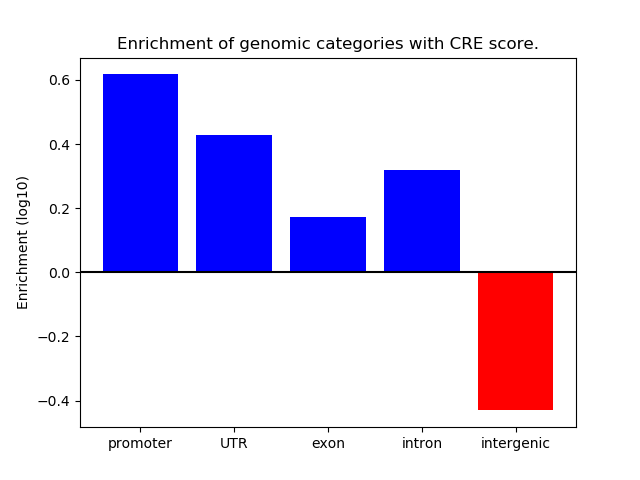
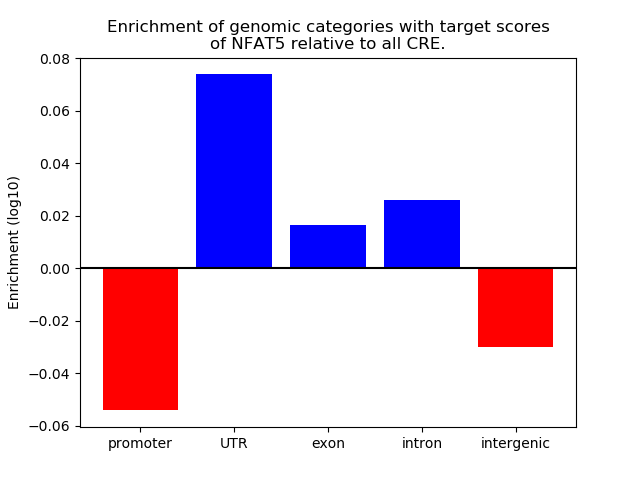
Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
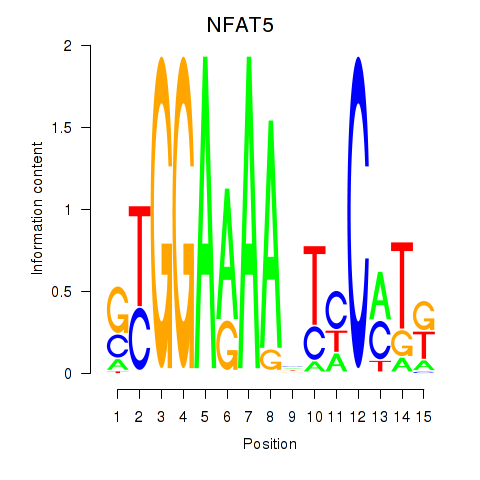
Results for NFAT5
Z-value: 1.03
Transcription factors associated with NFAT5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NFAT5
|
ENSG00000102908.16 | nuclear factor of activated T cells 5 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr16_69653321_69653472 | NFAT5 | 27639 | 0.149140 | 0.96 | 2.6e-05 | Click! |
| chr16_69653099_69653250 | NFAT5 | 27861 | 0.148694 | 0.95 | 7.6e-05 | Click! |
| chr16_69600802_69601252 | NFAT5 | 773 | 0.528857 | 0.80 | 1.0e-02 | Click! |
| chr16_69652942_69653093 | NFAT5 | 28018 | 0.148377 | 0.71 | 3.3e-02 | Click! |
| chr16_69568902_69569053 | NFAT5 | 30020 | 0.146465 | -0.70 | 3.6e-02 | Click! |
Activity of the NFAT5 motif across conditions
Conditions sorted by the z-value of the NFAT5 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
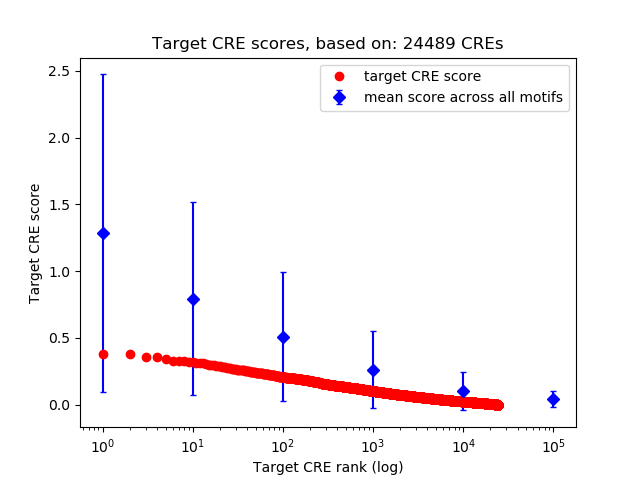
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr9_134546803_134547477 | 0.38 |
RAPGEF1 |
Rap guanine nucleotide exchange factor (GEF) 1 |
38089 |
0.16 |
| chrX_128904932_128905357 | 0.38 |
SASH3 |
SAM and SH3 domain containing 3 |
8816 |
0.2 |
| chr1_226913224_226913375 | 0.36 |
ITPKB |
inositol-trisphosphate 3-kinase B |
11860 |
0.22 |
| chr18_3060720_3060948 | 0.36 |
ENSG00000252353 |
. |
32732 |
0.14 |
| chr8_82053234_82053464 | 0.34 |
PAG1 |
phosphoprotein associated with glycosphingolipid microdomains 1 |
29046 |
0.23 |
| chr1_100908166_100908711 | 0.33 |
RP5-837M10.4 |
|
43115 |
0.14 |
| chr17_1775725_1776037 | 0.33 |
RPA1 |
replication protein A1, 70kDa |
6448 |
0.16 |
| chr6_143173861_143174012 | 0.33 |
HIVEP2 |
human immunodeficiency virus type I enhancer binding protein 2 |
15752 |
0.27 |
| chr1_206740198_206740524 | 0.32 |
RASSF5 |
Ras association (RalGDS/AF-6) domain family member 5 |
9868 |
0.16 |
| chr3_68135261_68135467 | 0.32 |
FAM19A1 |
family with sequence similarity 19 (chemokine (C-C motif)-like), member A1 |
79991 |
0.12 |
| chr14_63979608_63979759 | 0.31 |
PPP2R5E |
protein phosphatase 2, regulatory subunit B', epsilon isoform |
4728 |
0.18 |
| chr8_87359664_87359960 | 0.31 |
WWP1 |
WW domain containing E3 ubiquitin protein ligase 1 |
4845 |
0.3 |
| chr3_33087403_33087747 | 0.31 |
GLB1 |
galactosidase, beta 1 |
50709 |
0.11 |
| chr2_136969948_136970391 | 0.30 |
CXCR4 |
chemokine (C-X-C motif) receptor 4 |
94434 |
0.09 |
| chr2_207306958_207307129 | 0.30 |
ADAM23 |
ADAM metallopeptidase domain 23 |
1220 |
0.57 |
| chrX_152603176_152603363 | 0.30 |
ZNF275 |
zinc finger protein 275 |
3656 |
0.21 |
| chr10_111830389_111830580 | 0.30 |
ADD3 |
adducin 3 (gamma) |
62762 |
0.11 |
| chr5_95151283_95151599 | 0.29 |
GLRX |
glutaredoxin (thioltransferase) |
6974 |
0.16 |
| chr4_40190799_40190950 | 0.29 |
RHOH |
ras homolog family member H |
1799 |
0.37 |
| chr18_2961462_2961796 | 0.29 |
RP11-737O24.1 |
|
5387 |
0.16 |
| chr12_54893042_54893194 | 0.28 |
NCKAP1L |
NCK-associated protein 1-like |
550 |
0.7 |
| chr16_68107852_68108775 | 0.28 |
NFATC3 |
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 3 |
10934 |
0.1 |
| chr5_49659303_49659576 | 0.28 |
EMB |
embigin |
65122 |
0.15 |
| chr14_51279547_51280135 | 0.28 |
RP11-286O18.1 |
|
8757 |
0.15 |
| chr6_144682280_144682476 | 0.28 |
UTRN |
utrophin |
17141 |
0.24 |
| chr15_66110664_66110894 | 0.27 |
ENSG00000252715 |
. |
25043 |
0.14 |
| chr8_38790336_38790830 | 0.27 |
PLEKHA2 |
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 2 |
31721 |
0.12 |
| chr8_134538342_134538795 | 0.27 |
ST3GAL1 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
26942 |
0.25 |
| chr2_198189379_198189688 | 0.27 |
AC010746.3 |
|
13416 |
0.15 |
| chr7_144527992_144528143 | 0.26 |
TPK1 |
thiamin pyrophosphokinase 1 |
5079 |
0.28 |
| chr14_55178975_55179167 | 0.26 |
SAMD4A |
sterile alpha motif domain containing 4A |
42480 |
0.18 |
| chr7_138768806_138768957 | 0.26 |
ZC3HAV1 |
zinc finger CCCH-type, antiviral 1 |
4868 |
0.24 |
| chr11_35268427_35268939 | 0.26 |
SLC1A2 |
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
18609 |
0.14 |
| chr11_128645048_128645199 | 0.26 |
FLI1 |
Fli-1 proto-oncogene, ETS transcription factor |
10438 |
0.22 |
| chr12_9805985_9806454 | 0.26 |
RP11-705C15.2 |
|
60 |
0.96 |
| chr10_22902783_22903038 | 0.26 |
PIP4K2A |
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha |
22268 |
0.26 |
| chr6_138196299_138196450 | 0.26 |
RP11-356I2.4 |
|
7004 |
0.23 |
| chr10_4023787_4024265 | 0.26 |
KLF6 |
Kruppel-like factor 6 |
196553 |
0.03 |
| chr20_57695964_57696140 | 0.26 |
ZNF831 |
zinc finger protein 831 |
70023 |
0.09 |
| chr1_2206148_2206653 | 0.26 |
SKI |
v-ski avian sarcoma viral oncogene homolog |
46266 |
0.08 |
| chr17_17087488_17087777 | 0.25 |
RP11-45M22.3 |
|
1236 |
0.32 |
| chr2_204583285_204583561 | 0.25 |
CD28 |
CD28 molecule |
12007 |
0.23 |
| chr11_57443281_57443465 | 0.25 |
ZDHHC5 |
zinc finger, DHHC-type containing 5 |
2757 |
0.15 |
| chr1_66836429_66836594 | 0.25 |
PDE4B |
phosphodiesterase 4B, cAMP-specific |
16446 |
0.28 |
| chr3_16356399_16356550 | 0.25 |
RP11-415F23.2 |
|
528 |
0.76 |
| chr11_6770543_6770694 | 0.25 |
OR2AG2 |
olfactory receptor, family 2, subfamily AG, member 2 |
19668 |
0.11 |
| chr14_61763065_61763216 | 0.25 |
TMEM30B |
transmembrane protein 30B |
14582 |
0.17 |
| chr1_92876308_92876612 | 0.24 |
ENSG00000242764 |
. |
8218 |
0.25 |
| chr10_11241753_11242016 | 0.24 |
RP3-323N1.2 |
|
28545 |
0.19 |
| chr2_148257449_148257801 | 0.24 |
ENSG00000202074 |
. |
4517 |
0.24 |
| chr7_100147991_100148484 | 0.24 |
AGFG2 |
ArfGAP with FG repeats 2 |
5053 |
0.1 |
| chr2_223169089_223169702 | 0.24 |
CCDC140 |
coiled-coil domain containing 140 |
1698 |
0.35 |
| chr18_13615466_13615831 | 0.24 |
LDLRAD4 |
low density lipoprotein receptor class A domain containing 4 |
3030 |
0.17 |
| chr17_20179870_20180077 | 0.24 |
ENSG00000252971 |
. |
13597 |
0.17 |
| chr1_202134842_202135197 | 0.24 |
PTPN7 |
protein tyrosine phosphatase, non-receptor type 7 |
4303 |
0.16 |
| chr5_156650274_156650456 | 0.24 |
CTB-4E7.1 |
|
1025 |
0.44 |
| chr16_85481767_85482285 | 0.24 |
ENSG00000264203 |
. |
6928 |
0.27 |
| chr2_10520453_10520734 | 0.23 |
HPCAL1 |
hippocalcin-like 1 |
39554 |
0.14 |
| chr1_167453393_167453610 | 0.23 |
RP11-104L21.2 |
|
25603 |
0.17 |
| chr8_134578488_134578789 | 0.23 |
ST3GAL1 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
5454 |
0.32 |
| chr4_109081982_109082160 | 0.23 |
LEF1 |
lymphoid enhancer-binding factor 1 |
5386 |
0.25 |
