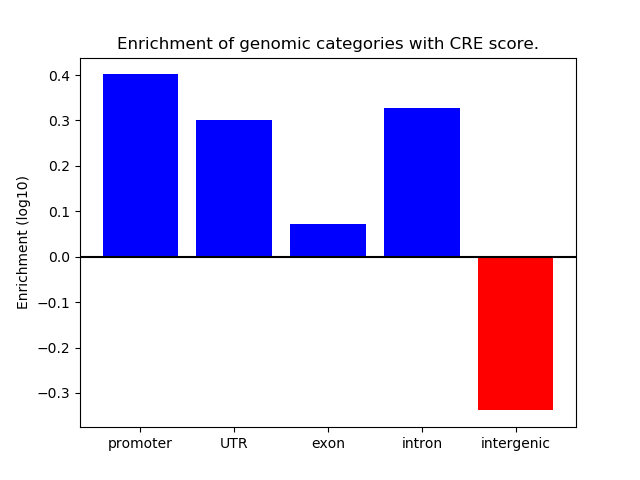
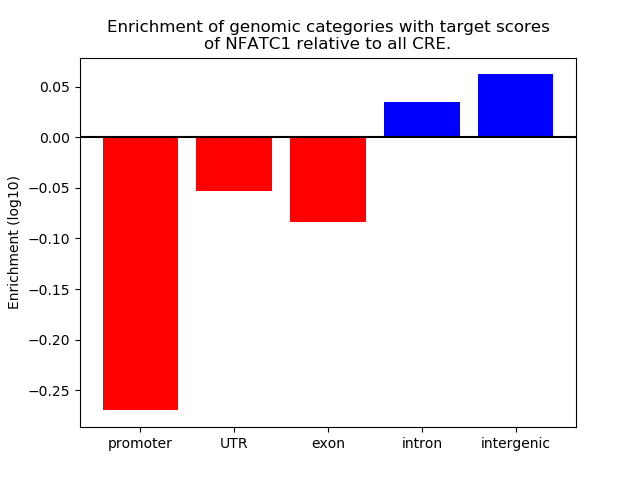
Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
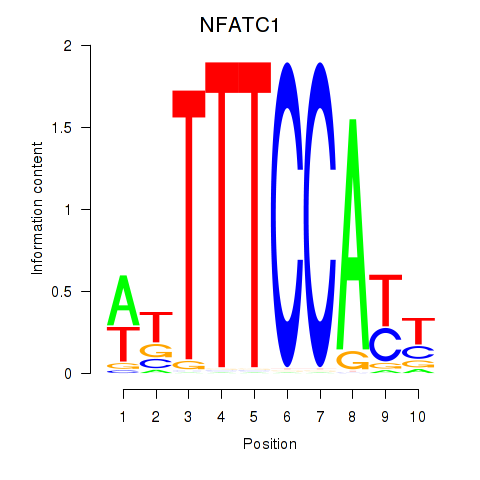
Results for NFATC1
Z-value: 1.61
Transcription factors associated with NFATC1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NFATC1
|
ENSG00000131196.13 | nuclear factor of activated T cells 1 |
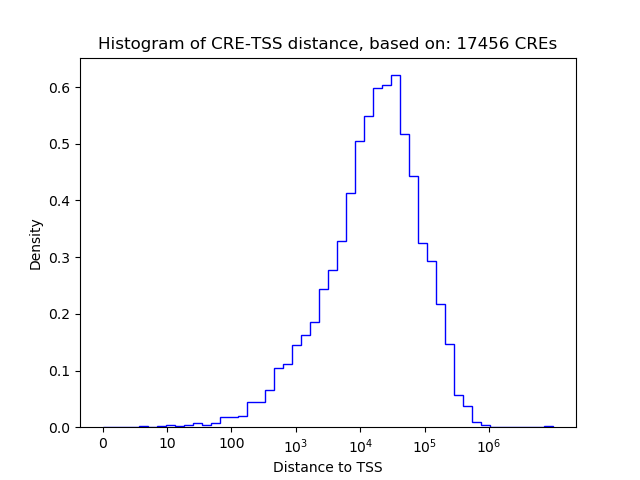
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr18_77159160_77159516 | NFATC1 | 984 | 0.631431 | -0.94 | 1.3e-04 | Click! |
| chr18_77164936_77165190 | NFATC1 | 4671 | 0.275487 | -0.88 | 1.6e-03 | Click! |
| chr18_77166940_77167091 | NFATC1 | 6623 | 0.257756 | -0.86 | 3.0e-03 | Click! |
| chr18_77154994_77155234 | NFATC1 | 742 | 0.728425 | -0.84 | 4.9e-03 | Click! |
| chr18_77165231_77165672 | NFATC1 | 5059 | 0.270449 | -0.84 | 5.1e-03 | Click! |
Activity of the NFATC1 motif across conditions
Conditions sorted by the z-value of the NFATC1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
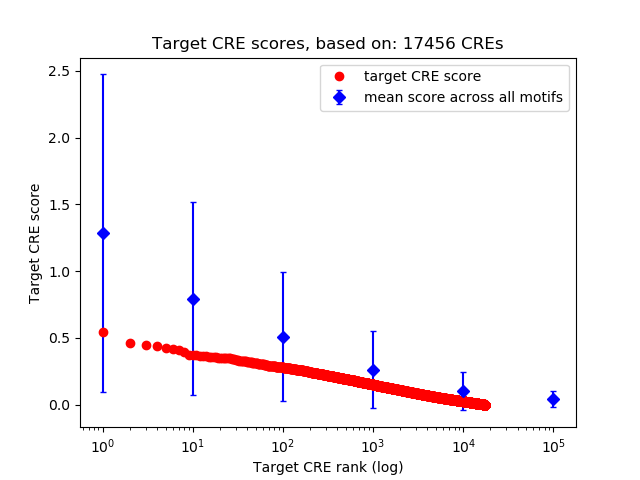
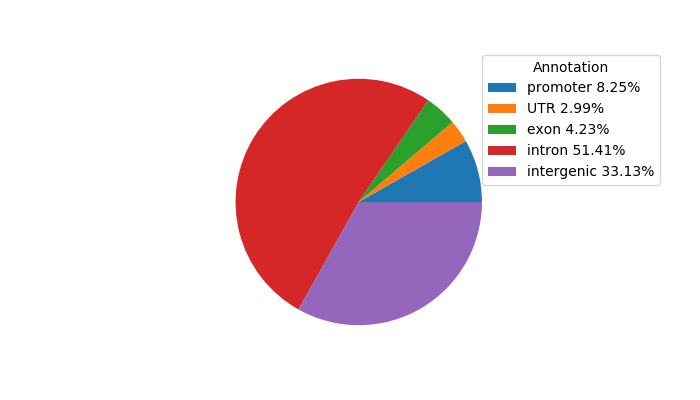
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr21_35902480_35902631 | 0.54 |
RCAN1 |
regulator of calcineurin 1 |
3294 |
0.26 |
| chr9_127104782_127105372 | 0.46 |
ENSG00000264237 |
. |
7733 |
0.2 |
| chr5_55164394_55164545 | 0.44 |
IL31RA |
interleukin 31 receptor A |
15111 |
0.19 |
| chr10_114863343_114863771 | 0.44 |
TCF7L2 |
transcription factor 7-like 2 (T-cell specific, HMG-box) |
40122 |
0.21 |
| chr11_121501091_121501242 | 0.43 |
SORL1 |
sortilin-related receptor, L(DLR class) A repeats containing |
40038 |
0.22 |
| chr20_10055682_10055833 | 0.41 |
ENSG00000264599 |
. |
18789 |
0.2 |
| chr1_6301763_6302398 | 0.41 |
HES3 |
hes family bHLH transcription factor 3 |
2172 |
0.18 |
| chr21_17944689_17944974 | 0.40 |
ENSG00000207863 |
. |
17726 |
0.2 |
| chr5_179246518_179247261 | 0.37 |
SQSTM1 |
sequestosome 1 |
870 |
0.42 |
| chr6_132511743_132511894 | 0.37 |
ENSG00000265669 |
. |
75415 |
0.11 |
| chr11_61690283_61690510 | 0.37 |
RAB3IL1 |
RAB3A interacting protein (rabin3)-like 1 |
2655 |
0.18 |
| chr14_69388884_69389440 | 0.37 |
ACTN1 |
actinin, alpha 1 |
25103 |
0.2 |
| chr11_19627998_19628149 | 0.36 |
ENSG00000207407 |
. |
15235 |
0.2 |
| chr8_141364670_141364821 | 0.36 |
TRAPPC9 |
trafficking protein particle complex 9 |
96284 |
0.08 |
| chr7_137353876_137354027 | 0.36 |
DGKI |
diacylglycerol kinase, iota |
177331 |
0.03 |
| chr7_151182024_151182303 | 0.36 |
ENSG00000241959 |
. |
20139 |
0.13 |
| chr15_33148725_33148876 | 0.35 |
FMN1 |
formin 1 |
31655 |
0.17 |
| chr2_47416519_47416670 | 0.35 |
CALM2 |
calmodulin 2 (phosphorylase kinase, delta) |
12854 |
0.21 |
| chr11_35448443_35448594 | 0.35 |
SLC1A2 |
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
6908 |
0.22 |
| chr6_82317027_82317178 | 0.35 |
ENSG00000212336 |
. |
14815 |
0.23 |
| chr6_56555787_56555938 | 0.35 |
DST |
dystonin |
48068 |
0.18 |
| chr1_219616434_219616585 | 0.35 |
ENSG00000252240 |
. |
220210 |
0.02 |
| chr2_216523684_216523835 | 0.35 |
ENSG00000212055 |
. |
219883 |
0.02 |
| chr16_15533494_15533711 | 0.35 |
C16orf45 |
chromosome 16 open reading frame 45 |
5270 |
0.21 |
| chr9_4303266_4303417 | 0.35 |
GLIS3 |
GLIS family zinc finger 3 |
3425 |
0.25 |
| chr6_148589578_148589729 | 0.35 |
SASH1 |
SAM and SH3 domain containing 1 |
3787 |
0.25 |
| chr6_128390231_128390382 | 0.34 |
PTPRK |
protein tyrosine phosphatase, receptor type, K |
4398 |
0.3 |
| chr4_71532339_71532558 | 0.34 |
IGJ |
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides |
71 |
0.96 |
| chr7_116804373_116804524 | 0.34 |
ST7-AS2 |
ST7 antisense RNA 2 |
17914 |
0.23 |
| chr1_78721233_78721384 | 0.34 |
ENSG00000237413 |
. |
26002 |
0.2 |
| chr4_157888355_157888628 | 0.33 |
PDGFC |
platelet derived growth factor C |
3564 |
0.27 |
| chr12_63155045_63155196 | 0.33 |
ENSG00000200296 |
. |
89561 |
0.08 |
| chr11_11995194_11995345 | 0.33 |
DKK3 |
dickkopf WNT signaling pathway inhibitor 3 |
34861 |
0.21 |
| chr8_108518687_108518838 | 0.33 |
ANGPT1 |
angiopoietin 1 |
8479 |
0.33 |
| chr3_149833178_149833329 | 0.33 |
RP11-167H9.4 |
|
17434 |
0.21 |
| chr6_128529618_128529908 | 0.33 |
PTPRK |
protein tyrosine phosphatase, receptor type, K |
113563 |
0.07 |
| chr18_52920557_52920708 | 0.33 |
TCF4 |
transcription factor 4 |
22270 |
0.26 |
| chr4_114304726_114304877 | 0.32 |
ANK2 |
ankyrin 2, neuronal |
10286 |
0.22 |
| chr8_32530949_32531215 | 0.32 |
NRG1 |
neuregulin 1 |
25725 |
0.27 |
| chr13_24617983_24618134 | 0.32 |
SPATA13 |
spermatogenesis associated 13 |
64114 |
0.11 |
| chr4_186578170_186578442 | 0.32 |
SORBS2 |
sorbin and SH3 domain containing 2 |
183 |
0.95 |
| chr8_120615500_120615651 | 0.32 |
ENPP2 |
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
10327 |
0.27 |
| chr5_15249746_15249921 | 0.32 |
ENSG00000202269 |
. |
138804 |
0.05 |
| chr4_79487146_79487361 | 0.32 |
ANXA3 |
annexin A3 |
12179 |
0.24 |
| chr2_69518306_69518457 | 0.32 |
ENSG00000252250 |
. |
43351 |
0.15 |
| chr13_75942001_75942152 | 0.32 |
TBC1D4-AS1 |
TBC1D4 antisense RNA 1 |
9354 |
0.24 |
| chr6_116685228_116685657 | 0.31 |
DSE |
dermatan sulfate epimerase |
5568 |
0.22 |
| chr13_74792422_74792795 | 0.31 |
ENSG00000206617 |
. |
70743 |
0.12 |
| chr11_121963341_121963492 | 0.31 |
ENSG00000207971 |
. |
7136 |
0.17 |
| chr2_177392067_177392218 | 0.31 |
ENSG00000252027 |
. |
137262 |
0.04 |
| chr5_43781026_43781177 | 0.31 |
NNT |
nicotinamide nucleotide transhydrogenase |
135642 |
0.05 |
| chr14_100240950_100241315 | 0.31 |
EML1 |
echinoderm microtubule associated protein like 1 |
1091 |
0.59 |
| chr9_79075654_79076239 | 0.31 |
GCNT1 |
glucosaminyl (N-acetyl) transferase 1, core 2 |
1800 |
0.46 |
| chr22_28625991_28626142 | 0.31 |
ENSG00000201209 |
. |
2745 |
0.37 |
| chr5_54157834_54157985 | 0.31 |
ENSG00000221073 |
. |
9133 |
0.26 |
| chr14_72142038_72142189 | 0.31 |
SIPA1L1 |
signal-induced proliferation-associated 1 like 1 |
56640 |
0.17 |
| chr6_132301065_132301216 | 0.30 |
CTGF |
connective tissue growth factor |
28627 |
0.18 |
| chr6_72121829_72121980 | 0.30 |
ENSG00000207827 |
. |
8580 |
0.23 |
| chr16_55520614_55520898 | 0.30 |
MMP2 |
matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase) |
1818 |
0.4 |
| chr1_96831244_96831395 | 0.30 |
ENSG00000200800 |
. |
139862 |
0.05 |
| chr9_103340467_103340618 | 0.30 |
MURC |
muscle-related coiled-coil protein |
181 |
0.95 |
| chr12_26430442_26430909 | 0.30 |
RP11-283G6.5 |
|
5822 |
0.22 |
| chr3_105510379_105510530 | 0.30 |
CBLB |
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
77433 |
0.13 |
| chr1_214647384_214647535 | 0.30 |
PTPN14 |
protein tyrosine phosphatase, non-receptor type 14 |
9313 |
0.29 |
| chr11_112112150_112112301 | 0.30 |
ENSG00000215954 |
. |
6328 |
0.12 |
| chr11_77313091_77313242 | 0.30 |
ENSG00000271767 |
. |
6942 |
0.16 |
| chr3_124827683_124827880 | 0.30 |
ENSG00000199327 |
. |
10263 |
0.17 |
| chr6_143670247_143670398 | 0.29 |
AL031320.1 |
|
89253 |
0.07 |
| chr9_21656575_21656726 | 0.29 |
ENSG00000244230 |
. |
42663 |
0.15 |
| chr11_102009263_102009414 | 0.29 |
YAP1 |
Yes-associated protein 1 |
26093 |
0.15 |
| chr2_223920021_223920251 | 0.29 |
KCNE4 |
potassium voltage-gated channel, Isk-related family, member 4 |
3274 |
0.35 |
| chr10_19026308_19026459 | 0.29 |
ARL5B |
ADP-ribosylation factor-like 5B |
78049 |
0.1 |
| chr20_6685250_6685401 | 0.29 |
BMP2 |
bone morphogenetic protein 2 |
62986 |
0.15 |
| chr2_9517769_9518003 | 0.29 |
ITGB1BP1 |
integrin beta 1 binding protein 1 |
42537 |
0.15 |
| chr12_107368695_107368994 | 0.29 |
C12orf23 |
chromosome 12 open reading frame 23 |
8131 |
0.17 |
| chr4_77725622_77725773 | 0.29 |
RP11-359D14.3 |
|
2580 |
0.3 |
| chr18_56247923_56248074 | 0.29 |
ENSG00000252284 |
. |
19865 |
0.14 |
| chr8_21766211_21767399 | 0.29 |
DOK2 |
docking protein 2, 56kDa |
4369 |
0.22 |
| chr5_71747550_71747979 | 0.29 |
RP11-389C8.2 |
|
9550 |
0.25 |
| chr8_131245327_131245615 | 0.29 |
ASAP1 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1 |
13817 |
0.26 |
| chr9_18479738_18479998 | 0.29 |
ADAMTSL1 |
ADAMTS-like 1 |
5637 |
0.32 |
| chr9_113639802_113639953 | 0.29 |
ENSG00000206923 |
. |
25740 |
0.21 |
| chr15_101711256_101711683 | 0.29 |
CHSY1 |
chondroitin sulfate synthase 1 |
80668 |
0.08 |
| chr10_17220959_17221110 | 0.28 |
TRDMT1 |
tRNA aspartic acid methyltransferase 1 |
4375 |
0.22 |
| chr5_158173853_158174004 | 0.28 |
CTD-2363C16.1 |
|
236086 |
0.02 |
| chr5_149519603_149519837 | 0.28 |
PDGFRB |
platelet-derived growth factor receptor, beta polypeptide |
2720 |
0.21 |
| chr2_109857650_109857801 | 0.28 |
ENSG00000265965 |
. |
72356 |
0.11 |
| chr5_39524488_39524639 | 0.28 |
DAB2 |
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
62161 |
0.15 |
| chr14_51228839_51228990 | 0.28 |
NIN |
ninein (GSK3B interacting protein) |
4599 |
0.2 |
| chr3_132324789_132324940 | 0.28 |
ACKR4 |
atypical chemokine receptor 4 |
8783 |
0.22 |
| chr2_100591017_100591168 | 0.28 |
AFF3 |
AF4/FMR2 family, member 3 |
129900 |
0.05 |
| chr11_73990200_73990857 | 0.28 |
ENSG00000206913 |
. |
26931 |
0.14 |
| chr22_40705760_40705911 | 0.28 |
ADSL |
adenylosuccinate lyase |
36672 |
0.14 |
| chr18_32382440_32382591 | 0.28 |
DTNA |
dystrobrevin, alpha |
15411 |
0.2 |
| chr1_175946929_175947183 | 0.28 |
ENSG00000252906 |
. |
9380 |
0.26 |
| chr4_40889441_40889724 | 0.28 |
APBB2 |
amyloid beta (A4) precursor protein-binding, family B, member 2 |
2860 |
0.27 |
| chr4_54717943_54718094 | 0.28 |
RP11-89B16.1 |
|
48020 |
0.17 |
| chr5_64487632_64487783 | 0.28 |
ENSG00000207439 |
. |
68511 |
0.14 |
| chr1_173310923_173311074 | 0.28 |
ENSG00000251817 |
. |
60782 |
0.13 |
| chr8_49431262_49431413 | 0.28 |
RP11-770E5.1 |
|
32790 |
0.24 |
| chr8_8537594_8537745 | 0.28 |
CLDN23 |
claudin 23 |
21779 |
0.21 |
| chr1_56181953_56182104 | 0.28 |
PIGQP1 |
phosphatidylinositol glycan anchor biosynthesis, class Q pseudogene 1 |
222925 |
0.02 |
| chr6_107978488_107978941 | 0.28 |
SCML4 |
sex comb on midleg-like 4 (Drosophila) |
74885 |
0.11 |
| chr10_15528350_15528501 | 0.28 |
FAM171A1 |
family with sequence similarity 171, member A1 |
115364 |
0.07 |
| chrX_13269027_13269605 | 0.28 |
GS1-600G8.3 |
|
59455 |
0.14 |
| chr7_137521422_137521598 | 0.28 |
DGKI |
diacylglycerol kinase, iota |
9772 |
0.27 |
| chr10_657520_657671 | 0.28 |
RP11-809C18.3 |
|
16983 |
0.15 |
| chr9_97703502_97703653 | 0.28 |
RP11-54O15.3 |
|
6349 |
0.2 |
| chr6_19698381_19698532 | 0.28 |
ENSG00000200957 |
. |
55696 |
0.16 |
| chr11_95838965_95839116 | 0.28 |
MTMR2 |
myotubularin related protein 2 |
181581 |
0.03 |
| chr2_36666145_36666296 | 0.28 |
CRIM1 |
cysteine rich transmembrane BMP regulator 1 (chordin-like) |
74613 |
0.1 |
| chr17_25642061_25642212 | 0.27 |
WSB1 |
WD repeat and SOCS box containing 1 |
20764 |
0.2 |
| chr11_123059894_123060045 | 0.27 |
CTD-2216M2.1 |
|
459 |
0.83 |
| chr3_148898157_148898308 | 0.27 |
CP |
ceruloplasmin (ferroxidase) |
824 |
0.65 |
| chr7_13782821_13782972 | 0.27 |
ETV1 |
ets variant 1 |
243170 |
0.02 |
| chr17_32580574_32580862 | 0.27 |
CCL2 |
chemokine (C-C motif) ligand 2 |
1586 |
0.24 |
| chr22_47426182_47426436 | 0.27 |
ENSG00000221672 |
. |
182506 |
0.03 |
| chr7_15841281_15841432 | 0.27 |
MEOX2 |
mesenchyme homeobox 2 |
114919 |
0.07 |
| chr7_90346858_90347009 | 0.27 |
CDK14 |
cyclin-dependent kinase 14 |
7757 |
0.3 |
| chr3_55191596_55191747 | 0.27 |
LRTM1 |
leucine-rich repeats and transmembrane domains 1 |
190556 |
0.03 |
| chr2_120996591_120996972 | 0.27 |
RALB |
v-ral simian leukemia viral oncogene homolog B |
859 |
0.59 |
| chr5_76463438_76463589 | 0.27 |
ZBED3-AS1 |
ZBED3 antisense RNA 1 |
28439 |
0.15 |
| chr5_169134550_169134701 | 0.27 |
DOCK2 |
dedicator of cytokinesis 2 |
3672 |
0.33 |
| chr10_74496065_74496270 | 0.27 |
ENSG00000266719 |
. |
15380 |
0.16 |
| chr2_237928848_237928999 | 0.27 |
ENSG00000202341 |
. |
4975 |
0.25 |
| chr13_76359048_76359199 | 0.27 |
LMO7 |
LIM domain 7 |
3851 |
0.31 |
| chr13_32914724_32915103 | 0.27 |
BRCA2 |
breast cancer 2, early onset |
25271 |
0.15 |
| chr12_26150271_26150549 | 0.27 |
RASSF8 |
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8 |
2473 |
0.34 |
| chr6_100715988_100716139 | 0.27 |
RP1-121G13.2 |
|
158931 |
0.04 |
| chr11_94526714_94526894 | 0.27 |
AMOTL1 |
angiomotin like 1 |
25267 |
0.2 |
| chr16_87891158_87892093 | 0.27 |
SLC7A5 |
solute carrier family 7 (amino acid transporter light chain, L system), member 5 |
5620 |
0.19 |
| chr22_47149537_47150247 | 0.27 |
CTA-29F11.1 |
|
8568 |
0.18 |
| chr2_161232105_161232912 | 0.27 |
ENSG00000252465 |
. |
20971 |
0.19 |
| chr5_71441214_71441365 | 0.27 |
ENSG00000264099 |
. |
24005 |
0.21 |
| chr15_45381136_45381287 | 0.27 |
RP11-109D20.2 |
|
14965 |
0.09 |
| chr2_151412177_151412328 | 0.27 |
RND3 |
Rho family GTPase 3 |
16727 |
0.31 |
| chr7_1499695_1500152 | 0.27 |
AC102953.4 |
|
350 |
0.72 |
| chr4_150087781_150087932 | 0.26 |
ENSG00000239168 |
. |
175071 |
0.04 |
| chr10_60838878_60839029 | 0.26 |
PHYHIPL |
phytanoyl-CoA 2-hydroxylase interacting protein-like |
97397 |
0.09 |
| chr2_158029538_158029689 | 0.26 |
GALNT5 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 5 (GalNAc-T5) |
84497 |
0.1 |
| chr2_191910102_191910403 | 0.26 |
ENSG00000231858 |
. |
24000 |
0.15 |
| chr5_39419337_39419488 | 0.26 |
DAB2 |
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
5558 |
0.3 |
| chr7_93954349_93954500 | 0.26 |
COL1A2 |
collagen, type I, alpha 2 |
69449 |
0.13 |
| chr4_7905515_7905666 | 0.26 |
AFAP1 |
actin filament associated protein 1 |
31550 |
0.17 |
| chr1_78769110_78769410 | 0.26 |
PTGFR |
prostaglandin F receptor (FP) |
308 |
0.93 |
| chr8_131244504_131244746 | 0.26 |
ASAP1 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1 |
14663 |
0.26 |
| chr12_115372372_115372523 | 0.26 |
ENSG00000252459 |
. |
196938 |
0.03 |
| chr3_124764370_124764521 | 0.26 |
HEG1 |
heart development protein with EGF-like domains 1 |
10357 |
0.19 |
| chr16_20891936_20892087 | 0.26 |
DCUN1D3 |
DCN1, defective in cullin neddylation 1, domain containing 3 |
11709 |
0.15 |
| chr2_85484950_85485101 | 0.26 |
ENSG00000221579 |
. |
41132 |
0.09 |
| chr17_41016311_41016609 | 0.26 |
AOC4P |
amine oxidase, copper containing 4, pseudogene |
2836 |
0.12 |
| chr1_235053615_235054079 | 0.26 |
ENSG00000239690 |
. |
13914 |
0.26 |
| chr1_66267521_66267672 | 0.26 |
PDE4B |
phosphodiesterase 4B, cAMP-specific |
8732 |
0.26 |
| chr18_21110127_21110312 | 0.26 |
NPC1 |
Niemann-Pick disease, type C1 |
3185 |
0.23 |
| chr2_208121057_208121376 | 0.26 |
AC007879.5 |
|
2240 |
0.34 |
| chr15_100051927_100052783 | 0.26 |
AC015660.1 |
HCG1993240; Serologically defined breast cancer antigen NY-BR-40; Uncharacterized protein |
13777 |
0.21 |
| chr9_73204265_73204416 | 0.26 |
ENSG00000272232 |
. |
9963 |
0.28 |
| chr16_9212818_9212969 | 0.26 |
RP11-473I1.10 |
|
11795 |
0.16 |
| chr16_84858002_84858153 | 0.26 |
RP11-254F19.2 |
|
3792 |
0.21 |
| chr16_66150653_66150937 | 0.26 |
ENSG00000201999 |
. |
184917 |
0.03 |
| chr2_45581409_45581560 | 0.26 |
SRBD1 |
S1 RNA binding domain 1 |
213670 |
0.02 |
| chr1_236827406_236827557 | 0.26 |
ACTN2 |
actinin, alpha 2 |
22273 |
0.21 |
| chr1_179322658_179322809 | 0.26 |
ENSG00000263633 |
. |
10715 |
0.19 |
| chr7_100067086_100067551 | 0.26 |
TSC22D4 |
TSC22 domain family, member 4 |
1619 |
0.17 |
| chr1_26113882_26114033 | 0.26 |
SEPN1 |
selenoprotein N, 1 |
12710 |
0.11 |
| chr22_43000472_43000822 | 0.26 |
POLDIP3 |
polymerase (DNA-directed), delta interacting protein 3 |
1675 |
0.23 |
| chr5_138164777_138164928 | 0.25 |
CTNNA1 |
catenin (cadherin-associated protein), alpha 1, 102kDa |
44875 |
0.12 |
| chr8_121275153_121275304 | 0.25 |
COL14A1 |
collagen, type XIV, alpha 1 |
66073 |
0.14 |
| chr2_218723877_218724028 | 0.25 |
TNS1 |
tensin 1 |
17069 |
0.25 |
| chr5_79887117_79887268 | 0.25 |
ANKRD34B |
ankyrin repeat domain 34B |
20885 |
0.16 |
| chr1_85809222_85809373 | 0.25 |
ENSG00000264380 |
. |
59004 |
0.09 |
| chr2_163053003_163053154 | 0.25 |
AC007750.5 |
|
36750 |
0.15 |
| chr5_111084347_111084498 | 0.25 |
NREP |
neuronal regeneration related protein |
7526 |
0.21 |
| chr11_130272299_130272590 | 0.25 |
ADAMTS8 |
ADAM metallopeptidase with thrombospondin type 1 motif, 8 |
26444 |
0.2 |
| chr4_72061057_72061208 | 0.25 |
SLC4A4 |
solute carrier family 4 (sodium bicarbonate cotransporter), member 4 |
8088 |
0.31 |
| chr13_30426801_30426952 | 0.25 |
UBL3 |
ubiquitin-like 3 |
2055 |
0.46 |
| chr6_85257015_85257166 | 0.25 |
RP11-132M7.3 |
|
142053 |
0.05 |
| chr6_75853592_75853743 | 0.25 |
COL12A1 |
collagen, type XII, alpha 1 |
8238 |
0.28 |
| chr15_32815666_32815817 | 0.25 |
AC135983.2 |
Protein LOC100996413 |
40919 |
0.1 |
| chr8_119757469_119757620 | 0.25 |
SAMD12 |
sterile alpha motif domain containing 12 |
123310 |
0.06 |
| chr3_73119570_73119721 | 0.25 |
PPP4R2 |
protein phosphatase 4, regulatory subunit 2 |
6480 |
0.15 |
| chr12_65926260_65926711 | 0.25 |
MSRB3 |
methionine sulfoxide reductase B3 |
205830 |
0.02 |
| chr12_116966140_116966291 | 0.25 |
MAP1LC3B2 |
microtubule-associated protein 1 light chain 3 beta 2 |
30971 |
0.22 |
| chr3_196870510_196870661 | 0.25 |
DLG1 |
discs, large homolog 1 (Drosophila) |
39920 |
0.16 |
| chr3_177186614_177186765 | 0.25 |
ENSG00000252028 |
. |
34651 |
0.24 |
| chr13_45478155_45478306 | 0.25 |
NUFIP1 |
nuclear fragile X mental retardation protein interacting protein 1 |
85388 |
0.08 |
| chr1_100056261_100056412 | 0.25 |
PALMD |
palmdelphin |
55163 |
0.15 |
| chr4_26022444_26022595 | 0.25 |
SMIM20 |
small integral membrane protein 20 |
106588 |
0.07 |
| chr3_187989530_187989681 | 0.25 |
LPP |
LIM domain containing preferred translocation partner in lipoma |
31953 |
0.22 |
| chr6_131360559_131360710 | 0.25 |
EPB41L2 |
erythrocyte membrane protein band 4.1-like 2 |
23082 |
0.26 |
| chr16_70736623_70736774 | 0.25 |
VAC14 |
Vac14 homolog (S. cerevisiae) |
2550 |
0.22 |
| chr3_72537234_72537495 | 0.25 |
RYBP |
RING1 and YY1 binding protein |
41295 |
0.18 |
| chr6_125583374_125583525 | 0.25 |
HDDC2 |
HD domain containing 2 |
39634 |
0.18 |
| chr15_48995639_48995790 | 0.25 |
FBN1 |
fibrillin 1 |
57668 |
0.12 |
| chr15_69596184_69596335 | 0.25 |
PAQR5 |
progestin and adipoQ receptor family member V |
4968 |
0.14 |
| chr15_67289212_67289363 | 0.25 |
SMAD3 |
SMAD family member 3 |
66814 |
0.13 |
| chr11_76050281_76050680 | 0.25 |
PRKRIR |
protein-kinase, interferon-inducible double stranded RNA dependent inhibitor, repressor of (P58 repressor) |
41506 |
0.11 |
| chr17_59295493_59295787 | 0.25 |
RP11-136H19.1 |
|
82315 |
0.08 |
| chr10_89300723_89300874 | 0.25 |
ENSG00000222192 |
. |
22145 |
0.19 |
| chr11_118789955_118791011 | 0.24 |
BCL9L |
B-cell CLL/lymphoma 9-like |
870 |
0.37 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.1 | 0.6 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.1 | 0.1 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.1 | 0.5 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) regulation of dendritic spine morphogenesis(GO:0061001) dendritic spine organization(GO:0097061) |
| 0.1 | 0.1 | GO:0043438 | acetoacetic acid metabolic process(GO:0043438) |
| 0.1 | 0.1 | GO:0010837 | regulation of keratinocyte proliferation(GO:0010837) |
| 0.1 | 0.3 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.1 | 0.3 | GO:0043129 | surfactant homeostasis(GO:0043129) chemical homeostasis within a tissue(GO:0048875) |
| 0.1 | 0.2 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.1 | 0.2 | GO:0071692 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.1 | 0.1 | GO:0072677 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.1 | 0.2 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.1 | 0.2 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.1 | 0.1 | GO:0061526 | acetylcholine secretion, neurotransmission(GO:0014055) regulation of acetylcholine secretion, neurotransmission(GO:0014056) acetylcholine secretion(GO:0061526) |
| 0.1 | 0.2 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.1 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 0.2 | GO:0060083 | smooth muscle contraction involved in micturition(GO:0060083) |
| 0.1 | 0.5 | GO:0007567 | parturition(GO:0007567) |
| 0.1 | 0.2 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.1 | 0.1 | GO:0060129 | thyroid-stimulating hormone-secreting cell differentiation(GO:0060129) |
| 0.1 | 0.2 | GO:0007132 | meiotic metaphase I(GO:0007132) |
| 0.0 | 0.1 | GO:0032353 | negative regulation of hormone metabolic process(GO:0032351) negative regulation of hormone biosynthetic process(GO:0032353) |
| 0.0 | 0.1 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.2 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.1 | GO:0051284 | negative regulation of release of sequestered calcium ion into cytosol(GO:0051280) positive regulation of sequestering of calcium ion(GO:0051284) negative regulation of calcium ion transmembrane transport(GO:1903170) |
| 0.0 | 0.1 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.2 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.0 | GO:0032344 | regulation of ketone biosynthetic process(GO:0010566) regulation of aldosterone metabolic process(GO:0032344) regulation of aldosterone biosynthetic process(GO:0032347) |
| 0.0 | 0.1 | GO:0060831 | smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:0060831) |
| 0.0 | 0.2 | GO:0046882 | negative regulation of follicle-stimulating hormone secretion(GO:0046882) |
| 0.0 | 0.2 | GO:0010799 | regulation of peptidyl-threonine phosphorylation(GO:0010799) positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.0 | 0.2 | GO:0001554 | luteolysis(GO:0001554) |
| 0.0 | 0.0 | GO:0032277 | negative regulation of gonadotropin secretion(GO:0032277) |
| 0.0 | 0.4 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.2 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.1 | GO:2000178 | negative regulation of neuroblast proliferation(GO:0007406) negative regulation of neural precursor cell proliferation(GO:2000178) |
| 0.0 | 0.1 | GO:0051834 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.0 | 0.2 | GO:0015840 | urea transport(GO:0015840) |
| 0.0 | 0.1 | GO:0033603 | positive regulation of dopamine secretion(GO:0033603) |
| 0.0 | 0.1 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.0 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.1 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.2 | GO:2000095 | regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.0 | 0.2 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.3 | GO:0006978 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) |
| 0.0 | 0.1 | GO:0002248 | connective tissue replacement involved in inflammatory response wound healing(GO:0002248) connective tissue replacement(GO:0097709) |
| 0.0 | 0.1 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.1 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.1 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.0 | 0.1 | GO:0060751 | branch elongation involved in mammary gland duct branching(GO:0060751) |
| 0.0 | 0.1 | GO:0060317 | cardiac epithelial to mesenchymal transition(GO:0060317) |
| 0.0 | 0.1 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 0.2 | GO:0002634 | regulation of germinal center formation(GO:0002634) |
| 0.0 | 0.2 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.1 | GO:1903053 | regulation of extracellular matrix disassembly(GO:0010715) negative regulation of extracellular matrix disassembly(GO:0010716) regulation of extracellular matrix organization(GO:1903053) negative regulation of extracellular matrix organization(GO:1903054) |
| 0.0 | 0.1 | GO:0009169 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.0 | 0.1 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.1 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.0 | 0.1 | GO:0060638 | mesenchymal-epithelial cell signaling(GO:0060638) |
| 0.0 | 0.1 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.2 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.0 | 0.1 | GO:0048319 | axial mesoderm morphogenesis(GO:0048319) |
| 0.0 | 0.0 | GO:0060510 | lung saccule development(GO:0060430) Type II pneumocyte differentiation(GO:0060510) |
| 0.0 | 0.1 | GO:0045650 | negative regulation of macrophage differentiation(GO:0045650) |
| 0.0 | 0.1 | GO:0019934 | cGMP-mediated signaling(GO:0019934) |
| 0.0 | 0.1 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.0 | 0.1 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.1 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.6 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.2 | GO:0045075 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.0 | 0.1 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.0 | 0.1 | GO:0061081 | positive regulation of myeloid leukocyte cytokine production involved in immune response(GO:0061081) |
| 0.0 | 0.2 | GO:0045749 | obsolete negative regulation of S phase of mitotic cell cycle(GO:0045749) |
| 0.0 | 0.1 | GO:0001766 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 0.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.1 | GO:0060900 | optic cup morphogenesis involved in camera-type eye development(GO:0002072) embryonic camera-type eye formation(GO:0060900) |
| 0.0 | 0.1 | GO:0016103 | diterpenoid catabolic process(GO:0016103) terpenoid catabolic process(GO:0016115) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.0 | GO:0006824 | cobalt ion transport(GO:0006824) |
| 0.0 | 0.1 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.0 | 0.6 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.1 | GO:0032506 | cytokinetic process(GO:0032506) |
| 0.0 | 0.1 | GO:0090273 | regulation of somatostatin secretion(GO:0090273) |
| 0.0 | 0.1 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.1 | GO:0070672 | response to interleukin-15(GO:0070672) |
| 0.0 | 0.0 | GO:0060242 | contact inhibition(GO:0060242) |
| 0.0 | 0.1 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.1 | GO:0009151 | purine deoxyribonucleotide metabolic process(GO:0009151) |
| 0.0 | 0.1 | GO:0031641 | regulation of myelination(GO:0031641) |
| 0.0 | 0.1 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 0.0 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.0 | 0.1 | GO:0051124 | synaptic growth at neuromuscular junction(GO:0051124) |
| 0.0 | 0.1 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.1 | GO:0070874 | negative regulation of glycogen metabolic process(GO:0070874) |
| 0.0 | 0.0 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.0 | 0.1 | GO:0032415 | regulation of sodium:proton antiporter activity(GO:0032415) regulation of sodium ion transmembrane transport(GO:1902305) regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.0 | 0.2 | GO:0034384 | high-density lipoprotein particle clearance(GO:0034384) |
| 0.0 | 0.2 | GO:0043030 | regulation of macrophage activation(GO:0043030) |
| 0.0 | 0.0 | GO:0010803 | regulation of tumor necrosis factor-mediated signaling pathway(GO:0010803) |
| 0.0 | 0.0 | GO:0055089 | fatty acid homeostasis(GO:0055089) |
| 0.0 | 0.0 | GO:0003308 | regulation of Wnt signaling pathway involved in heart development(GO:0003307) negative regulation of Wnt signaling pathway involved in heart development(GO:0003308) regulation of heart induction by regulation of canonical Wnt signaling pathway(GO:0090081) |
| 0.0 | 0.1 | GO:0042487 | regulation of odontogenesis of dentin-containing tooth(GO:0042487) |
| 0.0 | 0.1 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.0 | 0.1 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.1 | GO:0006930 | substrate-dependent cell migration, cell extension(GO:0006930) |
| 0.0 | 0.1 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.0 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.1 | GO:0006734 | NADH metabolic process(GO:0006734) |
| 0.0 | 0.0 | GO:0046639 | negative regulation of alpha-beta T cell differentiation(GO:0046639) |
| 0.0 | 0.0 | GO:0060664 | epithelial cell proliferation involved in salivary gland morphogenesis(GO:0060664) |
| 0.0 | 0.2 | GO:0046653 | tetrahydrofolate metabolic process(GO:0046653) |
| 0.0 | 0.2 | GO:0007520 | syncytium formation by plasma membrane fusion(GO:0000768) myoblast fusion(GO:0007520) |
| 0.0 | 0.1 | GO:0048311 | mitochondrion distribution(GO:0048311) |
| 0.0 | 0.1 | GO:0046607 | positive regulation of centrosome cycle(GO:0046607) |
| 0.0 | 0.0 | GO:0061318 | renal filtration cell differentiation(GO:0061318) glomerular epithelium development(GO:0072010) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.0 | 0.2 | GO:0048844 | artery morphogenesis(GO:0048844) |
| 0.0 | 0.1 | GO:0031394 | regulation of prostaglandin biosynthetic process(GO:0031392) positive regulation of prostaglandin biosynthetic process(GO:0031394) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.0 | 0.0 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.0 | 0.0 | GO:0009415 | response to water deprivation(GO:0009414) response to water(GO:0009415) |
| 0.0 | 0.1 | GO:0032825 | regulation of natural killer cell differentiation(GO:0032823) positive regulation of natural killer cell differentiation(GO:0032825) |
| 0.0 | 0.1 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.0 | 0.0 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
| 0.0 | 0.1 | GO:0002283 | neutrophil activation involved in immune response(GO:0002283) neutrophil degranulation(GO:0043312) |
| 0.0 | 0.1 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.0 | GO:0072193 | ureter smooth muscle development(GO:0072191) ureter smooth muscle cell differentiation(GO:0072193) |
| 0.0 | 0.3 | GO:0021846 | cell proliferation in forebrain(GO:0021846) |
| 0.0 | 0.1 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.0 | 0.0 | GO:0030202 | heparin metabolic process(GO:0030202) |
| 0.0 | 0.1 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.1 | GO:0000019 | regulation of mitotic recombination(GO:0000019) |
| 0.0 | 0.1 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.0 | 0.0 | GO:0060684 | epithelial-mesenchymal cell signaling(GO:0060684) |
| 0.0 | 0.1 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.1 | GO:0034393 | positive regulation of muscle cell apoptotic process(GO:0010661) positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.0 | 0.3 | GO:0048008 | platelet-derived growth factor receptor signaling pathway(GO:0048008) |
| 0.0 | 0.0 | GO:0021798 | forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.0 | 0.1 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.0 | 0.0 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) |
| 0.0 | 0.0 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.0 | 0.1 | GO:0071731 | response to nitric oxide(GO:0071731) |
| 0.0 | 0.3 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 0.0 | GO:0051541 | elastin metabolic process(GO:0051541) |
| 0.0 | 0.1 | GO:0001547 | antral ovarian follicle growth(GO:0001547) |
| 0.0 | 0.0 | GO:0001553 | luteinization(GO:0001553) |
| 0.0 | 0.0 | GO:0008215 | spermine metabolic process(GO:0008215) |
| 0.0 | 0.0 | GO:0048631 | regulation of skeletal muscle tissue growth(GO:0048631) |
| 0.0 | 0.0 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.0 | 0.1 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.0 | 0.0 | GO:0015791 | polyol transport(GO:0015791) |
| 0.0 | 0.1 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 0.0 | GO:1904251 | regulation of bile acid biosynthetic process(GO:0070857) regulation of bile acid metabolic process(GO:1904251) |
| 0.0 | 0.1 | GO:0032410 | negative regulation of transporter activity(GO:0032410) |
| 0.0 | 0.0 | GO:0060435 | bronchiole development(GO:0060435) |
| 0.0 | 0.1 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.0 | 0.0 | GO:0003078 | obsolete regulation of natriuresis(GO:0003078) |
| 0.0 | 0.0 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.0 | 0.1 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.1 | GO:0060390 | regulation of SMAD protein import into nucleus(GO:0060390) |
| 0.0 | 0.0 | GO:0009200 | deoxyribonucleoside triphosphate metabolic process(GO:0009200) |
| 0.0 | 0.0 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.0 | 0.0 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 0.0 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.0 | 0.1 | GO:0021930 | cell proliferation in hindbrain(GO:0021534) cell proliferation in external granule layer(GO:0021924) cerebellar granule cell precursor proliferation(GO:0021930) |
| 0.0 | 0.0 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.1 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.1 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 0.0 | GO:0045992 | negative regulation of embryonic development(GO:0045992) |
| 0.0 | 0.0 | GO:0046877 | regulation of saliva secretion(GO:0046877) |
| 0.0 | 0.1 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.1 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 0.0 | GO:0008300 | isoprenoid catabolic process(GO:0008300) |
| 0.0 | 0.0 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 0.0 | 0.1 | GO:0007350 | blastoderm segmentation(GO:0007350) |
| 0.0 | 0.0 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.0 | GO:0021978 | telencephalon regionalization(GO:0021978) |
| 0.0 | 0.1 | GO:0044253 | positive regulation of collagen metabolic process(GO:0010714) positive regulation of collagen biosynthetic process(GO:0032967) positive regulation of multicellular organismal metabolic process(GO:0044253) |
| 0.0 | 0.0 | GO:0007501 | mesodermal cell fate specification(GO:0007501) |
| 0.0 | 0.1 | GO:0009629 | response to gravity(GO:0009629) |
| 0.0 | 0.0 | GO:0002023 | reduction of food intake in response to dietary excess(GO:0002023) |
| 0.0 | 0.1 | GO:0046498 | S-adenosylhomocysteine metabolic process(GO:0046498) |
| 0.0 | 0.0 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.1 | GO:0065005 | protein-lipid complex assembly(GO:0065005) |
| 0.0 | 0.1 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.0 | GO:0033023 | mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) |
| 0.0 | 0.0 | GO:0060157 | urinary bladder development(GO:0060157) |
| 0.0 | 0.0 | GO:0046325 | negative regulation of glucose import(GO:0046325) |
| 0.0 | 0.0 | GO:0003032 | detection of oxygen(GO:0003032) |
| 0.0 | 0.0 | GO:0000492 | small nucleolar ribonucleoprotein complex assembly(GO:0000491) box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.1 | GO:0042364 | water-soluble vitamin biosynthetic process(GO:0042364) |
| 0.0 | 0.1 | GO:0046835 | carbohydrate phosphorylation(GO:0046835) |
| 0.0 | 0.2 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.0 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.0 | GO:0034201 | response to oleic acid(GO:0034201) |
| 0.0 | 0.2 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.0 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.0 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.0 | 0.0 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) |
| 0.0 | 0.0 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.0 | GO:0014834 | skeletal muscle satellite cell maintenance involved in skeletal muscle regeneration(GO:0014834) |
| 0.0 | 0.0 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.0 | 0.0 | GO:0070099 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) substrate-dependent cerebral cortex tangential migration(GO:0021825) postnatal olfactory bulb interneuron migration(GO:0021827) Roundabout signaling pathway(GO:0035385) regulation of chemokine-mediated signaling pathway(GO:0070099) negative regulation of chemokine-mediated signaling pathway(GO:0070100) |
| 0.0 | 0.0 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.0 | GO:0032222 | regulation of synaptic transmission, cholinergic(GO:0032222) |
| 0.0 | 0.1 | GO:0032332 | positive regulation of chondrocyte differentiation(GO:0032332) |
| 0.0 | 0.0 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.0 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.1 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.0 | 0.0 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.0 | GO:0003099 | positive regulation of the force of heart contraction by chemical signal(GO:0003099) |
| 0.0 | 0.1 | GO:0006693 | prostaglandin metabolic process(GO:0006693) |
| 0.0 | 0.2 | GO:0045104 | intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 | 0.0 | GO:0060433 | bronchus development(GO:0060433) |
| 0.0 | 0.1 | GO:0007199 | G-protein coupled receptor signaling pathway coupled to cGMP nucleotide second messenger(GO:0007199) |
| 0.0 | 0.0 | GO:0039656 | modulation by virus of host gene expression(GO:0039656) |
| 0.0 | 0.0 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.1 | 0.4 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 0.1 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.1 | 0.2 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.1 | 0.6 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 0.2 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 0.2 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 0.0 | 0.3 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.1 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.3 | GO:0098642 | network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.0 | 0.1 | GO:0070083 | clathrin-sculpted monoamine transport vesicle(GO:0070081) clathrin-sculpted monoamine transport vesicle membrane(GO:0070083) |
| 0.0 | 0.0 | GO:0098651 | basement membrane collagen trimer(GO:0098651) |
| 0.0 | 0.2 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.1 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.0 | 0.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.2 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.1 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.2 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.2 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.1 | GO:0060199 | clathrin-sculpted glutamate transport vesicle(GO:0060199) clathrin-sculpted glutamate transport vesicle membrane(GO:0060203) |
| 0.0 | 0.1 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.1 | GO:1903561 | extracellular exosome(GO:0070062) extracellular vesicle(GO:1903561) |
| 0.0 | 0.2 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.1 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.0 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.2 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.1 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.1 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.1 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.1 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.1 | GO:0044291 | intercalated disc(GO:0014704) cell-cell contact zone(GO:0044291) |
| 0.0 | 0.6 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.3 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.1 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.1 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.1 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.2 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.0 | 0.4 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.1 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 0.1 | GO:0005954 | calcium- and calmodulin-dependent protein kinase complex(GO:0005954) |
| 0.0 | 0.2 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.0 | 0.0 | GO:0043656 | host(GO:0018995) host cell part(GO:0033643) host intracellular part(GO:0033646) intracellular region of host(GO:0043656) host cell(GO:0043657) other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.1 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.0 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.0 | 0.1 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.1 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.2 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 0.0 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.0 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.1 | GO:0042581 | specific granule(GO:0042581) |
| 0.0 | 0.1 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.1 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.6 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.0 | 0.0 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.0 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.1 | GO:0071547 | piP-body(GO:0071547) |
| 0.0 | 0.1 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.4 | GO:0008305 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 0.1 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.0 | 0.2 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.0 | 0.1 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.0 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.1 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.2 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.3 | GO:0034704 | calcium channel complex(GO:0034704) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0001099 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.1 | 0.4 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.1 | 0.4 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 0.3 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 0.2 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.1 | 0.3 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.1 | 0.2 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.1 | 0.2 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 0.2 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.1 | 0.4 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 0.2 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.1 | 0.1 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.1 | 0.2 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 0.2 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.1 | 0.2 | GO:0042805 | actinin binding(GO:0042805) |
| 0.1 | 0.4 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.1 | 0.3 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.1 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.0 | 0.2 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.1 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.9 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.2 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.2 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.3 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.1 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.1 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.0 | 0.1 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.4 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.1 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.1 | GO:0004470 | malic enzyme activity(GO:0004470) |
| 0.0 | 1.1 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.2 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.1 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.0 | GO:0050543 | icosatetraenoic acid binding(GO:0050543) |
| 0.0 | 0.3 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.2 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.0 | 0.1 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.0 | 0.1 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.2 | GO:0070696 | transmembrane receptor protein serine/threonine kinase binding(GO:0070696) |
| 0.0 | 0.1 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.0 | 0.1 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.1 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.0 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.0 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.2 | GO:0022821 | potassium ion antiporter activity(GO:0022821) |
| 0.0 | 0.3 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.1 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.1 | GO:0004340 | glucokinase activity(GO:0004340) |
| 0.0 | 0.1 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.2 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.1 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.0 | 0.1 | GO:0004645 | phosphorylase activity(GO:0004645) |
| 0.0 | 0.3 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.1 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.1 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 0.1 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.0 | 0.1 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.1 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.0 | 0.1 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.0 | 0.1 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.1 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.2 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.1 | GO:0016972 | thiol oxidase activity(GO:0016972) |
| 0.0 | 0.1 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.0 | 0.1 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.1 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.2 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.1 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.1 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.1 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.1 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.1 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.0 | 0.1 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.0 | 0.1 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.1 | GO:0032296 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.0 | 0.1 | GO:0008035 | high-density lipoprotein particle binding(GO:0008035) |
| 0.0 | 0.1 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.2 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.0 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.0 | 0.0 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.1 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.1 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.0 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.1 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.0 | 0.0 | GO:0000404 | heteroduplex DNA loop binding(GO:0000404) |
| 0.0 | 0.0 | GO:0015199 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.0 | 0.0 | GO:0016623 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.0 | 0.1 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.1 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.3 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.0 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.2 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.0 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.0 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.0 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.1 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.2 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.1 | GO:0004083 | bisphosphoglycerate mutase activity(GO:0004082) bisphosphoglycerate 2-phosphatase activity(GO:0004083) phosphoglycerate mutase activity(GO:0004619) |
| 0.0 | 0.1 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.0 | 0.1 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.2 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.0 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.1 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.0 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.0 | 0.8 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.0 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.1 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.0 | 0.1 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.1 | GO:0048185 | activin binding(GO:0048185) |
| 0.0 | 0.0 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.0 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 0.0 | 0.0 | GO:0050698 | proteoglycan sulfotransferase activity(GO:0050698) |
| 0.0 | 0.1 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.1 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.1 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.1 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.0 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.2 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.0 | GO:0042910 | xenobiotic-transporting ATPase activity(GO:0008559) xenobiotic transporter activity(GO:0042910) |
| 0.0 | 0.0 | GO:0016668 | oxidoreductase activity, acting on a sulfur group of donors, NAD(P) as acceptor(GO:0016668) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.2 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.1 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.0 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.1 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.0 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.0 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.1 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.0 | 0.0 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.6 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.1 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.6 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 1.2 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.1 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 0.0 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.0 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.2 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.0 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 0.5 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.2 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.3 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.0 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.4 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.1 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 1.1 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.4 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.4 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.0 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.0 | 1.4 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.0 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.0 | 0.2 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.6 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.2 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.0 | 0.2 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.4 | REACTOME GRB2 EVENTS IN ERBB2 SIGNALING | Genes involved in GRB2 events in ERBB2 signaling |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.1 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.0 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS | Genes involved in Synthesis of bile acids and bile salts |
| 0.0 | 0.0 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.0 | 0.2 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.2 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.0 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.1 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 0.0 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.1 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.2 | REACTOME PEPTIDE HORMONE BIOSYNTHESIS | Genes involved in Peptide hormone biosynthesis |
| 0.0 | 0.1 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.1 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.0 | 0.0 | REACTOME NUCLEAR EVENTS KINASE AND TRANSCRIPTION FACTOR ACTIVATION | Genes involved in Nuclear Events (kinase and transcription factor activation) |
| 0.0 | 0.0 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 0.1 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.0 | 0.0 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.0 | 0.0 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |