Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
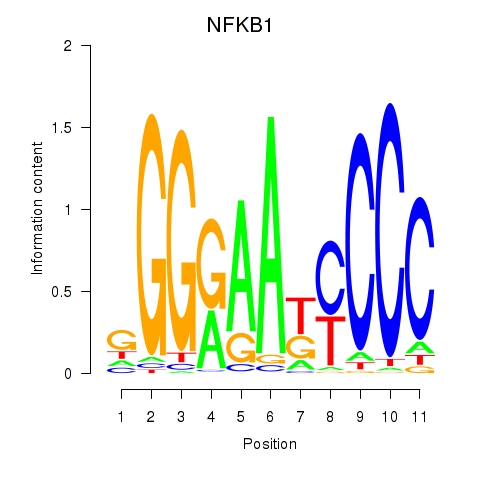
Results for NFKB1
Z-value: 1.23
Transcription factors associated with NFKB1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NFKB1
|
ENSG00000109320.7 | nuclear factor kappa B subunit 1 |
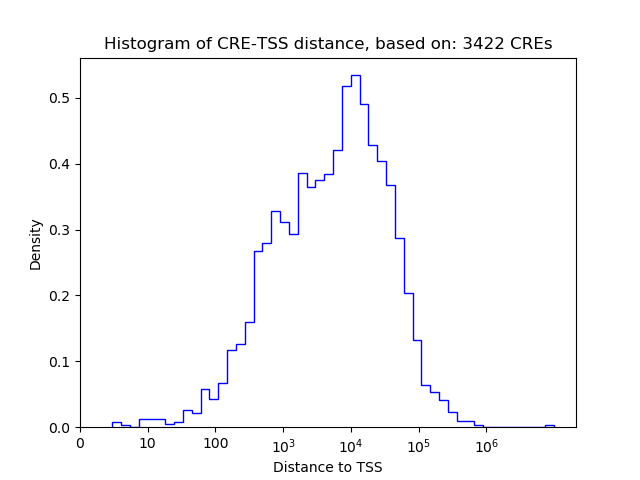
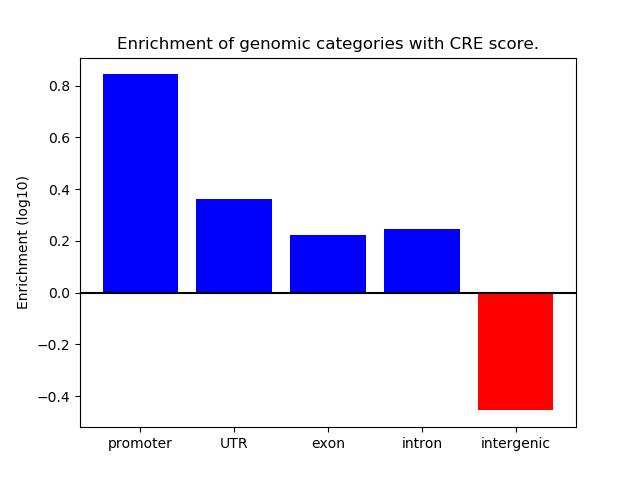
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr4_103441772_103441923 | NFKB1 | 17315 | 0.192702 | 0.94 | 1.4e-04 | Click! |
| chr4_103501773_103501924 | NFKB1 | 2827 | 0.300010 | 0.93 | 2.6e-04 | Click! |
| chr4_103508048_103508199 | NFKB1 | 9102 | 0.224562 | 0.93 | 2.9e-04 | Click! |
| chr4_103508537_103508688 | NFKB1 | 9591 | 0.223443 | 0.91 | 6.5e-04 | Click! |
| chr4_103439720_103439932 | NFKB1 | 15294 | 0.196873 | 0.90 | 8.3e-04 | Click! |
Activity of the NFKB1 motif across conditions
Conditions sorted by the z-value of the NFKB1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
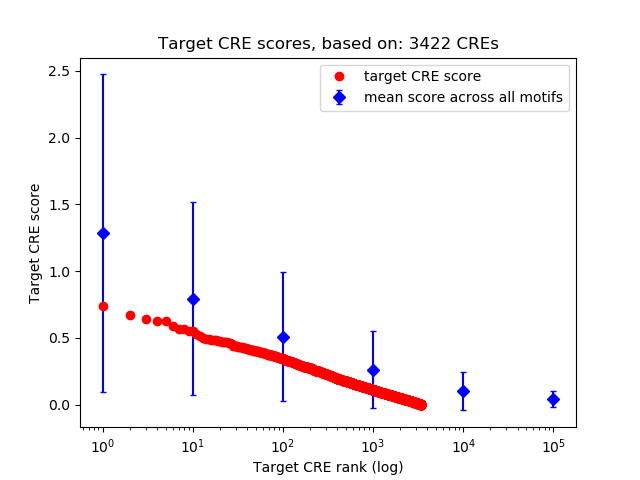
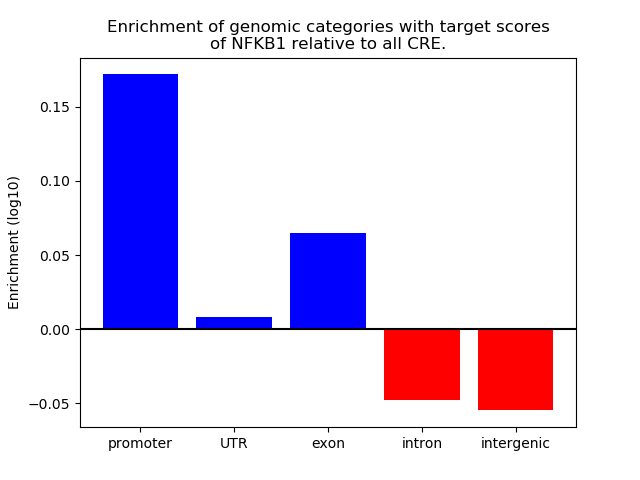
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr3_33090529_33090761 | 0.74 |
GLB1 |
galactosidase, beta 1 |
47639 |
0.11 |
| chr14_76006081_76006729 | 0.67 |
BATF |
basic leucine zipper transcription factor, ATF-like |
17502 |
0.16 |
| chr16_84640518_84640816 | 0.64 |
COTL1 |
coactosin-like 1 (Dictyostelium) |
10998 |
0.16 |
| chr5_156641312_156641556 | 0.63 |
CTB-4E7.1 |
|
9956 |
0.13 |
| chr22_42689321_42689472 | 0.63 |
TCF20 |
transcription factor 20 (AR1) |
50226 |
0.13 |
| chr17_40250698_40250849 | 0.59 |
DHX58 |
DEXH (Asp-Glu-X-His) box polypeptide 58 |
13708 |
0.09 |
| chr17_72276562_72276971 | 0.57 |
DNAI2 |
dynein, axonemal, intermediate chain 2 |
885 |
0.45 |
| chr1_26647230_26647381 | 0.56 |
UBXN11 |
UBX domain protein 11 |
2451 |
0.17 |
| chr3_18794151_18794302 | 0.55 |
ENSG00000228956 |
. |
6990 |
0.34 |
| chr11_60850304_60850538 | 0.55 |
CD5 |
CD5 molecule |
19446 |
0.16 |
| chr1_55264905_55265056 | 0.53 |
TTC22 |
tetratricopeptide repeat domain 22 |
1891 |
0.25 |
| chr10_64571462_64571613 | 0.51 |
EGR2 |
early growth response 2 |
4578 |
0.21 |
| chr14_21538883_21539166 | 0.50 |
NDRG2 |
NDRG family member 2 |
7 |
0.88 |
| chr15_66181382_66181533 | 0.49 |
RAB11A |
RAB11A, member RAS oncogene family |
19106 |
0.19 |
| chr11_286134_286286 | 0.49 |
RP11-326C3.2 |
|
1095 |
0.25 |
| chr11_13317651_13317839 | 0.48 |
ARNTL |
aryl hydrocarbon receptor nuclear translocator-like |
13788 |
0.26 |
| chr18_13377615_13378173 | 0.48 |
LDLRAD4 |
low density lipoprotein receptor class A domain containing 4 |
4659 |
0.18 |
| chr4_2489410_2489561 | 0.48 |
RNF4 |
ring finger protein 4 |
2053 |
0.31 |
| chr6_91107785_91108329 | 0.48 |
ENSG00000252676 |
. |
48434 |
0.16 |
| chr19_5965498_5965678 | 0.48 |
RANBP3 |
RAN binding protein 3 |
12502 |
0.1 |
| chr20_47419324_47419475 | 0.47 |
PREX1 |
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 |
25021 |
0.23 |
| chr15_70535119_70535350 | 0.47 |
ENSG00000200216 |
. |
49659 |
0.18 |
| chr4_100738820_100739031 | 0.47 |
DAPP1 |
dual adaptor of phosphotyrosine and 3-phosphoinositides |
922 |
0.66 |
| chr3_193821410_193821561 | 0.47 |
RP11-135A1.2 |
|
27505 |
0.18 |
| chr2_219757049_219757365 | 0.47 |
WNT10A |
wingless-type MMTV integration site family, member 10A |
10324 |
0.11 |
| chr10_72742471_72742709 | 0.46 |
PCBD1 |
pterin-4 alpha-carbinolamine dehydratase/dimerization cofactor of hepatocyte nuclear factor 1 alpha |
94049 |
0.08 |
| chr9_96362201_96362412 | 0.45 |
PHF2 |
PHD finger protein 2 |
23397 |
0.18 |
| chr14_99684918_99685081 | 0.44 |
AL109767.1 |
|
44286 |
0.15 |
| chr8_145806767_145806918 | 0.44 |
CTD-2517M22.9 |
|
2582 |
0.15 |
| chr16_30562550_30562810 | 0.44 |
ZNF764 |
zinc finger protein 764 |
6904 |
0.07 |
| chr11_118334021_118334172 | 0.44 |
KMT2A |
lysine (K)-specific methyltransferase 2A |
18450 |
0.1 |
| chr14_91876296_91876447 | 0.43 |
CCDC88C |
coiled-coil domain containing 88C |
7319 |
0.25 |
| chr19_46283041_46283318 | 0.43 |
DMPK |
dystrophia myotonica-protein kinase |
620 |
0.48 |
| chr6_157204193_157204560 | 0.43 |
ARID1B |
AT rich interactive domain 1B (SWI1-like) |
18131 |
0.27 |
| chr20_56193359_56193625 | 0.43 |
ZBP1 |
Z-DNA binding protein 1 |
1958 |
0.39 |
| chr7_36661111_36661262 | 0.43 |
ENSG00000221561 |
. |
20663 |
0.17 |
| chr9_131672255_131672406 | 0.43 |
PHYHD1 |
phytanoyl-CoA dioxygenase domain containing 1 |
10844 |
0.11 |
| chr5_96141131_96141340 | 0.42 |
CTD-2260A17.3 |
|
135 |
0.94 |
| chr17_71589495_71589717 | 0.42 |
RP11-277J6.2 |
|
47559 |
0.14 |
| chr16_57612267_57612540 | 0.42 |
GPR56 |
G protein-coupled receptor 56 |
32161 |
0.11 |
| chr19_49839198_49839350 | 0.42 |
CD37 |
CD37 molecule |
553 |
0.56 |
| chr9_134143505_134143979 | 0.41 |
FAM78A |
family with sequence similarity 78, member A |
2138 |
0.29 |
| chr13_46756767_46757026 | 0.41 |
LCP1 |
lymphocyte cytosolic protein 1 (L-plastin) |
437 |
0.8 |
| chr3_59924803_59924954 | 0.41 |
NPCDR1 |
nasopharyngeal carcinoma, down-regulated 1 |
32705 |
0.26 |
| chr11_1876364_1876520 | 0.41 |
LSP1 |
lymphocyte-specific protein 1 |
2242 |
0.16 |
| chr12_58153854_58154005 | 0.41 |
MARCH9 |
membrane-associated ring finger (C3HC4) 9 |
2659 |
0.09 |
| chr12_111022388_111022644 | 0.41 |
PPTC7 |
PTC7 protein phosphatase homolog (S. cerevisiae) |
1391 |
0.36 |
| chr14_24046924_24047247 | 0.41 |
JPH4 |
junctophilin 4 |
909 |
0.42 |
| chr2_202137440_202137733 | 0.41 |
CASP8 |
caspase 8, apoptosis-related cysteine peptidase |
225 |
0.94 |
| chr11_9862251_9862402 | 0.40 |
SBF2 |
SET binding factor 2 |
1176 |
0.37 |
| chr15_92027521_92027672 | 0.40 |
SV2B |
synaptic vesicle glycoprotein 2B |
258496 |
0.02 |
| chr15_69083939_69084090 | 0.40 |
ENSG00000265195 |
. |
10250 |
0.23 |
| chr7_44496571_44496722 | 0.40 |
NUDCD3 |
NudC domain containing 3 |
33833 |
0.15 |
| chr2_16081360_16081718 | 0.40 |
MYCNOS |
MYCN opposite strand |
165 |
0.89 |
| chr5_156652968_156653188 | 0.40 |
CTB-4E7.1 |
|
1688 |
0.28 |
| chr11_59378216_59378367 | 0.39 |
OSBP |
oxysterol binding protein |
5326 |
0.13 |
| chr2_113735290_113735441 | 0.39 |
IL36G |
interleukin 36, gamma |
231 |
0.92 |
| chr17_76343735_76343886 | 0.39 |
SOCS3 |
suppressor of cytokine signaling 3 |
12345 |
0.14 |
| chr8_144051895_144052046 | 0.39 |
LY6E |
lymphocyte antigen 6 complex, locus E |
47429 |
0.1 |
| chr6_37481071_37481281 | 0.39 |
CCDC167 |
coiled-coil domain containing 167 |
13478 |
0.2 |
| chr14_69584826_69584977 | 0.39 |
ENSG00000206768 |
. |
25769 |
0.18 |
| chr11_114217659_114217947 | 0.39 |
RP11-64D24.2 |
|
9490 |
0.18 |
| chr4_4387520_4387671 | 0.39 |
NSG1 |
Homo sapiens neuron specific gene family member 1 (NSG1), transcript variant 3, mRNA. |
388 |
0.89 |
| chr5_96215523_96215674 | 0.38 |
ERAP2 |
endoplasmic reticulum aminopeptidase 2 |
330 |
0.87 |
| chr2_8468501_8468695 | 0.38 |
AC011747.7 |
|
347298 |
0.01 |
| chr3_17064504_17064655 | 0.38 |
PLCL2 |
phospholipase C-like 2 |
12593 |
0.22 |
| chr11_86004444_86004595 | 0.38 |
C11orf73 |
chromosome 11 open reading frame 73 |
8734 |
0.19 |
| chr9_100860283_100860503 | 0.38 |
TRIM14 |
tripartite motif containing 14 |
5550 |
0.2 |
| chr4_37894193_37894870 | 0.38 |
TBC1D1 |
TBC1 (tre-2/USP6, BUB2, cdc16) domain family, member 1 |
1811 |
0.44 |
| chr12_68177291_68177442 | 0.37 |
RP11-335O4.3 |
|
112667 |
0.07 |
| chr19_42382114_42382373 | 0.37 |
CD79A |
CD79a molecule, immunoglobulin-associated alpha |
868 |
0.44 |
| chr19_38908290_38908478 | 0.37 |
RASGRP4 |
RAS guanyl releasing protein 4 |
8418 |
0.09 |
| chr17_65409882_65410033 | 0.37 |
ENSG00000201547 |
. |
4960 |
0.16 |
| chr11_73721865_73722409 | 0.37 |
UCP3 |
uncoupling protein 3 (mitochondrial, proton carrier) |
1657 |
0.32 |
| chr14_65139760_65139972 | 0.37 |
PLEKHG3 |
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
30954 |
0.17 |
| chr1_9182292_9182443 | 0.37 |
GPR157 |
G protein-coupled receptor 157 |
6816 |
0.17 |
| chr1_27961026_27961296 | 0.37 |
FGR |
feline Gardner-Rasheed sarcoma viral oncogene homolog |
566 |
0.68 |
| chr12_105063980_105064272 | 0.37 |
ENSG00000264295 |
. |
78715 |
0.1 |
| chr16_68115229_68115482 | 0.37 |
NFATC3 |
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 3 |
3892 |
0.13 |
| chr10_90590706_90591168 | 0.36 |
ANKRD22 |
ankyrin repeat domain 22 |
20638 |
0.15 |
| chr12_4082516_4082813 | 0.36 |
RP11-664D1.1 |
|
68278 |
0.12 |
| chr6_159457792_159457943 | 0.36 |
TAGAP |
T-cell activation RhoGTPase activating protein |
8183 |
0.19 |
| chr12_68385106_68385257 | 0.36 |
IFNG-AS1 |
IFNG antisense RNA 1 |
1872 |
0.5 |
| chr9_92035991_92036147 | 0.36 |
SEMA4D |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D |
2321 |
0.37 |
| chr14_91863332_91863575 | 0.36 |
CCDC88C |
coiled-coil domain containing 88C |
20237 |
0.21 |
| chr19_2052263_2052418 | 0.36 |
MKNK2 |
MAP kinase interacting serine/threonine kinase 2 |
1097 |
0.34 |
| chr2_198076609_198076760 | 0.36 |
ANKRD44 |
ankyrin repeat domain 44 |
13922 |
0.2 |
| chr10_126424854_126425234 | 0.36 |
FAM53B |
family with sequence similarity 53, member B |
6495 |
0.18 |
| chr3_14470267_14470741 | 0.36 |
SLC6A6 |
solute carrier family 6 (neurotransmitter transporter), member 6 |
3674 |
0.28 |
| chr15_31555674_31556263 | 0.35 |
KLF13 |
Kruppel-like factor 13 |
63090 |
0.13 |
| chr18_3003132_3003283 | 0.35 |
LPIN2 |
lipin 2 |
8738 |
0.17 |
| chr10_73454547_73455031 | 0.35 |
C10orf105 |
chromosome 10 open reading frame 105 |
24789 |
0.19 |
| chr10_112602586_112603108 | 0.35 |
PDCD4 |
programmed cell death 4 (neoplastic transformation inhibitor) |
28718 |
0.13 |
| chr10_9336383_9336663 | 0.35 |
ENSG00000212505 |
. |
637729 |
0.0 |
| chr16_67813577_67813770 | 0.35 |
RANBP10 |
RAN binding protein 10 |
26787 |
0.09 |
| chr12_111115979_111116149 | 0.35 |
HVCN1 |
hydrogen voltage-gated channel 1 |
10450 |
0.16 |
| chr3_121373642_121373829 | 0.35 |
ENSG00000243544 |
. |
592 |
0.48 |
| chr15_40601498_40601800 | 0.35 |
PLCB2 |
phospholipase C, beta 2 |
1526 |
0.21 |
| chr16_3146077_3146254 | 0.35 |
ZSCAN10 |
zinc finger and SCAN domain containing 10 |
3064 |
0.09 |
| chr17_46517090_46517319 | 0.35 |
SKAP1 |
src kinase associated phosphoprotein 1 |
9623 |
0.14 |
| chr1_181286343_181286494 | 0.35 |
CACNA1E |
calcium channel, voltage-dependent, R type, alpha 1E subunit |
95820 |
0.09 |
| chr9_98444198_98444349 | 0.35 |
DKFZP434H0512 |
Protein LOC100506667; Putative uncharacterized protein DKFZp434H0512 |
90332 |
0.08 |
| chr6_37123484_37123718 | 0.34 |
PIM1 |
pim-1 oncogene |
14378 |
0.18 |
| chr18_47486716_47486867 | 0.34 |
RP11-813F20.4 |
|
10536 |
0.19 |
| chr3_43236443_43236646 | 0.34 |
ENSG00000222331 |
. |
15819 |
0.22 |
| chr10_124136204_124136523 | 0.34 |
PLEKHA1 |
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1 |
2013 |
0.32 |
| chr13_24848573_24849154 | 0.33 |
SPATA13 |
spermatogenesis associated 13 |
2036 |
0.28 |
| chr17_46484714_46484933 | 0.33 |
SKAP1 |
src kinase associated phosphoprotein 1 |
22729 |
0.12 |
| chr12_3858731_3858882 | 0.33 |
EFCAB4B |
EF-hand calcium binding domain 4B |
3457 |
0.29 |
| chr20_2722524_2722808 | 0.33 |
EBF4 |
early B-cell factor 4 |
35779 |
0.09 |
| chr13_99216536_99217042 | 0.33 |
STK24 |
serine/threonine kinase 24 |
12328 |
0.21 |
| chr22_31730141_31730401 | 0.33 |
RP3-400N23.6 |
|
1064 |
0.37 |
| chr9_108165581_108165732 | 0.33 |
FSD1L |
fibronectin type III and SPRY domain containing 1-like |
44421 |
0.16 |
| chr9_73021961_73022182 | 0.33 |
KLF9 |
Kruppel-like factor 9 |
7469 |
0.28 |
| chr9_33137738_33137889 | 0.33 |
B4GALT1 |
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 1 |
9616 |
0.16 |
| chr17_27943593_27943795 | 0.33 |
CORO6 |
coronin 6 |
1441 |
0.2 |
| chr1_9774574_9774847 | 0.33 |
PIK3CD |
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta |
3417 |
0.22 |
| chr20_5296649_5296800 | 0.32 |
PROKR2 |
prokineticin receptor 2 |
654 |
0.76 |
| chr3_196379983_196380199 | 0.32 |
PIGX |
phosphatidylinositol glycan anchor biosynthesis, class X |
13445 |
0.12 |
| chr17_17087488_17087777 | 0.32 |
RP11-45M22.3 |
|
1236 |
0.32 |
| chr14_65404309_65404460 | 0.32 |
GPX2 |
glutathione peroxidase 2 (gastrointestinal) |
5054 |
0.12 |
| chr3_46735633_46736482 | 0.32 |
ALS2CL |
ALS2 C-terminal like |
866 |
0.49 |
| chr11_60873950_60874231 | 0.32 |
CD5 |
CD5 molecule |
4109 |
0.21 |
| chr12_113513206_113513364 | 0.32 |
DTX1 |
deltex homolog 1 (Drosophila) |
17790 |
0.15 |
| chr19_8583523_8583746 | 0.32 |
ZNF414 |
zinc finger protein 414 |
4590 |
0.13 |
| chr4_40229105_40229274 | 0.32 |
RHOH |
ras homolog family member H |
27225 |
0.19 |
| chr16_50730890_50731308 | 0.32 |
NOD2 |
nucleotide-binding oligomerization domain containing 2 |
49 |
0.96 |
| chr3_195919316_195919605 | 0.32 |
ENSG00000222335 |
. |
9760 |
0.14 |
| chr10_64493160_64493418 | 0.32 |
ADO |
2-aminoethanethiol (cysteamine) dioxygenase |
71227 |
0.09 |
| chr7_36325933_36326467 | 0.32 |
EEPD1 |
endonuclease/exonuclease/phosphatase family domain containing 1 |
10553 |
0.18 |
| chr2_136814589_136814740 | 0.32 |
AC093391.2 |
|
44629 |
0.15 |
| chr15_67204936_67205097 | 0.32 |
SMAD3 |
SMAD family member 3 |
151085 |
0.04 |
| chr2_235332578_235332755 | 0.31 |
ARL4C |
ADP-ribosylation factor-like 4C |
72578 |
0.13 |
| chr21_46337804_46338132 | 0.31 |
ITGB2 |
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
2802 |
0.14 |
| chr4_6137198_6137349 | 0.31 |
JAKMIP1 |
janus kinase and microtubule interacting protein 1 |
59071 |
0.14 |
| chr12_4262653_4262869 | 0.31 |
CCND2 |
cyclin D2 |
120177 |
0.05 |
| chr14_99692121_99692272 | 0.31 |
AL109767.1 |
|
37089 |
0.16 |
| chr13_49077988_49078168 | 0.31 |
RCBTB2 |
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 2 |
29135 |
0.22 |
| chr2_109234348_109234509 | 0.31 |
LIMS1 |
LIM and senescent cell antigen-like domains 1 |
3294 |
0.3 |
| chr5_141386076_141386227 | 0.31 |
GNPDA1 |
glucosamine-6-phosphate deaminase 1 |
6208 |
0.17 |
| chr20_2734143_2734294 | 0.31 |
CPXM1 |
carboxypeptidase X (M14 family), member 1 |
47065 |
0.08 |
| chr1_26038694_26038845 | 0.31 |
MAN1C1 |
mannosidase, alpha, class 1C, member 1 |
2676 |
0.26 |
| chr9_116112694_116113190 | 0.31 |
BSPRY |
B-box and SPRY domain containing |
1121 |
0.39 |
| chr12_121848379_121848530 | 0.31 |
RNF34 |
ring finger protein 34, E3 ubiquitin protein ligase |
1176 |
0.51 |
| chr17_35298519_35298670 | 0.30 |
RP11-445F12.2 |
|
2161 |
0.21 |
| chr1_166459519_166459725 | 0.30 |
FAM78B |
family with sequence similarity 78, member B |
323416 |
0.01 |
| chr11_64821448_64821599 | 0.30 |
ENSG00000266123 |
. |
4012 |
0.08 |
| chr3_42059799_42059950 | 0.30 |
ULK4 |
unc-51 like kinase 4 |
55952 |
0.13 |
| chrX_19833042_19833193 | 0.30 |
SH3KBP1 |
SH3-domain kinase binding protein 1 |
15248 |
0.27 |
| chr5_40406098_40406422 | 0.30 |
ENSG00000265615 |
. |
85840 |
0.1 |
| chr11_5719864_5720015 | 0.30 |
TRIM22 |
tripartite motif containing 22 |
7682 |
0.12 |
| chr8_144287595_144287746 | 0.30 |
GPIHBP1 |
glycosylphosphatidylinositol anchored high density lipoprotein binding protein 1 |
7398 |
0.15 |
| chr3_186252123_186252274 | 0.30 |
CRYGS |
crystallin, gamma S |
10042 |
0.17 |
| chr15_29330394_29330545 | 0.30 |
APBA2 |
amyloid beta (A4) precursor protein-binding, family A, member 2 |
6058 |
0.28 |
| chr16_50733857_50734060 | 0.30 |
NOD2 |
nucleotide-binding oligomerization domain containing 2 |
2908 |
0.16 |
| chr6_150322514_150322665 | 0.30 |
RAET1K |
retinoic acid early transcript 1K pseudogene |
3704 |
0.19 |
| chr8_74375126_74375277 | 0.30 |
RP11-434I12.2 |
|
106505 |
0.07 |
| chr10_126367388_126367539 | 0.29 |
FAM53B-AS1 |
FAM53B antisense RNA 1 |
24731 |
0.19 |
| chr2_197066450_197066719 | 0.29 |
ENSG00000239161 |
. |
13462 |
0.18 |
| chr17_75321389_75321609 | 0.29 |
SEPT9 |
septin 9 |
2807 |
0.3 |
| chrX_48792186_48792645 | 0.29 |
PIM2 |
pim-2 oncogene |
16114 |
0.08 |
| chr14_21777648_21777955 | 0.29 |
RPGRIP1 |
retinitis pigmentosa GTPase regulator interacting protein 1 |
7897 |
0.16 |
| chr10_14625977_14626446 | 0.29 |
FAM107B |
family with sequence similarity 107, member B |
11873 |
0.24 |
| chr20_44994972_44995158 | 0.29 |
SLC35C2 |
solute carrier family 35 (GDP-fucose transporter), member C2 |
2022 |
0.33 |
| chr5_141478725_141478876 | 0.29 |
NDFIP1 |
Nedd4 family interacting protein 1 |
9270 |
0.24 |
| chr22_23583322_23583546 | 0.29 |
BCR |
breakpoint cluster region |
12713 |
0.16 |
| chr17_47296320_47296793 | 0.29 |
ABI3 |
ABI family, member 3 |
369 |
0.83 |
| chr4_147379710_147379861 | 0.29 |
SLC10A7 |
solute carrier family 10, member 7 |
63084 |
0.13 |
| chr10_7233238_7233389 | 0.29 |
SFMBT2 |
Scm-like with four mbt domains 2 |
217394 |
0.02 |
| chr7_99221023_99221247 | 0.29 |
ZSCAN25 |
zinc finger and SCAN domain containing 25 |
3905 |
0.17 |
| chr5_176857213_176857619 | 0.28 |
GRK6 |
G protein-coupled receptor kinase 6 |
3561 |
0.12 |
| chr6_144740266_144740510 | 0.28 |
UTRN |
utrophin |
75151 |
0.11 |
| chr22_20229537_20229756 | 0.28 |
RTN4R |
reticulon 4 receptor |
1561 |
0.29 |
| chrX_19763111_19763463 | 0.28 |
SH3KBP1 |
SH3-domain kinase binding protein 1 |
1242 |
0.62 |
| chr11_2720199_2720447 | 0.28 |
KCNQ1OT1 |
KCNQ1 opposite strand/antisense transcript 1 (non-protein coding) |
901 |
0.68 |
| chr9_97347238_97347389 | 0.28 |
FBP2 |
fructose-1,6-bisphosphatase 2 |
8762 |
0.24 |
| chr6_10656054_10656229 | 0.28 |
ENSG00000201048 |
. |
18860 |
0.13 |
| chr19_40459440_40459607 | 0.28 |
PSMC4 |
proteasome (prosome, macropain) 26S subunit, ATPase, 4 |
17389 |
0.14 |
| chr21_45715633_45715784 | 0.28 |
PFKL |
phosphofructokinase, liver |
4226 |
0.11 |
| chr13_96109760_96109911 | 0.28 |
CLDN10 |
claudin 10 |
23977 |
0.22 |
| chr18_46362885_46363036 | 0.28 |
RP11-484L8.1 |
|
1819 |
0.46 |
| chr17_56064502_56064792 | 0.28 |
VEZF1 |
vascular endothelial zinc finger 1 |
866 |
0.54 |
| chr16_88921173_88921341 | 0.28 |
TRAPPC2L |
trafficking protein particle complex 2-like |
1371 |
0.23 |
| chr10_74016888_74017047 | 0.28 |
DDIT4 |
DNA-damage-inducible transcript 4 |
16711 |
0.14 |
| chr22_29976535_29976734 | 0.28 |
NIPSNAP1 |
nipsnap homolog 1 (C. elegans) |
429 |
0.79 |
| chr13_101177691_101177842 | 0.28 |
GGACT |
gamma-glutamylamine cyclotransferase |
8141 |
0.18 |
| chr2_191883690_191883911 | 0.28 |
AC067945.2 |
|
1120 |
0.35 |
| chr3_58102043_58102194 | 0.28 |
FLNB |
filamin B, beta |
33475 |
0.17 |
| chr5_156451707_156451858 | 0.28 |
HAVCR1 |
hepatitis A virus cellular receptor 1 |
33625 |
0.15 |
| chr10_8444237_8444486 | 0.28 |
ENSG00000212505 |
. |
254433 |
0.02 |
| chr16_57170035_57170227 | 0.28 |
CPNE2 |
copine II |
17020 |
0.13 |
| chr12_122087386_122087583 | 0.28 |
MORN3 |
MORN repeat containing 3 |
20076 |
0.15 |
| chr12_54390196_54390380 | 0.28 |
HOXC-AS2 |
HOXC cluster antisense RNA 2 |
281 |
0.66 |
| chr19_10450885_10451036 | 0.28 |
ICAM3 |
intercellular adhesion molecule 3 |
461 |
0.63 |
| chr10_88299612_88299763 | 0.28 |
RP11-77P6.2 |
|
17985 |
0.18 |
| chr4_123749245_123749396 | 0.28 |
ENSG00000253069 |
. |
1151 |
0.39 |
| chr19_16476504_16477086 | 0.27 |
EPS15L1 |
epidermal growth factor receptor pathway substrate 15-like 1 |
4031 |
0.18 |
| chr1_36859369_36859520 | 0.27 |
LSM10 |
LSM10, U7 small nuclear RNA associated |
4049 |
0.14 |
| chr12_111108052_111108203 | 0.27 |
HVCN1 |
hydrogen voltage-gated channel 1 |
18387 |
0.14 |
| chr12_56554135_56554286 | 0.27 |
MYL6 |
myosin, light chain 6, alkali, smooth muscle and non-muscle |
2030 |
0.11 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0002385 | innate immune response in mucosa(GO:0002227) mucosal immune response(GO:0002385) |
| 0.1 | 0.3 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.1 | 0.2 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.1 | 0.2 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.1 | 0.2 | GO:0032928 | regulation of superoxide anion generation(GO:0032928) |
| 0.1 | 0.2 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.1 | 0.4 | GO:0002483 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.1 | 0.1 | GO:0002251 | organ or tissue specific immune response(GO:0002251) |
| 0.1 | 0.3 | GO:0051797 | regulation of hair follicle development(GO:0051797) |
| 0.1 | 0.2 | GO:0036037 | CD8-positive, alpha-beta T cell activation(GO:0036037) CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.1 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.0 | 0.2 | GO:0014819 | regulation of skeletal muscle contraction(GO:0014819) |
| 0.0 | 0.1 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.1 | GO:0045341 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) |
| 0.0 | 0.3 | GO:0008631 | intrinsic apoptotic signaling pathway in response to oxidative stress(GO:0008631) cell death in response to oxidative stress(GO:0036473) |
| 0.0 | 0.2 | GO:1901339 | activation of store-operated calcium channel activity(GO:0032237) regulation of store-operated calcium channel activity(GO:1901339) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.0 | GO:0010535 | regulation of activation of Janus kinase activity(GO:0010533) regulation of activation of JAK2 kinase activity(GO:0010534) positive regulation of activation of JAK2 kinase activity(GO:0010535) positive regulation of activation of Janus kinase activity(GO:0010536) |
| 0.0 | 0.1 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.0 | 0.1 | GO:0048293 | isotype switching to IgE isotypes(GO:0048289) regulation of isotype switching to IgE isotypes(GO:0048293) |
| 0.0 | 0.1 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.3 | GO:0060484 | lung-associated mesenchyme development(GO:0060484) |
| 0.0 | 0.1 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.1 | GO:0060836 | lymphatic endothelial cell differentiation(GO:0060836) |
| 0.0 | 0.1 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.0 | 0.2 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) |
| 0.0 | 0.1 | GO:2000008 | regulation of protein localization to cell surface(GO:2000008) |
| 0.0 | 0.2 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.1 | GO:0090224 | regulation of spindle organization(GO:0090224) |
| 0.0 | 0.6 | GO:0002639 | positive regulation of immunoglobulin production(GO:0002639) |
| 0.0 | 0.1 | GO:0002323 | natural killer cell activation involved in immune response(GO:0002323) natural killer cell degranulation(GO:0043320) |
| 0.0 | 0.1 | GO:0021610 | cranial nerve structural organization(GO:0021604) facial nerve morphogenesis(GO:0021610) facial nerve structural organization(GO:0021612) |
| 0.0 | 0.1 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.0 | 0.1 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.0 | 0.1 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 0.2 | GO:1901019 | regulation of ryanodine-sensitive calcium-release channel activity(GO:0060314) regulation of calcium ion transmembrane transporter activity(GO:1901019) regulation of cation channel activity(GO:2001257) |
| 0.0 | 0.1 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) ribonucleoside diphosphate catabolic process(GO:0009191) |
| 0.0 | 0.1 | GO:0002085 | inhibition of neuroepithelial cell differentiation(GO:0002085) |
| 0.0 | 0.1 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.0 | 0.1 | GO:0060129 | thyroid-stimulating hormone-secreting cell differentiation(GO:0060129) |
| 0.0 | 0.2 | GO:0042104 | positive regulation of activated T cell proliferation(GO:0042104) |
| 0.0 | 0.1 | GO:0010716 | regulation of extracellular matrix disassembly(GO:0010715) negative regulation of extracellular matrix disassembly(GO:0010716) regulation of extracellular matrix organization(GO:1903053) negative regulation of extracellular matrix organization(GO:1903054) |
| 0.0 | 0.2 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.1 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.0 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 | 0.1 | GO:2000344 | positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.1 | GO:0042415 | norepinephrine metabolic process(GO:0042415) |
| 0.0 | 0.1 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.1 | GO:0002689 | negative regulation of leukocyte chemotaxis(GO:0002689) |
| 0.0 | 0.1 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.0 | 0.1 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 0.0 | 0.1 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.0 | 0.1 | GO:0070306 | lens fiber cell differentiation(GO:0070306) |
| 0.0 | 0.0 | GO:0045112 | integrin biosynthetic process(GO:0045112) |
| 0.0 | 0.1 | GO:0090009 | primitive streak formation(GO:0090009) |
| 0.0 | 0.3 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.0 | 0.4 | GO:0046834 | lipid phosphorylation(GO:0046834) |
| 0.0 | 0.1 | GO:0001885 | endothelial cell development(GO:0001885) |
| 0.0 | 0.1 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.0 | GO:2000516 | positive regulation of CD4-positive, alpha-beta T cell differentiation(GO:0043372) positive regulation of CD4-positive, alpha-beta T cell activation(GO:2000516) |
| 0.0 | 0.0 | GO:0021783 | preganglionic parasympathetic fiber development(GO:0021783) |
| 0.0 | 0.0 | GO:0060789 | hair follicle placode formation(GO:0060789) |
| 0.0 | 0.5 | GO:0002286 | T cell activation involved in immune response(GO:0002286) |
| 0.0 | 0.1 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.0 | 0.0 | GO:0032914 | transforming growth factor beta1 production(GO:0032905) regulation of transforming growth factor beta1 production(GO:0032908) positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.0 | 0.0 | GO:0010831 | positive regulation of myotube differentiation(GO:0010831) |
| 0.0 | 0.1 | GO:0034227 | tRNA wobble base modification(GO:0002097) tRNA wobble uridine modification(GO:0002098) tRNA thio-modification(GO:0034227) |
| 0.0 | 0.1 | GO:0010956 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.0 | 0.1 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.1 | GO:0009713 | catechol-containing compound biosynthetic process(GO:0009713) catecholamine biosynthetic process(GO:0042423) |
| 0.0 | 0.0 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.0 | 0.1 | GO:0071503 | response to heparin(GO:0071503) |
| 0.0 | 0.0 | GO:0071635 | negative regulation of transforming growth factor beta production(GO:0071635) |
| 0.0 | 0.1 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.1 | GO:0034625 | fatty acid elongation, monounsaturated fatty acid(GO:0034625) |
| 0.0 | 0.1 | GO:0060396 | growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.0 | 0.2 | GO:0046348 | amino sugar catabolic process(GO:0046348) glucosamine-containing compound catabolic process(GO:1901072) |
| 0.0 | 0.0 | GO:0031943 | regulation of glucocorticoid metabolic process(GO:0031943) |
| 0.0 | 0.4 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.0 | 0.1 | GO:0043268 | positive regulation of potassium ion transport(GO:0043268) |
| 0.0 | 0.0 | GO:0002669 | lymphocyte anergy(GO:0002249) regulation of T cell anergy(GO:0002667) positive regulation of T cell anergy(GO:0002669) T cell anergy(GO:0002870) regulation of lymphocyte anergy(GO:0002911) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.0 | 0.1 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.1 | GO:0039656 | modulation by virus of host gene expression(GO:0039656) |
| 0.0 | 0.0 | GO:0001705 | ectoderm formation(GO:0001705) |
| 0.0 | 0.1 | GO:0046831 | regulation of RNA export from nucleus(GO:0046831) |
| 0.0 | 0.1 | GO:0051709 | regulation of killing of cells of other organism(GO:0051709) |
| 0.0 | 0.1 | GO:0006801 | superoxide metabolic process(GO:0006801) |
| 0.0 | 0.0 | GO:0010882 | regulation of cardiac muscle contraction by calcium ion signaling(GO:0010882) |
| 0.0 | 0.1 | GO:0042816 | pyridoxine metabolic process(GO:0008614) pyridoxine biosynthetic process(GO:0008615) vitamin B6 metabolic process(GO:0042816) vitamin B6 biosynthetic process(GO:0042819) |
| 0.0 | 0.0 | GO:0042940 | D-amino acid transport(GO:0042940) |
| 0.0 | 0.1 | GO:0051583 | dopamine uptake involved in synaptic transmission(GO:0051583) catecholamine uptake involved in synaptic transmission(GO:0051934) catecholamine uptake(GO:0090493) dopamine uptake(GO:0090494) |
| 0.0 | 0.1 | GO:0021873 | forebrain neuroblast division(GO:0021873) |
| 0.0 | 0.0 | GO:0032785 | negative regulation of DNA-templated transcription, elongation(GO:0032785) negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 0.1 | GO:0097576 | vacuole fusion(GO:0097576) |
| 0.0 | 0.1 | GO:0000966 | RNA 5'-end processing(GO:0000966) |
| 0.0 | 0.1 | GO:0051657 | maintenance of organelle location(GO:0051657) |
| 0.0 | 0.0 | GO:0070102 | interleukin-6-mediated signaling pathway(GO:0070102) |
| 0.0 | 0.0 | GO:0046512 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.0 | 0.1 | GO:0034650 | cortisol metabolic process(GO:0034650) cortisol biosynthetic process(GO:0034651) |
| 0.0 | 0.1 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.0 | GO:0070296 | sarcoplasmic reticulum calcium ion transport(GO:0070296) |
| 0.0 | 0.1 | GO:0006388 | RNA splicing, via endonucleolytic cleavage and ligation(GO:0000394) tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.0 | GO:0002093 | auditory receptor cell morphogenesis(GO:0002093) auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.0 | GO:0071921 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.0 | 0.1 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.0 | 0.0 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.0 | 0.1 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 0.1 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 0.0 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.0 | 0.1 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 0.0 | GO:0009448 | gamma-aminobutyric acid metabolic process(GO:0009448) |
| 0.0 | 0.2 | GO:0060079 | excitatory postsynaptic potential(GO:0060079) |
| 0.0 | 0.1 | GO:0008089 | anterograde axonal transport(GO:0008089) |
| 0.0 | 0.0 | GO:0098754 | detoxification of inorganic compound(GO:0061687) stress response to metal ion(GO:0097501) detoxification(GO:0098754) |
| 0.0 | 0.0 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.0 | 0.1 | GO:0000303 | response to superoxide(GO:0000303) |
| 0.0 | 0.1 | GO:0007090 | obsolete regulation of S phase of mitotic cell cycle(GO:0007090) |
| 0.0 | 0.2 | GO:0071445 | obsolete cellular response to protein stimulus(GO:0071445) |
| 0.0 | 0.0 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 0.0 | 0.0 | GO:0045066 | regulatory T cell differentiation(GO:0045066) |
| 0.0 | 0.1 | GO:0008608 | attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.0 | 0.2 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.0 | 0.0 | GO:0032693 | negative regulation of interleukin-10 production(GO:0032693) interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.0 | 0.1 | GO:0030146 | obsolete diuresis(GO:0030146) |
| 0.0 | 0.1 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.0 | GO:0033194 | response to hydroperoxide(GO:0033194) |
| 0.0 | 0.1 | GO:0031935 | regulation of chromatin silencing(GO:0031935) |
| 0.0 | 0.1 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.1 | GO:0019317 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.0 | 0.2 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.0 | 0.1 | GO:0006349 | regulation of gene expression by genetic imprinting(GO:0006349) |
| 0.0 | 0.0 | GO:0002827 | positive regulation of T-helper 1 type immune response(GO:0002827) |
| 0.0 | 0.1 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.0 | 0.1 | GO:0006337 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.0 | 0.0 | GO:0006772 | thiamine metabolic process(GO:0006772) |
| 0.0 | 0.1 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.2 | GO:0045646 | regulation of erythrocyte differentiation(GO:0045646) |
| 0.0 | 0.0 | GO:0032801 | receptor catabolic process(GO:0032801) |
| 0.0 | 0.0 | GO:0051446 | positive regulation of meiotic nuclear division(GO:0045836) positive regulation of meiotic cell cycle(GO:0051446) |
| 0.0 | 0.1 | GO:0006000 | fructose metabolic process(GO:0006000) |
| 0.0 | 0.1 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 0.0 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.0 | GO:0006007 | glucose catabolic process(GO:0006007) |
| 0.0 | 0.3 | GO:0050771 | negative regulation of axonogenesis(GO:0050771) |
| 0.0 | 0.0 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.0 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.0 | 0.0 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.0 | 0.0 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.1 | GO:0006554 | lysine metabolic process(GO:0006553) lysine catabolic process(GO:0006554) |
| 0.0 | 0.1 | GO:0002418 | immune response to tumor cell(GO:0002418) |
| 0.0 | 0.0 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.0 | 0.0 | GO:0046337 | phosphatidylethanolamine biosynthetic process(GO:0006646) phosphatidylethanolamine metabolic process(GO:0046337) |
| 0.0 | 0.0 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.0 | 0.0 | GO:0043137 | DNA replication, Okazaki fragment processing(GO:0033567) DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.0 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 0.0 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.1 | GO:0090231 | regulation of spindle checkpoint(GO:0090231) |
| 0.0 | 0.1 | GO:0010759 | regulation of macrophage chemotaxis(GO:0010758) positive regulation of macrophage chemotaxis(GO:0010759) mononuclear cell migration(GO:0071674) regulation of mononuclear cell migration(GO:0071675) |
| 0.0 | 0.3 | GO:0008633 | obsolete activation of pro-apoptotic gene products(GO:0008633) |
| 0.0 | 0.1 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.0 | 0.0 | GO:0009822 | alkaloid catabolic process(GO:0009822) |
| 0.0 | 0.0 | GO:0002903 | negative regulation of B cell apoptotic process(GO:0002903) |
| 0.0 | 0.0 | GO:0031282 | regulation of guanylate cyclase activity(GO:0031282) |
| 0.0 | 0.1 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.0 | GO:0000281 | mitotic cytokinesis(GO:0000281) |
| 0.0 | 0.0 | GO:0072201 | negative regulation of mesenchymal cell proliferation(GO:0072201) |
| 0.0 | 0.4 | GO:0030049 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.0 | 0.1 | GO:0016556 | mRNA modification(GO:0016556) |
| 0.0 | 0.2 | GO:0050830 | defense response to Gram-positive bacterium(GO:0050830) |
| 0.0 | 0.0 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.0 | 0.0 | GO:0048859 | formation of anatomical boundary(GO:0048859) |
| 0.0 | 0.0 | GO:0010939 | regulation of necrotic cell death(GO:0010939) |
| 0.0 | 0.0 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.0 | 0.1 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.1 | GO:0008653 | lipopolysaccharide metabolic process(GO:0008653) |
| 0.0 | 0.0 | GO:0046083 | adenine salvage(GO:0006168) purine nucleobase salvage(GO:0043096) adenine metabolic process(GO:0046083) adenine biosynthetic process(GO:0046084) |
| 0.0 | 0.0 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.4 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 0.0 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.0 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.1 | GO:0048169 | regulation of long-term neuronal synaptic plasticity(GO:0048169) |
| 0.0 | 0.0 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.0 | GO:0051532 | NFAT protein import into nucleus(GO:0051531) regulation of NFAT protein import into nucleus(GO:0051532) |
| 0.0 | 0.0 | GO:0045188 | circadian sleep/wake cycle, non-REM sleep(GO:0042748) regulation of circadian sleep/wake cycle, non-REM sleep(GO:0045188) |
| 0.0 | 0.0 | GO:0072583 | synaptic vesicle recycling(GO:0036465) synaptic vesicle endocytosis(GO:0048488) clathrin-mediated endocytosis(GO:0072583) |
| 0.0 | 0.0 | GO:0070193 | synaptonemal complex organization(GO:0070193) |
| 0.0 | 0.1 | GO:0046579 | positive regulation of Ras protein signal transduction(GO:0046579) |
| 0.0 | 0.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0031264 | death-inducing signaling complex(GO:0031264) |
| 0.1 | 0.3 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.1 | 0.4 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 0.4 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.2 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.1 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 0.1 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.4 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.1 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.1 | GO:0001652 | granular component(GO:0001652) |
| 0.0 | 0.1 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.1 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.1 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.0 | 0.2 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 0.1 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.3 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.1 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.2 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.1 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.0 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.2 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.1 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.1 | GO:0000791 | euchromatin(GO:0000791) |
| 0.0 | 0.1 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.1 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.2 | GO:0005858 | axonemal dynein complex(GO:0005858) |
| 0.0 | 0.3 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.0 | 0.1 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.0 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.0 | 0.1 | GO:0030312 | external encapsulating structure(GO:0030312) |
| 0.0 | 0.0 | GO:0032449 | CBM complex(GO:0032449) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.1 | GO:0090576 | RNA polymerase III transcription factor complex(GO:0090576) |
| 0.0 | 0.2 | GO:0045120 | pronucleus(GO:0045120) |
| 0.0 | 0.2 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.0 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.0 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.0 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 0.0 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 0.0 | GO:0090533 | cation-transporting ATPase complex(GO:0090533) |
| 0.0 | 0.2 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.1 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.0 | GO:0060199 | clathrin-sculpted glutamate transport vesicle(GO:0060199) clathrin-sculpted glutamate transport vesicle membrane(GO:0060203) |
| 0.0 | 0.1 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.0 | 0.1 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.0 | GO:0042583 | chromaffin granule(GO:0042583) chromaffin granule membrane(GO:0042584) |
| 0.0 | 0.0 | GO:0030288 | outer membrane-bounded periplasmic space(GO:0030288) periplasmic space(GO:0042597) |
| 0.0 | 0.0 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.1 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.1 | GO:0090568 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) nuclear transcriptional repressor complex(GO:0090568) |
| 0.0 | 0.0 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.1 | 0.4 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.1 | 0.2 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.1 | 0.4 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.1 | 0.2 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.1 | 0.2 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.1 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.0 | 0.4 | GO:0050780 | dopamine receptor binding(GO:0050780) |
| 0.0 | 0.1 | GO:0042156 | obsolete zinc-mediated transcriptional activator activity(GO:0042156) |
| 0.0 | 0.2 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.3 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.2 | GO:0015925 | galactosidase activity(GO:0015925) |
| 0.0 | 0.1 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.2 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.1 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.2 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.0 | 0.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.2 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.2 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.2 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.1 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.0 | 0.2 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.2 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.2 | GO:0050656 | 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.0 | 0.2 | GO:0016595 | glutamate binding(GO:0016595) |
| 0.0 | 0.2 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 0.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.1 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 0.0 | 0.1 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.3 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.1 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.1 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.1 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.0 | 0.1 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.0 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.1 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.0 | 0.1 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.2 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.1 | GO:0052659 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) inositol-1,3,4,5-tetrakisphosphate 5-phosphatase activity(GO:0052659) |
| 0.0 | 0.2 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.1 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.1 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) |
| 0.0 | 0.1 | GO:0030955 | potassium ion binding(GO:0030955) |
| 0.0 | 0.1 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.2 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.1 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.1 | GO:0017060 | 3-galactosyl-N-acetylglucosaminide 4-alpha-L-fucosyltransferase activity(GO:0017060) |
| 0.0 | 0.1 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 0.1 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.2 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.4 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.1 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 0.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.4 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.0 | 0.1 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.0 | 0.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.1 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.1 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.0 | 0.3 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 0.0 | GO:0030151 | molybdenum ion binding(GO:0030151) |
| 0.0 | 0.1 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.3 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.1 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.1 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 0.1 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 0.1 | GO:0003840 | gamma-glutamyltransferase activity(GO:0003840) |
| 0.0 | 0.1 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 0.1 | GO:0022897 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 0.0 | 0.1 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 0.0 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.1 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.0 | GO:0070061 | fructose binding(GO:0070061) |
| 0.0 | 0.0 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.0 | 0.1 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.0 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.0 | GO:0052743 | inositol tetrakisphosphate phosphatase activity(GO:0052743) |
| 0.0 | 0.1 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.1 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.1 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.1 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.1 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.0 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) xenobiotic transporter activity(GO:0042910) |
| 0.0 | 0.2 | GO:0016411 | acylglycerol O-acyltransferase activity(GO:0016411) |
| 0.0 | 0.1 | GO:0004308 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) |
| 0.0 | 0.3 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.1 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.0 | GO:0046980 | tapasin binding(GO:0046980) |
| 0.0 | 0.0 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.1 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.2 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.0 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.0 | 0.1 | GO:0015929 | hexosaminidase activity(GO:0015929) |
| 0.0 | 0.1 | GO:0031432 | titin binding(GO:0031432) |
| 0.0 | 0.1 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.0 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.0 | 0.1 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.1 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.0 | 0.0 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.0 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.0 | 0.1 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.1 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.0 | 0.0 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.1 | GO:0034593 | phosphatidylinositol bisphosphate phosphatase activity(GO:0034593) |
| 0.0 | 0.1 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.1 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.1 | GO:0003709 | obsolete RNA polymerase III transcription factor activity(GO:0003709) |
| 0.0 | 0.1 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.0 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.0 | 0.0 | GO:0015924 | mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.0 | 0.0 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.0 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.0 | 0.0 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.0 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.0 | 0.0 | GO:0005346 | purine ribonucleotide transmembrane transporter activity(GO:0005346) |
| 0.0 | 0.1 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.0 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 1.3 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.2 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.5 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 0.4 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.0 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.1 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.4 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.1 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.1 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.4 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.5 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.0 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.1 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 0.0 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.3 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.1 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.2 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.5 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 0.1 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 0.1 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.2 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 0.1 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 0.2 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.1 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 0.0 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.0 | 0.1 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 0.1 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.4 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.4 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.0 | 0.5 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.0 | 0.7 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.4 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.0 | 0.2 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.2 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.0 | 0.2 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.7 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.0 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 0.1 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.0 | 0.2 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF RAS | Genes involved in CREB phosphorylation through the activation of Ras |
| 0.0 | 0.0 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 0.4 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.2 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 0.3 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.2 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.0 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.1 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.1 | REACTOME IL 2 SIGNALING | Genes involved in Interleukin-2 signaling |
| 0.0 | 0.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.1 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.2 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.1 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.2 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 0.1 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.7 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.0 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.1 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.0 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.0 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.0 | 0.1 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.1 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.3 | REACTOME TCR SIGNALING | Genes involved in TCR signaling |