| chr5_66453399_66453643 | 0.23 |
MAST4 |
microtubule associated serine/threonine kinase family member 4 |
15259 |
0.25 |
| chr5_85334298_85334449 | 0.23 |
NBPF22P |
neuroblastoma breakpoint family, member 22, pseudogene |
244211 |
0.02 |
| chr12_132092633_132092991 | 0.23 |
ENSG00000212154 |
. |
52796 |
0.15 |
| chr3_16353015_16353233 | 0.23 |
RP11-415F23.2 |
|
2822 |
0.24 |
| chr1_120684562_120684908 | 0.23 |
ENSG00000207149 |
. |
15173 |
0.24 |
| chr18_46311251_46311510 | 0.23 |
RP11-484L8.1 |
|
49761 |
0.15 |
| chr3_33058315_33058653 | 0.23 |
CCR4 |
chemokine (C-C motif) receptor 4 |
65418 |
0.1 |
| chr12_57868027_57868178 | 0.23 |
ARHGAP9 |
Rho GTPase activating protein 9 |
615 |
0.5 |
| chr8_60010060_60010211 | 0.22 |
RP11-25K19.1 |
|
21464 |
0.19 |
| chr11_36472075_36472226 | 0.22 |
RP11-219O3.2 |
|
24478 |
0.18 |
| chr17_37251149_37251404 | 0.22 |
PLXDC1 |
plexin domain containing 1 |
12464 |
0.15 |
| chr2_30559818_30559976 | 0.22 |
ENSG00000221377 |
. |
95417 |
0.07 |
| chrX_128921294_128921545 | 0.22 |
SASH3 |
SAM and SH3 domain containing 3 |
7459 |
0.2 |
| chr3_3213200_3213369 | 0.22 |
CRBN |
cereblon |
8074 |
0.19 |
| chr12_112440617_112440858 | 0.22 |
TMEM116 |
transmembrane protein 116 |
3052 |
0.22 |
| chr14_53773006_53773157 | 0.22 |
RP11-547D23.1 |
|
153009 |
0.04 |
| chr16_80625275_80625613 | 0.22 |
RP11-525K10.3 |
|
28412 |
0.14 |
| chr8_26171540_26171691 | 0.22 |
PPP2R2A |
protein phosphatase 2, regulatory subunit B, alpha |
20616 |
0.23 |
| chr12_28404046_28404197 | 0.22 |
CCDC91 |
coiled-coil domain containing 91 |
5537 |
0.28 |
| chr2_42523029_42523463 | 0.22 |
EML4 |
echinoderm microtubule associated protein like 4 |
5147 |
0.28 |
| chr6_152011555_152011706 | 0.22 |
ESR1 |
estrogen receptor 1 |
1 |
0.99 |
| chr16_88759405_88759694 | 0.22 |
RP5-1142A6.5 |
|
3360 |
0.09 |
| chr6_108043823_108043992 | 0.22 |
SCML4 |
sex comb on midleg-like 4 (Drosophila) |
9692 |
0.29 |
| chrX_49148626_49148900 | 0.22 |
GAGE10 |
G antigen 10 |
11385 |
0.08 |
| chr1_198590725_198590954 | 0.21 |
PTPRC |
protein tyrosine phosphatase, receptor type, C |
16962 |
0.24 |
| chr13_51464563_51464756 | 0.21 |
RNASEH2B-AS1 |
RNASEH2B antisense RNA 1 |
1119 |
0.51 |
| chr19_41807008_41807159 | 0.21 |
CCDC97 |
coiled-coil domain containing 97 |
9011 |
0.11 |
| chr7_44681338_44681649 | 0.21 |
OGDH |
oxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide) |
17432 |
0.18 |
| chr20_47391272_47391546 | 0.21 |
ENSG00000251876 |
. |
35424 |
0.19 |
| chr10_14625977_14626446 | 0.21 |
FAM107B |
family with sequence similarity 107, member B |
11873 |
0.24 |
| chr12_92899152_92899351 | 0.21 |
ENSG00000238865 |
. |
39512 |
0.17 |
| chr3_33472190_33472341 | 0.21 |
UBP1 |
upstream binding protein 1 (LBP-1a) |
9237 |
0.21 |
| chr4_3203376_3203582 | 0.21 |
HTT |
huntingtin |
41390 |
0.15 |
| chr6_11211149_11211363 | 0.21 |
NEDD9 |
neural precursor cell expressed, developmentally down-regulated 9 |
21635 |
0.19 |
| chr14_91769117_91769483 | 0.21 |
CCDC88C |
coiled-coil domain containing 88C |
17288 |
0.17 |
| chr13_100037229_100037523 | 0.21 |
ENSG00000207719 |
. |
28991 |
0.17 |
| chr2_158327174_158327325 | 0.21 |
CYTIP |
cytohesin 1 interacting protein |
18092 |
0.22 |
| chr12_123712658_123712809 | 0.21 |
MPHOSPH9 |
M-phase phosphoprotein 9 |
2260 |
0.21 |
| chr2_231457387_231457706 | 0.21 |
ENSG00000199791 |
. |
5710 |
0.22 |
| chr10_17725949_17726205 | 0.21 |
ENSG00000251959 |
. |
4890 |
0.18 |
| chr16_53948312_53948589 | 0.21 |
FTO |
fat mass and obesity associated |
3282 |
0.32 |
| chrX_134517228_134517850 | 0.21 |
ZNF449 |
zinc finger protein 449 |
38818 |
0.14 |
| chr16_29826201_29826393 | 0.20 |
PAGR1 |
PAXIP1 associated glutamate-rich protein 1 |
988 |
0.24 |
| chr13_52527282_52527433 | 0.20 |
ATP7B |
ATPase, Cu++ transporting, beta polypeptide |
8694 |
0.22 |
| chr3_150923497_150923774 | 0.20 |
GPR171 |
G protein-coupled receptor 171 |
2647 |
0.22 |
| chr12_47758069_47758220 | 0.20 |
ENSG00000264906 |
. |
92 |
0.98 |
| chr9_92137029_92137304 | 0.20 |
SEMA4D |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D |
24121 |
0.2 |
| chr7_37405451_37405685 | 0.20 |
ELMO1 |
engulfment and cell motility 1 |
12296 |
0.2 |
| chr7_151049124_151049275 | 0.20 |
NUB1 |
negative regulator of ubiquitin-like proteins 1 |
6768 |
0.16 |
| chr1_169659993_169660655 | 0.20 |
SELL |
selectin L |
20515 |
0.18 |
| chr7_50321524_50321798 | 0.20 |
IKZF1 |
IKAROS family zinc finger 1 (Ikaros) |
22663 |
0.25 |
| chr22_23132033_23132342 | 0.20 |
ENSG00000207833 |
. |
2790 |
0.07 |
| chr14_66394986_66395728 | 0.20 |
CTD-2014B16.3 |
Uncharacterized protein |
75884 |
0.11 |
| chr6_37476350_37476501 | 0.20 |
CCDC167 |
coiled-coil domain containing 167 |
8727 |
0.21 |
| chr5_54333803_54333961 | 0.20 |
ENSG00000207229 |
. |
2793 |
0.19 |
| chr2_106755042_106755216 | 0.20 |
UXS1 |
UDP-glucuronate decarboxylase 1 |
113 |
0.98 |
| chr3_71755190_71755341 | 0.20 |
EIF4E3 |
eukaryotic translation initiation factor 4E family member 3 |
19261 |
0.21 |
| chr1_151797364_151797736 | 0.20 |
RORC |
RAR-related orphan receptor C |
1017 |
0.29 |
| chr5_156451707_156451858 | 0.20 |
HAVCR1 |
hepatitis A virus cellular receptor 1 |
33625 |
0.15 |
| chr17_77803360_77803511 | 0.20 |
CBX4 |
chromobox homolog 4 |
8245 |
0.15 |
| chr19_42945492_42946028 | 0.20 |
CXCL17 |
chemokine (C-X-C motif) ligand 17 |
1440 |
0.28 |
| chr6_11717864_11718015 | 0.20 |
ENSG00000207419 |
. |
7887 |
0.25 |
| chr17_48986753_48987319 | 0.20 |
TOB1 |
transducer of ERBB2, 1 |
41697 |
0.12 |
| chr1_40850590_40850842 | 0.20 |
SMAP2 |
small ArfGAP2 |
10396 |
0.17 |
| chr6_31740986_31741137 | 0.20 |
VWA7 |
von Willebrand factor A domain containing 7 |
4010 |
0.06 |
| chr11_95951199_95951370 | 0.20 |
ENSG00000266192 |
. |
123318 |
0.06 |
| chr8_117133446_117133597 | 0.20 |
ENSG00000221793 |
. |
15477 |
0.28 |
| chr11_128293469_128293620 | 0.20 |
ETS1 |
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
81745 |
0.1 |
| chr3_28337245_28337396 | 0.20 |
AZI2 |
5-azacytidine induced 2 |
36578 |
0.17 |
| chr6_16421963_16422331 | 0.20 |
ENSG00000265642 |
. |
6607 |
0.32 |
| chr6_151693582_151694001 | 0.20 |
ENSG00000252615 |
. |
5443 |
0.15 |
| chr16_3554613_3554882 | 0.20 |
CLUAP1 |
clusterin associated protein 1 |
2703 |
0.18 |
| chr8_134512123_134512433 | 0.20 |
ST3GAL1 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
652 |
0.83 |
| chr6_130040950_130041101 | 0.20 |
ARHGAP18 |
Rho GTPase activating protein 18 |
9655 |
0.25 |
| chr12_110787824_110788162 | 0.20 |
ATP2A2 |
ATPase, Ca++ transporting, cardiac muscle, slow twitch 2 |
3987 |
0.2 |
| chr21_15894298_15894574 | 0.20 |
SAMSN1 |
SAM domain, SH3 domain and nuclear localization signals 1 |
24226 |
0.22 |
| chr3_16355269_16355595 | 0.20 |
RP11-415F23.2 |
|
514 |
0.77 |
| chr18_60981487_60981741 | 0.20 |
RP11-28F1.2 |
|
299 |
0.89 |
| chr21_19155131_19155430 | 0.20 |
AL109761.5 |
|
10525 |
0.23 |
| chrX_135960364_135960605 | 0.20 |
ENSG00000206979 |
. |
946 |
0.43 |
| chr12_121333840_121333991 | 0.19 |
SPPL3 |
signal peptide peptidase like 3 |
7258 |
0.19 |
| chr12_121677712_121677919 | 0.19 |
CAMKK2 |
calcium/calmodulin-dependent protein kinase kinase 2, beta |
20530 |
0.16 |
| chr14_91798820_91799198 | 0.19 |
ENSG00000265856 |
. |
1048 |
0.58 |
| chr10_73839065_73839273 | 0.19 |
SPOCK2 |
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2 |
8917 |
0.24 |
| chr5_50010543_50010746 | 0.19 |
PARP8 |
poly (ADP-ribose) polymerase family, member 8 |
47253 |
0.2 |
| chr21_46686004_46686155 | 0.19 |
POFUT2 |
protein O-fucosyltransferase 2 |
19527 |
0.17 |
| chr4_76860371_76860572 | 0.19 |
NAAA |
N-acylethanolamine acid amidase |
942 |
0.47 |
| chr10_112602586_112603108 | 0.19 |
PDCD4 |
programmed cell death 4 (neoplastic transformation inhibitor) |
28718 |
0.13 |
| chr1_108327675_108328746 | 0.19 |
ENSG00000265536 |
. |
9332 |
0.26 |
| chrX_41641506_41641657 | 0.19 |
GPR82 |
G protein-coupled receptor 82 |
58173 |
0.13 |
| chr3_183067564_183068011 | 0.19 |
MCF2L2 |
MCF.2 cell line derived transforming sequence-like 2 |
77978 |
0.07 |
| chr9_14314790_14315056 | 0.19 |
NFIB |
nuclear factor I/B |
342 |
0.89 |
| chr10_22030567_22030830 | 0.19 |
ENSG00000252634 |
. |
43523 |
0.16 |
| chr12_68025335_68025706 | 0.19 |
DYRK2 |
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 2 |
16598 |
0.25 |
| chr7_149565914_149566574 | 0.19 |
ATP6V0E2 |
ATPase, H+ transporting V0 subunit e2 |
3813 |
0.19 |
| chr10_126384949_126385100 | 0.19 |
FAM53B-AS1 |
FAM53B antisense RNA 1 |
7170 |
0.22 |
| chr3_33096251_33096868 | 0.19 |
GLB1 |
galactosidase, beta 1 |
41725 |
0.12 |
| chr10_11241276_11241580 | 0.19 |
RP3-323N1.2 |
|
28089 |
0.19 |
| chr15_61135175_61135326 | 0.19 |
RP11-554D20.1 |
|
78311 |
0.11 |
| chr18_2958940_2959410 | 0.19 |
RP11-737O24.1 |
|
7841 |
0.15 |
| chr6_27122996_27123197 | 0.19 |
ENSG00000265565 |
. |
7691 |
0.12 |
| chrX_154492108_154492373 | 0.19 |
RAB39B |
RAB39B, member RAS oncogene family |
1634 |
0.36 |
| chr4_120202212_120202363 | 0.19 |
C4orf3 |
chromosome 4 open reading frame 3 |
19789 |
0.17 |
| chrX_128912114_128912265 | 0.19 |
SASH3 |
SAM and SH3 domain containing 3 |
1771 |
0.37 |
| chr2_84743044_84743233 | 0.19 |
DNAH6 |
dynein, axonemal, heavy chain 6 |
441 |
0.89 |
| chr14_99688948_99689139 | 0.19 |
AL109767.1 |
|
40242 |
0.16 |
| chr18_18673078_18673254 | 0.19 |
ROCK1 |
Rho-associated, coiled-coil containing protein kinase 1 |
18646 |
0.19 |
| chr16_79311831_79311982 | 0.19 |
ENSG00000222244 |
. |
13555 |
0.29 |
| chr14_76017369_76017520 | 0.19 |
FLVCR2 |
feline leukemia virus subgroup C cellular receptor family, member 2 |
27516 |
0.13 |
| chr14_92469799_92469950 | 0.19 |
TRIP11 |
thyroid hormone receptor interactor 11 |
12221 |
0.18 |
| chr15_44963811_44964098 | 0.19 |
PATL2 |
protein associated with topoisomerase II homolog 2 (yeast) |
3271 |
0.19 |
| chr10_104411938_104412104 | 0.19 |
TRIM8 |
tripartite motif containing 8 |
7377 |
0.18 |
| chr12_9853577_9853728 | 0.18 |
CLEC2D |
C-type lectin domain family 2, member D |
13155 |
0.13 |
| chr2_65255557_65255797 | 0.18 |
AC007386.4 |
|
8954 |
0.17 |
| chr19_54876287_54876508 | 0.18 |
LAIR1 |
leukocyte-associated immunoglobulin-like receptor 1 |
17 |
0.96 |
| chr13_41188048_41188199 | 0.18 |
FOXO1 |
forkhead box O1 |
52611 |
0.14 |
| chr1_151593899_151594050 | 0.18 |
ENSG00000206635 |
. |
7826 |
0.11 |
| chr3_56836593_56836843 | 0.18 |
ARHGEF3 |
Rho guanine nucleotide exchange factor (GEF) 3 |
723 |
0.78 |
| chr14_92329341_92329492 | 0.18 |
TC2N |
tandem C2 domains, nuclear |
4457 |
0.23 |
| chr1_24851023_24851224 | 0.18 |
ENSG00000266551 |
. |
5081 |
0.18 |
| chr4_38513354_38513505 | 0.18 |
RP11-617D20.1 |
|
112767 |
0.06 |
| chr2_158277456_158277614 | 0.18 |
CYTIP |
cytohesin 1 interacting protein |
18391 |
0.2 |
| chr15_86243586_86243737 | 0.18 |
RP11-815J21.1 |
|
679 |
0.57 |
| chr17_15847097_15847248 | 0.18 |
ADORA2B |
adenosine A2b receptor |
1059 |
0.53 |
| chr3_45989617_45989903 | 0.18 |
CXCR6 |
chemokine (C-X-C motif) receptor 6 |
3219 |
0.21 |
| chr20_37503628_37504045 | 0.18 |
ENSG00000240474 |
. |
2423 |
0.3 |
| chr6_11714882_11715244 | 0.18 |
ENSG00000207419 |
. |
5011 |
0.27 |
| chr11_69078676_69078863 | 0.18 |
MYEOV |
myeloma overexpressed |
17144 |
0.27 |
| chr8_126661069_126661220 | 0.18 |
ENSG00000266452 |
. |
204337 |
0.03 |
| chr1_213245297_213245448 | 0.18 |
RPS6KC1 |
ribosomal protein S6 kinase, 52kDa, polypeptide 1 |
20768 |
0.23 |
| chr8_23107497_23107648 | 0.18 |
CHMP7 |
charged multivesicular body protein 7 |
3400 |
0.16 |
| chr17_46912636_46912982 | 0.18 |
CALCOCO2 |
calcium binding and coiled-coil domain 2 |
4219 |
0.14 |
| chr3_150600490_150600641 | 0.18 |
RP11-166N6.2 |
|
7858 |
0.16 |
| chr1_26003341_26003560 | 0.18 |
RP1-187B23.1 |
|
17848 |
0.17 |
| chr8_129595182_129595552 | 0.18 |
ENSG00000221351 |
. |
236673 |
0.02 |
| chr1_84902550_84902701 | 0.18 |
DNASE2B |
deoxyribonuclease II beta |
28712 |
0.16 |
| chr13_48762180_48762745 | 0.18 |
ITM2B |
integral membrane protein 2B |
44832 |
0.16 |
| chr15_99600402_99600553 | 0.18 |
SYNM |
synemin, intermediate filament protein |
37943 |
0.12 |
| chr3_152018760_152019009 | 0.18 |
MBNL1 |
muscleblind-like splicing regulator 1 |
897 |
0.64 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.1 | 0.3 | GO:0045589 | regulation of regulatory T cell differentiation(GO:0045589) |
| 0.1 | 0.3 | GO:0001911 | negative regulation of leukocyte mediated cytotoxicity(GO:0001911) |
| 0.1 | 0.2 | GO:0030011 | maintenance of cell polarity(GO:0030011) |
| 0.1 | 0.2 | GO:0060750 | epithelial cell proliferation involved in mammary gland duct elongation(GO:0060750) |
| 0.1 | 0.1 | GO:0002713 | negative regulation of B cell mediated immunity(GO:0002713) negative regulation of immunoglobulin mediated immune response(GO:0002890) |
| 0.1 | 0.2 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.1 | 0.2 | GO:0045066 | regulatory T cell differentiation(GO:0045066) |
| 0.1 | 0.6 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.1 | 0.4 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.1 | 0.3 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 0.1 | 0.1 | GO:0048537 | mucosal-associated lymphoid tissue development(GO:0048537) |
| 0.1 | 0.2 | GO:0090292 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.1 | GO:0006772 | thiamine metabolic process(GO:0006772) |
| 0.0 | 0.1 | GO:0006154 | adenosine catabolic process(GO:0006154) |
| 0.0 | 0.1 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.1 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.1 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 0.1 | GO:0002677 | negative regulation of chronic inflammatory response(GO:0002677) |
| 0.0 | 0.0 | GO:0043368 | positive T cell selection(GO:0043368) |
| 0.0 | 0.2 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.0 | 0.2 | GO:0002313 | mature B cell differentiation involved in immune response(GO:0002313) mature B cell differentiation(GO:0002335) |
| 0.0 | 0.2 | GO:0070670 | response to interleukin-4(GO:0070670) |
| 0.0 | 0.2 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.1 | GO:0042091 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.0 | 0.2 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.0 | 0.1 | GO:0000042 | protein targeting to Golgi(GO:0000042) establishment of protein localization to Golgi(GO:0072600) |
| 0.0 | 0.1 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 0.2 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.0 | GO:0070669 | response to interleukin-2(GO:0070669) |
| 0.0 | 0.1 | GO:0060413 | atrial septum development(GO:0003283) atrial septum morphogenesis(GO:0060413) |
| 0.0 | 0.1 | GO:0033262 | regulation of nuclear cell cycle DNA replication(GO:0033262) |
| 0.0 | 0.1 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 0.2 | GO:0016553 | base conversion or substitution editing(GO:0016553) |
| 0.0 | 0.1 | GO:0060437 | lung growth(GO:0060437) |
| 0.0 | 0.1 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 0.1 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) regulation of store-operated calcium channel activity(GO:1901339) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.3 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.1 | GO:0002678 | positive regulation of chronic inflammatory response(GO:0002678) |
| 0.0 | 0.0 | GO:0060557 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 0.0 | 0.1 | GO:0060676 | ureteric bud formation(GO:0060676) |
| 0.0 | 0.1 | GO:0044332 | Wnt signaling pathway involved in dorsal/ventral axis specification(GO:0044332) regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000053) |
| 0.0 | 0.1 | GO:0010536 | regulation of activation of Janus kinase activity(GO:0010533) positive regulation of activation of Janus kinase activity(GO:0010536) |
| 0.0 | 0.1 | GO:0034135 | regulation of toll-like receptor 2 signaling pathway(GO:0034135) |
| 0.0 | 0.1 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.1 | GO:0046398 | UDP-glucuronate biosynthetic process(GO:0006065) UDP-glucuronate metabolic process(GO:0046398) |
| 0.0 | 0.0 | GO:0071801 | regulation of podosome assembly(GO:0071801) positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.2 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) leukocyte adhesion to vascular endothelial cell(GO:0061756) |
| 0.0 | 0.0 | GO:0034776 | response to histamine(GO:0034776) |
| 0.0 | 0.1 | GO:0046137 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.0 | 0.3 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) |
| 0.0 | 0.1 | GO:0090322 | regulation of superoxide metabolic process(GO:0090322) |
| 0.0 | 0.2 | GO:0002360 | T cell lineage commitment(GO:0002360) |
| 0.0 | 0.1 | GO:0060594 | mammary gland specification(GO:0060594) |
| 0.0 | 0.2 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.1 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.1 | GO:0009143 | nucleoside triphosphate catabolic process(GO:0009143) |
| 0.0 | 0.1 | GO:1901985 | positive regulation of histone acetylation(GO:0035066) positive regulation of protein acetylation(GO:1901985) positive regulation of peptidyl-lysine acetylation(GO:2000758) |
| 0.0 | 0.1 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.1 | GO:0006922 | obsolete cleavage of lamin involved in execution phase of apoptosis(GO:0006922) |
| 0.0 | 0.1 | GO:0019987 | obsolete negative regulation of anti-apoptosis(GO:0019987) |
| 0.0 | 0.1 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.2 | GO:0045885 | obsolete positive regulation of survival gene product expression(GO:0045885) |
| 0.0 | 0.1 | GO:0002407 | dendritic cell chemotaxis(GO:0002407) dendritic cell migration(GO:0036336) |
| 0.0 | 0.0 | GO:0090047 | obsolete positive regulation of transcription regulator activity(GO:0090047) |
| 0.0 | 0.1 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.0 | GO:0071637 | heterotypic cell-cell adhesion(GO:0034113) regulation of heterotypic cell-cell adhesion(GO:0034114) monocyte chemotactic protein-1 production(GO:0071605) regulation of monocyte chemotactic protein-1 production(GO:0071637) |
| 0.0 | 0.2 | GO:0019059 | obsolete initiation of viral infection(GO:0019059) |
| 0.0 | 0.0 | GO:0032713 | negative regulation of interleukin-4 production(GO:0032713) |
| 0.0 | 0.1 | GO:0072531 | pyrimidine-containing compound transmembrane transport(GO:0072531) |
| 0.0 | 0.0 | GO:0042695 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.0 | 0.1 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.0 | GO:0031223 | auditory behavior(GO:0031223) |
| 0.0 | 0.1 | GO:0010829 | negative regulation of glucose transport(GO:0010829) negative regulation of glucose import(GO:0046325) |
| 0.0 | 0.1 | GO:0010614 | negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.0 | 0.1 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) negative regulation of signal transduction by p53 class mediator(GO:1901797) |
| 0.0 | 0.1 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.0 | 0.2 | GO:0006554 | lysine metabolic process(GO:0006553) lysine catabolic process(GO:0006554) |
| 0.0 | 0.0 | GO:0045341 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) |
| 0.0 | 0.2 | GO:0001782 | B cell homeostasis(GO:0001782) |
| 0.0 | 0.1 | GO:0031062 | positive regulation of histone methylation(GO:0031062) |
| 0.0 | 0.1 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.0 | 0.2 | GO:0002837 | regulation of response to tumor cell(GO:0002834) positive regulation of response to tumor cell(GO:0002836) regulation of immune response to tumor cell(GO:0002837) positive regulation of immune response to tumor cell(GO:0002839) |
| 0.0 | 0.1 | GO:0009048 | dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 0.0 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.0 | GO:0033622 | integrin activation(GO:0033622) |
| 0.0 | 0.1 | GO:0071364 | cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.0 | 0.1 | GO:0032695 | negative regulation of interleukin-12 production(GO:0032695) |
| 0.0 | 1.2 | GO:0031294 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.0 | 0.0 | GO:0001916 | regulation of T cell mediated cytotoxicity(GO:0001914) positive regulation of T cell mediated cytotoxicity(GO:0001916) |
| 0.0 | 0.0 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.2 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.0 | 0.0 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) positive regulation of lamellipodium organization(GO:1902745) |
| 0.0 | 0.1 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.1 | GO:0008216 | spermidine metabolic process(GO:0008216) |
| 0.0 | 0.1 | GO:0015801 | aromatic amino acid transport(GO:0015801) |
| 0.0 | 0.1 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.0 | 0.0 | GO:0007549 | dosage compensation(GO:0007549) |
| 0.0 | 0.1 | GO:0018119 | protein nitrosylation(GO:0017014) peptidyl-cysteine S-nitrosylation(GO:0018119) |
| 0.0 | 0.2 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 0.1 | GO:0022038 | corpus callosum development(GO:0022038) |
| 0.0 | 0.0 | GO:0030202 | heparin metabolic process(GO:0030202) |
| 0.0 | 0.1 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.0 | 0.1 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.0 | 0.1 | GO:0032897 | negative regulation of viral transcription(GO:0032897) |
| 0.0 | 0.0 | GO:0071360 | cellular response to exogenous dsRNA(GO:0071360) |
| 0.0 | 0.1 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.1 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
| 0.0 | 0.1 | GO:0003323 | type B pancreatic cell development(GO:0003323) |
| 0.0 | 0.1 | GO:0021570 | rhombomere 4 development(GO:0021570) |
| 0.0 | 0.1 | GO:0030187 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.0 | 0.1 | GO:0035511 | oxidative DNA demethylation(GO:0035511) |
| 0.0 | 0.1 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.0 | GO:0010511 | regulation of phosphatidylinositol biosynthetic process(GO:0010511) |
| 0.0 | 0.0 | GO:0072161 | mesenchymal cell differentiation involved in kidney development(GO:0072161) metanephric mesenchymal cell differentiation(GO:0072162) mesenchymal cell differentiation involved in renal system development(GO:2001012) |
| 0.0 | 0.1 | GO:0032236 | obsolete positive regulation of calcium ion transport via store-operated calcium channel activity(GO:0032236) |
| 0.0 | 0.0 | GO:0021561 | facial nerve development(GO:0021561) |
| 0.0 | 0.1 | GO:0009698 | phenylpropanoid metabolic process(GO:0009698) coumarin metabolic process(GO:0009804) |
| 0.0 | 0.1 | GO:0002507 | tolerance induction(GO:0002507) |
| 0.0 | 0.1 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 0.0 | 0.0 | GO:0002227 | innate immune response in mucosa(GO:0002227) mucosal immune response(GO:0002385) |
| 0.0 | 0.1 | GO:0090382 | phagolysosome assembly(GO:0001845) phagosome maturation(GO:0090382) |
| 0.0 | 0.1 | GO:0034616 | response to laminar fluid shear stress(GO:0034616) |
| 0.0 | 0.1 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 0.0 | GO:0016556 | mRNA modification(GO:0016556) |
| 0.0 | 0.1 | GO:0003056 | regulation of vascular smooth muscle contraction(GO:0003056) |
| 0.0 | 0.1 | GO:0000730 | DNA recombinase assembly(GO:0000730) double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.0 | 0.1 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) chloride ion homeostasis(GO:0055064) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.0 | GO:0009151 | purine deoxyribonucleotide metabolic process(GO:0009151) |
| 0.0 | 0.2 | GO:0007512 | adult heart development(GO:0007512) |
| 0.0 | 0.0 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.1 | GO:0043267 | negative regulation of potassium ion transport(GO:0043267) |
| 0.0 | 0.1 | GO:0019228 | neuronal action potential(GO:0019228) |
| 0.0 | 0.1 | GO:0070059 | intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress(GO:0070059) |
| 0.0 | 0.1 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.0 | GO:0032776 | DNA methylation on cytosine(GO:0032776) C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 0.0 | GO:0052251 | induction by organism of defense response of other organism involved in symbiotic interaction(GO:0052251) |
| 0.0 | 0.1 | GO:0090502 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) cleavage involved in rRNA processing(GO:0000469) endonucleolytic cleavage involved in rRNA processing(GO:0000478) endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000479) RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.0 | 0.0 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.0 | 0.1 | GO:0072180 | mesonephric duct morphogenesis(GO:0072180) |
| 0.0 | 0.0 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.1 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.0 | 0.0 | GO:0003161 | cardiac conduction system development(GO:0003161) |
| 0.0 | 0.0 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 0.0 | GO:0010903 | negative regulation of very-low-density lipoprotein particle remodeling(GO:0010903) |
| 0.0 | 0.1 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.0 | 0.2 | GO:0021904 | dorsal/ventral neural tube patterning(GO:0021904) |
| 0.0 | 0.0 | GO:0045906 | negative regulation of vasoconstriction(GO:0045906) |
| 0.0 | 0.0 | GO:0090400 | stress-induced premature senescence(GO:0090400) |
| 0.0 | 0.0 | GO:0006924 | activation-induced cell death of T cells(GO:0006924) |
| 0.0 | 0.0 | GO:0051132 | NK T cell proliferation(GO:0001866) NK T cell activation(GO:0051132) |
| 0.0 | 0.0 | GO:0051573 | negative regulation of histone H3-K9 methylation(GO:0051573) |
| 0.0 | 0.1 | GO:0051788 | response to misfolded protein(GO:0051788) |
| 0.0 | 0.1 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.0 | GO:0034201 | response to oleic acid(GO:0034201) |
| 0.0 | 0.1 | GO:0002483 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 0.0 | GO:0001787 | natural killer cell proliferation(GO:0001787) |
| 0.0 | 0.0 | GO:0070340 | detection of bacterial lipoprotein(GO:0042494) detection of bacterial lipopeptide(GO:0070340) |
| 0.0 | 0.0 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.1 | GO:2000344 | positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.0 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.0 | 0.0 | GO:0030889 | negative regulation of B cell proliferation(GO:0030889) |
| 0.0 | 0.2 | GO:0060334 | regulation of response to interferon-gamma(GO:0060330) regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.0 | 0.1 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.1 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.3 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.0 | GO:0048532 | anatomical structure arrangement(GO:0048532) |
| 0.0 | 0.1 | GO:0048341 | paraxial mesoderm formation(GO:0048341) |
| 0.0 | 0.2 | GO:0034199 | activation of protein kinase A activity(GO:0034199) |
| 0.0 | 0.0 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.1 | GO:0090659 | adult walking behavior(GO:0007628) walking behavior(GO:0090659) |
| 0.0 | 0.1 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.0 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.1 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.0 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 0.0 | 0.0 | GO:0002328 | pro-B cell differentiation(GO:0002328) |
| 0.0 | 0.0 | GO:0060261 | positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 0.1 | GO:0002921 | negative regulation of humoral immune response(GO:0002921) |
| 0.0 | 0.1 | GO:0008291 | acetylcholine metabolic process(GO:0008291) acetate ester metabolic process(GO:1900619) |
| 0.0 | 0.1 | GO:0015840 | urea transport(GO:0015840) |
| 0.0 | 0.0 | GO:0002093 | auditory receptor cell morphogenesis(GO:0002093) auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.0 | GO:0015959 | diadenosine polyphosphate metabolic process(GO:0015959) |
| 0.0 | 0.0 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.2 | GO:0042274 | ribosomal small subunit biogenesis(GO:0042274) |
| 0.0 | 0.0 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.0 | 0.0 | GO:0048289 | isotype switching to IgE isotypes(GO:0048289) regulation of isotype switching to IgE isotypes(GO:0048293) |
| 0.0 | 0.2 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.0 | 0.0 | GO:0072193 | ureter smooth muscle development(GO:0072191) ureter smooth muscle cell differentiation(GO:0072193) |
| 0.0 | 0.0 | GO:0010560 | positive regulation of glycoprotein biosynthetic process(GO:0010560) positive regulation of glycoprotein metabolic process(GO:1903020) |
| 0.0 | 0.0 | GO:0070120 | ciliary neurotrophic factor-mediated signaling pathway(GO:0070120) |
| 0.0 | 0.0 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 | 0.0 | GO:0034695 | response to prostaglandin E(GO:0034695) |
| 0.0 | 0.1 | GO:0070664 | negative regulation of leukocyte proliferation(GO:0070664) |
| 0.0 | 0.0 | GO:0042996 | regulation of Golgi to plasma membrane protein transport(GO:0042996) |
| 0.0 | 0.0 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.0 | 0.0 | GO:0051790 | short-chain fatty acid biosynthetic process(GO:0051790) |
| 0.0 | 0.0 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.0 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.4 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.0 | 0.0 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.1 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.0 | 0.0 | GO:0060760 | positive regulation of response to cytokine stimulus(GO:0060760) |
| 0.0 | 0.1 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.0 | 0.0 | GO:2001259 | positive regulation of calcium ion transmembrane transporter activity(GO:1901021) positive regulation of cation channel activity(GO:2001259) |
| 0.0 | 0.1 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.0 | GO:0048643 | positive regulation of skeletal muscle tissue development(GO:0048643) |
| 0.0 | 0.0 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.0 | 0.0 | GO:0014874 | response to stimulus involved in regulation of muscle adaptation(GO:0014874) |
| 0.0 | 0.0 | GO:0010759 | positive regulation of macrophage chemotaxis(GO:0010759) |
| 0.0 | 0.0 | GO:0071071 | regulation of phospholipid biosynthetic process(GO:0071071) negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 0.0 | 0.1 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 | 0.0 | GO:0060287 | epithelial cilium movement(GO:0003351) epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.2 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.0 | GO:0043374 | CD8-positive, alpha-beta T cell activation(GO:0036037) CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.0 | GO:0051533 | NFAT protein import into nucleus(GO:0051531) regulation of NFAT protein import into nucleus(GO:0051532) positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.0 | 0.1 | GO:0000303 | response to superoxide(GO:0000303) |
| 0.0 | 0.0 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.1 | GO:0006895 | Golgi to endosome transport(GO:0006895) Golgi to vacuole transport(GO:0006896) |
| 0.0 | 0.0 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.0 | 0.0 | GO:0000281 | mitotic cytokinesis(GO:0000281) |
| 0.0 | 0.1 | GO:0007263 | nitric oxide mediated signal transduction(GO:0007263) |
| 0.0 | 0.0 | GO:0070293 | renal absorption(GO:0070293) |
| 0.0 | 0.0 | GO:0032070 | regulation of deoxyribonuclease activity(GO:0032070) |
| 0.0 | 0.0 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.0 | 0.0 | GO:0046877 | regulation of saliva secretion(GO:0046877) |
| 0.0 | 0.1 | GO:0034375 | high-density lipoprotein particle remodeling(GO:0034375) |
| 0.0 | 0.0 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.0 | GO:0002691 | regulation of cellular extravasation(GO:0002691) |
| 0.0 | 0.2 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 0.0 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.0 | 0.0 | GO:0001661 | conditioned taste aversion(GO:0001661) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 0.2 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.1 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.3 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.5 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.4 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.1 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.1 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.0 | 0.2 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.1 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.0 | 0.1 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.1 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.1 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.2 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.1 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.3 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.1 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.1 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.7 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.1 | GO:0001740 | Barr body(GO:0001740) |
| 0.0 | 0.2 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.5 | GO:0005626 | obsolete insoluble fraction(GO:0005626) |
| 0.0 | 0.0 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.1 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.0 | 0.1 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.0 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.1 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.0 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.1 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.1 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.0 | 0.1 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.0 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.1 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.1 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.0 | GO:0001652 | granular component(GO:0001652) |
| 0.0 | 0.1 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.0 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.2 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.1 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.4 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.0 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.1 | GO:0030062 | mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.0 | 0.1 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:0045179 | apical cortex(GO:0045179) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.1 | 0.5 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.1 | 0.2 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.1 | 0.2 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.1 | 0.2 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 0.2 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.1 | 0.2 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.1 | 0.2 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.1 | 0.2 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.2 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.2 | GO:0015925 | galactosidase activity(GO:0015925) |
| 0.0 | 0.1 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.1 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.2 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.2 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.4 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.3 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.2 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.1 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.0 | 0.2 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.3 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.2 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.3 | GO:0034593 | phosphatidylinositol bisphosphate phosphatase activity(GO:0034593) |
| 0.0 | 0.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.2 | GO:0051766 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) inositol trisphosphate kinase activity(GO:0051766) |
| 0.0 | 0.7 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.4 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.1 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.1 | GO:0001637 | G-protein coupled chemoattractant receptor activity(GO:0001637) chemokine receptor activity(GO:0004950) |
| 0.0 | 0.0 | GO:0001098 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.0 | 0.1 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.0 | 0.1 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.1 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 1.2 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.2 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.1 | GO:0003909 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.1 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.1 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.1 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.0 | 0.2 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.1 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.3 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.1 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.1 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 0.2 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.2 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.1 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.0 | 0.1 | GO:0004476 | mannose-6-phosphate isomerase activity(GO:0004476) |
| 0.0 | 0.1 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.0 | 0.2 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) |
| 0.0 | 0.1 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.0 | 0.1 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.0 | 0.1 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.0 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.0 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.1 | GO:0043734 | DNA demethylase activity(GO:0035514) DNA-N1-methyladenine dioxygenase activity(GO:0043734) |
| 0.0 | 0.1 | GO:0046404 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.0 | 0.1 | GO:0016778 | diphosphotransferase activity(GO:0016778) |
| 0.0 | 0.1 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.1 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.1 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.0 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.0 | 0.0 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.3 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.0 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.0 | 0.1 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.0 | GO:0043175 | RNA polymerase core enzyme binding(GO:0043175) |
| 0.0 | 0.0 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.0 | 0.3 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.1 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.0 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.2 | GO:0005416 | cation:amino acid symporter activity(GO:0005416) |
| 0.0 | 0.1 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.1 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.2 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.1 | GO:0099604 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.0 | 0.0 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) xenobiotic transporter activity(GO:0042910) |
| 0.0 | 0.1 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.1 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.0 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.0 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.0 | 0.0 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 0.0 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.0 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.0 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.1 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.0 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.0 | 0.4 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.0 | GO:0031177 | phosphopantetheine binding(GO:0031177) |
| 0.0 | 0.1 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 0.0 | GO:0004844 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.0 | 0.1 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.0 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.0 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.0 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.1 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.1 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.1 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.0 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.0 | 0.3 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.2 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.0 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.0 | GO:0000832 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) |
| 0.0 | 0.1 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.0 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.0 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.1 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.0 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.0 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.0 | 0.0 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 0.0 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.0 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.0 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.0 | 0.2 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.1 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.0 | 0.0 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.1 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.0 | 0.0 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.3 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.0 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 2.2 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.4 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.2 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.1 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 1.3 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.1 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.6 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.0 | 0.1 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.5 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 0.5 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.1 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 0.1 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.1 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.4 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 0.1 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.2 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.1 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.0 | 0.1 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.0 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.1 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.0 | 0.1 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 0.4 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.3 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.2 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 0.0 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.3 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.7 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 0.6 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.1 | 0.2 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.6 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.4 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.6 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.2 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.2 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.3 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.2 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.3 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.1 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 0.2 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.0 | 0.4 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.5 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.2 | REACTOME IL 2 SIGNALING | Genes involved in Interleukin-2 signaling |
| 0.0 | 0.5 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.2 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.4 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.3 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.0 | 0.4 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.1 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.1 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.2 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.4 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.0 | 0.6 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.2 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.0 | REACTOME APC CDC20 MEDIATED DEGRADATION OF NEK2A | Genes involved in APC-Cdc20 mediated degradation of Nek2A |
| 0.0 | 0.1 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.1 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 0.2 | REACTOME COSTIMULATION BY THE CD28 FAMILY | Genes involved in Costimulation by the CD28 family |
| 0.0 | 0.1 | REACTOME ADP SIGNALLING THROUGH P2RY1 | Genes involved in ADP signalling through P2Y purinoceptor 1 |
| 0.0 | 0.2 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.3 | REACTOME POST NMDA RECEPTOR ACTIVATION EVENTS | Genes involved in Post NMDA receptor activation events |
| 0.0 | 0.1 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.0 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.2 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.0 | REACTOME ABORTIVE ELONGATION OF HIV1 TRANSCRIPT IN THE ABSENCE OF TAT | Genes involved in Abortive elongation of HIV-1 transcript in the absence of Tat |
| 0.0 | 0.0 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 0.0 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.0 | 0.1 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.1 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |