 | 0.20 |
PRKCB |
protein kinase C, beta |
59091 |
0.13 |
| chr12_12013798_12013949 | 0.20 |
ETV6 |
ets variant 6 |
24998 |
0.26 |
| chr19_17790984_17791135 | 0.19 |
UNC13A |
unc-13 homolog A (C. elegans) |
4081 |
0.18 |
| chr3_168931111_168931360 | 0.19 |
MECOM |
MDS1 and EVI1 complex locus |
55791 |
0.17 |
| chr18_46580939_46581171 | 0.19 |
ENSG00000263849 |
. |
4917 |
0.22 |
| chr3_148477195_148477534 | 0.19 |
AGTR1 |
angiotensin II receptor, type 1 |
19779 |
0.22 |
| chr9_95823092_95823243 | 0.19 |
SUSD3 |
sushi domain containing 3 |
2106 |
0.29 |
| chr18_13268737_13268888 | 0.19 |
LDLRAD4 |
low density lipoprotein receptor class A domain containing 4 |
9290 |
0.2 |
| chr14_52782524_52782774 | 0.19 |
PTGER2 |
prostaglandin E receptor 2 (subtype EP2), 53kDa |
1536 |
0.5 |
| chr16_46782782_46782933 | 0.19 |
MYLK3 |
myosin light chain kinase 3 |
636 |
0.74 |
| chr2_6920801_6920998 | 0.19 |
AC017076.5 |
|
52763 |
0.14 |
| chr16_81868234_81868705 | 0.19 |
PLCG2 |
phospholipase C, gamma 2 (phosphatidylinositol-specific) |
48405 |
0.17 |
| chr19_11433238_11433389 | 0.19 |
CTC-510F12.6 |
|
481 |
0.63 |
| chr2_200527098_200527249 | 0.19 |
FTCDNL1 |
formiminotransferase cyclodeaminase N-terminal like |
188423 |
0.03 |
| chr1_171624147_171624306 | 0.18 |
MYOC |
myocilin, trabecular meshwork inducible glucocorticoid response |
2403 |
0.34 |
| chr15_66938215_66938421 | 0.18 |
SMAD6 |
SMAD family member 6 |
56248 |
0.1 |
| chr11_109966413_109966654 | 0.18 |
ZC3H12C |
zinc finger CCCH-type containing 12C |
2446 |
0.45 |
| chr13_21620980_21621217 | 0.18 |
ENSG00000201821 |
. |
8292 |
0.15 |
| chr12_2723257_2723602 | 0.18 |
CACNA1C-AS3 |
CACNA1C antisense RNA 3 |
6823 |
0.24 |
| chr17_72542253_72542431 | 0.18 |
CD300C |
CD300c molecule |
60 |
0.96 |
| chr19_12800165_12800409 | 0.18 |
CTD-2659N19.2 |
|
551 |
0.5 |
| chr2_145462594_145462835 | 0.18 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
184093 |
0.03 |
| chr22_26047284_26047507 | 0.18 |
ENSG00000222585 |
. |
63741 |
0.09 |
| chr1_153941559_153941710 | 0.18 |
CREB3L4 |
cAMP responsive element binding protein 3-like 4 |
848 |
0.32 |
| chr3_42068290_42068573 | 0.17 |
TRAK1 |
trafficking protein, kinesin binding 1 |
64131 |
0.11 |
| chr3_193564257_193564433 | 0.17 |
ENSG00000243991 |
. |
105835 |
0.07 |
| chr14_63672742_63672893 | 0.17 |
RHOJ |
ras homolog family member J |
1240 |
0.55 |
| chr22_17739441_17739786 | 0.17 |
CECR1 |
cat eye syndrome chromosome region, candidate 1 |
36734 |
0.16 |
| chr16_6914698_6914849 | 0.17 |
ENSG00000200869 |
. |
9127 |
0.29 |
| chr10_4927599_4927750 | 0.17 |
AKR1C1 |
aldo-keto reductase family 1, member C1 |
7122 |
0.22 |
| chr22_23933547_23933698 | 0.17 |
IGLL1 |
immunoglobulin lambda-like polypeptide 1 |
11127 |
0.15 |
| chr13_43919735_43919886 | 0.17 |
ENOX1 |
ecto-NOX disulfide-thiol exchanger 1 |
15403 |
0.31 |
| chr15_39687457_39687608 | 0.17 |
RP11-624L4.1 |
|
25378 |
0.25 |
| chr6_56194231_56194392 | 0.17 |
RP3-445N2.1 |
|
2604 |
0.38 |
| chr4_109454301_109454616 | 0.17 |
ENSG00000266046 |
. |
43838 |
0.17 |
| chr5_177667409_177667628 | 0.17 |
PHYKPL |
5-phosphohydroxy-L-lysine phospho-lyase |
7732 |
0.21 |
| chr1_117040601_117040786 | 0.17 |
ENSG00000200547 |
. |
45535 |
0.11 |
| chr14_75295916_75296067 | 0.17 |
YLPM1 |
YLP motif containing 1 |
12050 |
0.14 |
| chr5_139036948_139037177 | 0.17 |
CXXC5 |
CXXC finger protein 5 |
2078 |
0.35 |
| chr5_179588041_179588247 | 0.17 |
RASGEF1C |
RasGEF domain family, member 1C |
22795 |
0.21 |
| chr4_10706517_10706811 | 0.17 |
CLNK |
cytokine-dependent hematopoietic cell linker |
20175 |
0.29 |
| chr2_29415943_29416138 | 0.17 |
ALK |
anaplastic lymphoma receptor tyrosine kinase |
30220 |
0.18 |
| chr3_51555997_51556148 | 0.17 |
RAD54L2 |
RAD54-like 2 (S. cerevisiae) |
19524 |
0.18 |
| chr7_142430889_142431059 | 0.17 |
PRSS1 |
protease, serine, 1 (trypsin 1) |
26345 |
0.17 |
| chr5_134718436_134718820 | 0.16 |
H2AFY |
H2A histone family, member Y |
16273 |
0.15 |
| chr3_188139152_188139752 | 0.16 |
LPP-AS1 |
LPP antisense RNA 1 |
147002 |
0.04 |
| chr3_71976245_71976396 | 0.16 |
ENSG00000239250 |
. |
99992 |
0.08 |
| chr1_205206275_205206426 | 0.16 |
RP11-383G10.5 |
|
355 |
0.86 |
| chr1_213499256_213499407 | 0.16 |
RPS6KC1 |
ribosomal protein S6 kinase, 52kDa, polypeptide 1 |
274727 |
0.01 |
| chr6_1850485_1850731 | 0.16 |
FOXC1 |
forkhead box C1 |
239927 |
0.02 |
| chr8_17004029_17004180 | 0.16 |
ZDHHC2 |
zinc finger, DHHC-type containing 2 |
9434 |
0.25 |
| chr6_169615307_169615476 | 0.16 |
XXyac-YX65C7_A.2 |
|
2042 |
0.43 |
| chr6_136748361_136748512 | 0.16 |
MAP7 |
microtubule-associated protein 7 |
39577 |
0.18 |
| chr19_19635191_19635342 | 0.16 |
YJEFN3 |
YjeF N-terminal domain containing 3 |
4404 |
0.12 |
| chr1_109934550_109935130 | 0.16 |
SORT1 |
sortilin 1 |
1139 |
0.45 |
| chr21_32641465_32641616 | 0.16 |
TIAM1 |
T-cell lymphoma invasion and metastasis 1 |
7543 |
0.31 |
| chr4_88628609_88628760 | 0.16 |
RP11-742B18.1 |
|
24838 |
0.15 |
| chr14_56880037_56880188 | 0.16 |
TMEM260 |
transmembrane protein 260 |
74960 |
0.12 |
| chr2_218776477_218776674 | 0.16 |
TNS1 |
tensin 1 |
6391 |
0.26 |
| chr4_16034919_16035070 | 0.16 |
ENSG00000251758 |
. |
24584 |
0.18 |
| chr2_122572651_122572909 | 0.16 |
TSN |
translin |
59033 |
0.13 |
| chr3_150805055_150805642 | 0.16 |
MED12L |
mediator complex subunit 12-like |
667 |
0.76 |
| chr17_72387513_72387690 | 0.16 |
GPR142 |
G protein-coupled receptor 142 |
24004 |
0.1 |
| chr1_235055137_235055288 | 0.16 |
ENSG00000239690 |
. |
15279 |
0.26 |
| chr12_102170928_102171079 | 0.16 |
ENSG00000201168 |
. |
2589 |
0.16 |
| chr17_55082561_55082712 | 0.16 |
ENSG00000265809 |
. |
2292 |
0.23 |
| chr7_150549245_150549396 | 0.16 |
AOC1 |
amine oxidase, copper containing 1 |
223 |
0.93 |
| chr16_70456809_70457112 | 0.16 |
ST3GAL2 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 2 |
16031 |
0.11 |
| chr15_45927791_45927942 | 0.16 |
SQRDL |
sulfide quinone reductase-like (yeast) |
928 |
0.56 |
| chr3_18022579_18022730 | 0.15 |
TBC1D5 |
TBC1 domain family, member 5 |
238510 |
0.02 |
| chr8_110032427_110032578 | 0.15 |
RP11-1084E5.1 |
|
43144 |
0.18 |
| chr3_37601222_37601385 | 0.15 |
ENSG00000239105 |
. |
13401 |
0.26 |
| chr12_53026253_53026404 | 0.15 |
KRT73 |
keratin 73 |
13985 |
0.1 |
| chr12_26303323_26303715 | 0.15 |
SSPN |
sarcospan |
15619 |
0.18 |
| chr18_10354698_10354849 | 0.15 |
ENSG00000239031 |
. |
35224 |
0.21 |
| chr5_82672926_82673088 | 0.15 |
VCAN |
versican |
94277 |
0.09 |
| chr9_92236942_92237093 | 0.15 |
GADD45G |
growth arrest and DNA-damage-inducible, gamma |
17064 |
0.27 |
| chr1_108484035_108484186 | 0.15 |
VAV3-AS1 |
VAV3 antisense RNA 1 |
22955 |
0.2 |
| chr12_48111947_48112151 | 0.15 |
ENDOU |
endonuclease, polyU-specific |
617 |
0.69 |
| chr5_142076865_142077016 | 0.15 |
FGF1 |
fibroblast growth factor 1 (acidic) |
646 |
0.77 |
| chr6_44537708_44537867 | 0.15 |
ENSG00000266619 |
. |
134409 |
0.05 |
| chr12_6727682_6727833 | 0.15 |
CHD4 |
chromodomain helicase DNA binding protein 4 |
11115 |
0.08 |
| chr7_158505623_158505780 | 0.15 |
NCAPG2 |
non-SMC condensin II complex, subunit G2 |
8181 |
0.23 |
| chr6_6587145_6587296 | 0.15 |
LY86-AS1 |
LY86 antisense RNA 1 |
206 |
0.91 |
| chr1_244481954_244482146 | 0.15 |
C1orf100 |
chromosome 1 open reading frame 100 |
33887 |
0.2 |
| chr14_24041929_24042080 | 0.15 |
JPH4 |
junctophilin 4 |
206 |
0.88 |
| chr14_70171005_70171156 | 0.15 |
SRSF5 |
serine/arginine-rich splicing factor 5 |
22537 |
0.21 |
| chr17_76875045_76875748 | 0.15 |
TIMP2 |
TIMP metallopeptidase inhibitor 2 |
5164 |
0.16 |
| chr8_134038596_134039031 | 0.15 |
TG |
thyroglobulin |
8869 |
0.25 |
| chr3_58060016_58060182 | 0.15 |
FLNB |
filamin B, beta |
3957 |
0.31 |
| chr19_44038735_44038963 | 0.15 |
ZNF575 |
zinc finger protein 575 |
1284 |
0.27 |
| chr12_121136482_121136633 | 0.15 |
RP11-173P15.3 |
|
747 |
0.48 |
| chr22_42095499_42095650 | 0.15 |
MEI1 |
meiosis inhibitor 1 |
56 |
0.95 |
| chr19_7720613_7720936 | 0.15 |
STXBP2 |
syntaxin binding protein 2 |
9929 |
0.08 |
| chr9_124048116_124048398 | 0.15 |
GSN-AS1 |
GSN antisense RNA 1 |
449 |
0.54 |
| chr17_36582123_36582274 | 0.15 |
ARHGAP23 |
Rho GTPase activating protein 23 |
2464 |
0.24 |
| chr14_54810734_54810960 | 0.15 |
CDKN3 |
cyclin-dependent kinase inhibitor 3 |
52826 |
0.16 |
| chr13_101259048_101259199 | 0.15 |
GGACT |
gamma-glutamylamine cyclotransferase |
17341 |
0.19 |
| chrX_123512646_123512832 | 0.15 |
SH2D1A |
SH2 domain containing 1A |
32300 |
0.24 |
| chr12_111836596_111836747 | 0.15 |
SH2B3 |
SH2B adaptor protein 3 |
7081 |
0.19 |
| chr13_24612924_24613153 | 0.14 |
SPATA13 |
spermatogenesis associated 13 |
59094 |
0.12 |
| chr17_63367349_63367500 | 0.14 |
ENSG00000265189 |
. |
49354 |
0.18 |
| chr5_111089659_111089857 | 0.14 |
NREP |
neuronal regeneration related protein |
2190 |
0.34 |
| chr8_125663658_125664035 | 0.14 |
RP11-532M24.1 |
|
71974 |
0.09 |
| chr4_139327703_139328022 | 0.14 |
SLC7A11 |
solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11 |
164359 |
0.04 |
| chr15_65871486_65871637 | 0.14 |
ENSG00000238715 |
. |
8150 |
0.13 |
| chr2_223733795_223733969 | 0.14 |
ACSL3 |
acyl-CoA synthetase long-chain family member 3 |
7601 |
0.24 |
| chr3_13056298_13056451 | 0.14 |
IQSEC1 |
IQ motif and Sec7 domain 1 |
27838 |
0.23 |
| chr17_26846701_26846852 | 0.14 |
FOXN1 |
forkhead box N1 |
4183 |
0.11 |
| chr21_43728792_43729093 | 0.14 |
TFF3 |
trefoil factor 3 (intestinal) |
6504 |
0.15 |
| chr11_73083087_73083238 | 0.14 |
RELT |
RELT tumor necrosis factor receptor |
4147 |
0.17 |
| chr6_26485880_26486031 | 0.14 |
BTN1A1 |
butyrophilin, subfamily 1, member A1 |
15494 |
0.11 |
| chr8_59583399_59583550 | 0.14 |
ENSG00000238433 |
. |
8571 |
0.23 |
| chr1_159173730_159173881 | 0.14 |
DARC |
Duffy blood group, atypical chemokine receptor |
266 |
0.86 |
| chr5_485358_485509 | 0.14 |
ENSG00000225138 |
. |
8073 |
0.13 |
| chr1_167073679_167073835 | 0.14 |
DUSP27 |
dual specificity phosphatase 27 (putative) |
9670 |
0.17 |
| chr3_29452909_29453060 | 0.14 |
ENSG00000216169 |
. |
42072 |
0.2 |
| chr3_69789465_69789616 | 0.14 |
MITF |
microphthalmia-associated transcription factor |
918 |
0.68 |
| chr1_161038891_161039092 | 0.14 |
ARHGAP30 |
Rho GTPase activating protein 30 |
465 |
0.61 |
| chr9_134752156_134752307 | 0.14 |
ENSG00000212395 |
. |
93886 |
0.08 |
| chr4_144296712_144296863 | 0.14 |
GAB1 |
GRB2-associated binding protein 1 |
1191 |
0.54 |
| chr1_46032186_46032337 | 0.14 |
AKR1A1 |
aldo-keto reductase family 1, member A1 (aldehyde reductase) |
15521 |
0.14 |
| chr1_56942791_56942942 | 0.14 |
ENSG00000223307 |
. |
99776 |
0.08 |
| chr22_25128279_25128430 | 0.14 |
PIWIL3 |
piwi-like RNA-mediated gene silencing 3 |
42090 |
0.15 |
| chr4_8404089_8404323 | 0.14 |
ENSG00000202054 |
. |
9464 |
0.18 |
| chr10_72271972_72272123 | 0.14 |
PALD1 |
phosphatase domain containing, paladin 1 |
33470 |
0.16 |
| chr13_111169859_111170091 | 0.14 |
COL4A2-AS1 |
COL4A2 antisense RNA 1 |
9449 |
0.21 |
| chr20_40072604_40072755 | 0.14 |
CHD6 |
chromodomain helicase DNA binding protein 6 |
55278 |
0.13 |
| chr10_125826370_125826646 | 0.14 |
CHST15 |
carbohydrate (N-acetylgalactosamine 4-sulfate 6-O) sulfotransferase 15 |
20267 |
0.26 |
| chr2_145237384_145237678 | 0.14 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
37584 |
0.19 |
| chr19_55142266_55142451 | 0.14 |
LILRB1 |
leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 1 |
167 |
0.89 |
| chr3_185463236_185463533 | 0.14 |
ENSG00000265470 |
. |
22308 |
0.19 |
| chr18_22677589_22677740 | 0.14 |
ZNF521 |
zinc finger protein 521 |
127089 |
0.06 |
| chr2_46574762_46574949 | 0.14 |
EPAS1 |
endothelial PAS domain protein 1 |
50314 |
0.15 |
| chr12_48554479_48554630 | 0.14 |
ASB8 |
ankyrin repeat and SOCS box containing 8 |
3176 |
0.17 |
| chr2_182284878_182285029 | 0.14 |
ITGA4 |
integrin, alpha 4 (antigen CD49D, alpha 4 subunit of VLA-4 receptor) |
36981 |
0.21 |
| chr8_105800425_105800620 | 0.14 |
ENSG00000263732 |
. |
33119 |
0.24 |
| chr20_19373243_19373394 | 0.14 |
ENSG00000221748 |
. |
51378 |
0.16 |
| chrX_77374906_77375064 | 0.14 |
PGK1 |
phosphoglycerate kinase 1 |
13126 |
0.23 |
| chr6_6588189_6588340 | 0.14 |
LY86 |
lymphocyte antigen 86 |
77 |
0.94 |
| chr18_46199452_46199714 | 0.14 |
ENSG00000266276 |
. |
2612 |
0.32 |
| chr5_373044_373258 | 0.14 |
AHRR |
aryl-hydrocarbon receptor repressor |
29508 |
0.14 |
| chr10_97995296_97995447 | 0.14 |
BLNK |
B-cell linker |
35784 |
0.16 |
| chr10_130978330_130978481 | 0.14 |
MGMT |
O-6-methylguanine-DNA methyltransferase |
287043 |
0.01 |
| chr10_98524301_98524550 | 0.14 |
ENSG00000207287 |
. |
21501 |
0.18 |
| chr7_99797920_99798071 | 0.14 |
STAG3 |
stromal antigen 3 |
4901 |
0.09 |
| chr11_2935428_2935683 | 0.14 |
SLC22A18AS |
solute carrier family 22 (organic cation transporter), member 18 antisense |
10585 |
0.11 |
| chr18_60561889_60562040 | 0.14 |
PHLPP1 |
PH domain and leucine rich repeat protein phosphatase 1 |
8419 |
0.25 |
| chr7_151542313_151542464 | 0.14 |
PRKAG2 |
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
30462 |
0.13 |
| chr14_100481690_100481841 | 0.14 |
EVL |
Enah/Vasp-like |
3954 |
0.23 |
| chr22_27530704_27530861 | 0.13 |
ENSG00000200443 |
. |
98932 |
0.09 |
| chr2_136577229_136577614 | 0.13 |
AC011893.3 |
|
340 |
0.86 |
| chr1_183880847_183880998 | 0.13 |
COLGALT2 |
collagen beta(1-O)galactosyltransferase 2 |
34830 |
0.2 |
| chr18_35302652_35302803 | 0.13 |
ENSG00000266530 |
. |
65629 |
0.15 |
| chr5_39400930_39401193 | 0.13 |
DAB2 |
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
6559 |
0.27 |
| chr9_94065201_94065395 | 0.13 |
AUH |
AU RNA binding protein/enoyl-CoA hydratase |
58879 |
0.16 |
| chr7_132139041_132139287 | 0.13 |
AC011625.1 |
|
102071 |
0.08 |
| chr1_27626590_27626805 | 0.13 |
TMEM222 |
transmembrane protein 222 |
21954 |
0.11 |
| chr18_53780471_53780622 | 0.13 |
ENSG00000201816 |
. |
33721 |
0.25 |
| chr16_86632416_86632567 | 0.13 |
FOXL1 |
forkhead box L1 |
20376 |
0.17 |
| chr11_71844321_71844472 | 0.13 |
FOLR3 |
folate receptor 3 (gamma) |
2360 |
0.15 |
| chr5_82297884_82298035 | 0.13 |
ENSG00000238835 |
. |
62197 |
0.13 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0032641 | lymphotoxin A production(GO:0032641) lymphotoxin A biosynthetic process(GO:0042109) |
| 0.0 | 0.2 | GO:0045542 | positive regulation of cholesterol biosynthetic process(GO:0045542) |
| 0.0 | 0.1 | GO:0072311 | renal filtration cell differentiation(GO:0061318) glomerular epithelium development(GO:0072010) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.0 | 0.1 | GO:0060298 | positive regulation of sarcomere organization(GO:0060298) |
| 0.0 | 0.1 | GO:0051712 | positive regulation of killing of cells of other organism(GO:0051712) |
| 0.0 | 0.0 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.0 | 0.1 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.0 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 0.1 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.0 | 0.1 | GO:0007028 | cytoplasm organization(GO:0007028) |
| 0.0 | 0.1 | GO:0009648 | photoperiodism(GO:0009648) |
| 0.0 | 0.1 | GO:0006562 | proline catabolic process(GO:0006562) |
| 0.0 | 0.1 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.0 | GO:0051709 | regulation of killing of cells of other organism(GO:0051709) |
| 0.0 | 0.1 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.1 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.0 | 0.1 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.2 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.0 | 0.1 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.1 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.1 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.0 | 0.1 | GO:0031558 | obsolete induction of apoptosis in response to chemical stimulus(GO:0031558) |
| 0.0 | 0.0 | GO:0003078 | obsolete regulation of natriuresis(GO:0003078) |
| 0.0 | 0.1 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.0 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.0 | 0.1 | GO:0060314 | regulation of ryanodine-sensitive calcium-release channel activity(GO:0060314) |
| 0.0 | 0.0 | GO:0061030 | epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.0 | 0.0 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.0 | GO:0009138 | pyrimidine nucleoside diphosphate metabolic process(GO:0009138) |
| 0.0 | 0.1 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.1 | GO:0046882 | negative regulation of follicle-stimulating hormone secretion(GO:0046882) |
| 0.0 | 0.0 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.0 | 0.0 | GO:0032959 | inositol trisphosphate biosynthetic process(GO:0032959) |
| 0.0 | 0.0 | GO:0050651 | dermatan sulfate proteoglycan biosynthetic process(GO:0050651) |
| 0.0 | 0.0 | GO:0071605 | monocyte chemotactic protein-1 production(GO:0071605) regulation of monocyte chemotactic protein-1 production(GO:0071637) |
| 0.0 | 0.0 | GO:0032682 | negative regulation of chemokine production(GO:0032682) negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.0 | 0.0 | GO:0042726 | flavin-containing compound metabolic process(GO:0042726) |
| 0.0 | 0.0 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 | 0.1 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.0 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.0 | GO:0042996 | regulation of Golgi to plasma membrane protein transport(GO:0042996) |
| 0.0 | 0.0 | GO:0032071 | regulation of endodeoxyribonuclease activity(GO:0032071) |
| 0.0 | 0.1 | GO:0051883 | disruption of cells of other organism involved in symbiotic interaction(GO:0051818) killing of cells in other organism involved in symbiotic interaction(GO:0051883) |
| 0.0 | 0.0 | GO:0031223 | auditory behavior(GO:0031223) |
| 0.0 | 0.0 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.1 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.1 | GO:0030949 | positive regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030949) |
| 0.0 | 0.0 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.0 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.1 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.1 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.1 | GO:0031312 | extrinsic component of organelle membrane(GO:0031312) |
| 0.0 | 0.0 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 0.1 | GO:0071437 | invadopodium(GO:0071437) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.2 | GO:0071553 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.0 | 0.1 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.0 | 0.2 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.1 | GO:0019956 | chemokine binding(GO:0019956) |
| 0.0 | 0.1 | GO:0019863 | IgE binding(GO:0019863) |
| 0.0 | 0.1 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.2 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.1 | GO:0034648 | histone demethylase activity (H3-K4 specific)(GO:0032453) histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.0 | 0.1 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.1 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.1 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.0 | GO:0046980 | tapasin binding(GO:0046980) |
| 0.0 | 0.1 | GO:0016653 | oxidoreductase activity, acting on NAD(P)H, heme protein as acceptor(GO:0016653) |
| 0.0 | 0.2 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.0 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.0 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
| 0.0 | 0.1 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.1 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.0 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.0 | 0.1 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.0 | GO:0000987 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.0 | 0.0 | GO:0042910 | xenobiotic-transporting ATPase activity(GO:0008559) xenobiotic transporter activity(GO:0042910) |
| 0.0 | 0.1 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.0 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.0 | 0.0 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.0 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.0 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.0 | 0.0 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 0.1 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.1 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.1 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.0 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.0 | GO:0047115 | trans-1,2-dihydrobenzene-1,2-diol dehydrogenase activity(GO:0047115) |
| 0.0 | 0.1 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.0 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.0 | GO:0005346 | purine ribonucleotide transmembrane transporter activity(GO:0005346) |
| 0.0 | 0.0 | GO:0016717 | oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water(GO:0016717) |
| 0.0 | 0.0 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.2 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.2 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.1 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.0 | GO:0031685 | adenosine receptor binding(GO:0031685) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 0.0 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.1 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.0 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME PLC BETA MEDIATED EVENTS | Genes involved in PLC beta mediated events |
| 0.0 | 0.2 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.2 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 0.1 | REACTOME PHOSPHOLIPASE C MEDIATED CASCADE | Genes involved in Phospholipase C-mediated cascade |
| 0.0 | 0.2 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.0 | 0.2 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.1 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.0 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |