Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
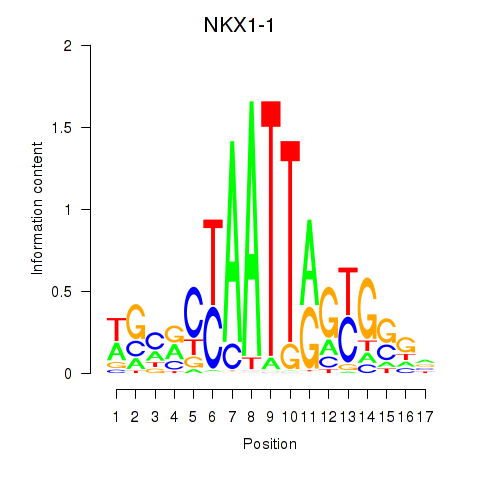
Results for NKX1-1
Z-value: 0.47
Transcription factors associated with NKX1-1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NKX1-1
|
ENSG00000235608.1 | NK1 homeobox 1 |
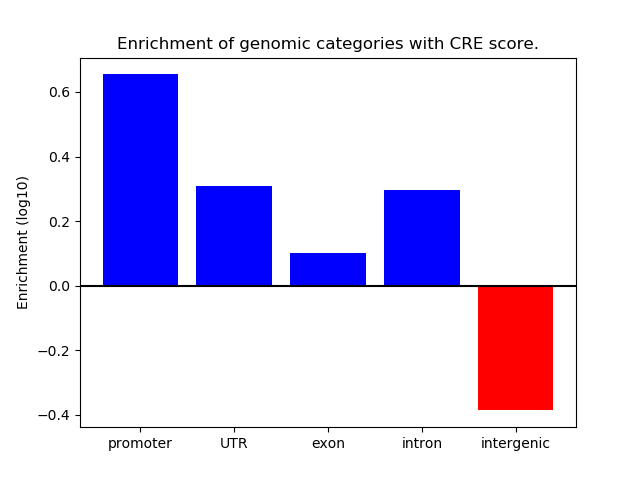
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr4_1397108_1397259 | NKX1-1 | 2936 | 0.204502 | 0.78 | 1.2e-02 | Click! |
| chr4_1523048_1523199 | NKX1-1 | 123004 | 0.041655 | 0.73 | 2.5e-02 | Click! |
| chr4_1396773_1396924 | NKX1-1 | 3271 | 0.192971 | 0.66 | 5.5e-02 | Click! |
| chr4_1523224_1523438 | NKX1-1 | 123212 | 0.041541 | 0.50 | 1.7e-01 | Click! |
| chr4_1396296_1396451 | NKX1-1 | 3746 | 0.181079 | 0.31 | 4.2e-01 | Click! |
Activity of the NKX1-1 motif across conditions
Conditions sorted by the z-value of the NKX1-1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
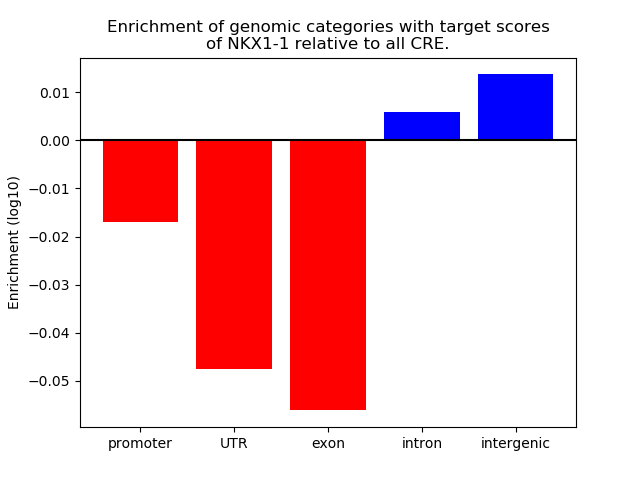
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr10_11155205_11156000 | 0.30 |
CELF2-AS2 |
CELF2 antisense RNA 2 |
15166 |
0.21 |
| chr10_6642170_6642420 | 0.22 |
PRKCQ |
protein kinase C, theta |
20032 |
0.28 |
| chr10_6087007_6087203 | 0.21 |
IL2RA |
interleukin 2 receptor, alpha |
17148 |
0.13 |
| chr4_68440895_68441046 | 0.21 |
STAP1 |
signal transducing adaptor family member 1 |
16524 |
0.2 |
| chr2_202077121_202077490 | 0.19 |
CASP8 |
caspase 8, apoptosis-related cysteine peptidase |
20861 |
0.15 |
| chr5_61873952_61874158 | 0.18 |
LRRC70 |
leucine rich repeat containing 70 |
507 |
0.52 |
| chr5_49959500_49959784 | 0.18 |
PARP8 |
poly (ADP-ribose) polymerase family, member 8 |
2091 |
0.49 |
| chr11_118097693_118098499 | 0.17 |
AMICA1 |
adhesion molecule, interacts with CXADR antigen 1 |
2287 |
0.23 |
| chr9_71643128_71643290 | 0.17 |
FXN |
frataxin |
6966 |
0.23 |
| chr9_134852338_134852532 | 0.16 |
MED27 |
mediator complex subunit 27 |
102818 |
0.07 |
| chr12_104863294_104863607 | 0.16 |
CHST11 |
carbohydrate (chondroitin 4) sulfotransferase 11 |
12671 |
0.27 |
| chr10_44277312_44277574 | 0.16 |
ZNF32 |
zinc finger protein 32 |
133139 |
0.05 |
| chr3_121990121_121990409 | 0.16 |
ENSG00000221474 |
. |
10435 |
0.16 |
| chr9_130623880_130624317 | 0.16 |
ENG |
endoglin |
7063 |
0.08 |
| chr1_173487659_173487906 | 0.15 |
RP3-436N22.3 |
|
36608 |
0.14 |
| chr8_90733427_90733578 | 0.15 |
RIPK2 |
receptor-interacting serine-threonine kinase 2 |
36473 |
0.23 |
| chr18_74872232_74872572 | 0.15 |
MBP |
myelin basic protein |
27602 |
0.24 |
| chr16_72887246_72887397 | 0.15 |
ENSG00000251868 |
. |
31430 |
0.16 |
| chr17_33850293_33850718 | 0.15 |
RP11-1094M14.5 |
|
13216 |
0.11 |
| chr8_19552777_19552928 | 0.15 |
CSGALNACT1 |
chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
12580 |
0.3 |
| chr18_77128932_77129101 | 0.15 |
RP11-800A18.4 |
|
25044 |
0.17 |
| chr12_109290560_109290711 | 0.15 |
DAO |
D-amino-acid oxidase |
16508 |
0.15 |
| chr10_87985596_87985747 | 0.14 |
ENSG00000199104 |
. |
38874 |
0.19 |
| chr10_5858173_5858324 | 0.14 |
GDI2 |
GDP dissociation inhibitor 2 |
1956 |
0.34 |
| chr1_172666866_172667065 | 0.14 |
FASLG |
Fas ligand (TNF superfamily, member 6) |
38807 |
0.19 |
| chr3_47367800_47368066 | 0.14 |
KIF9 |
kinesin family member 9 |
42992 |
0.11 |
| chr3_5469487_5469638 | 0.14 |
ENSG00000241227 |
. |
174654 |
0.03 |
| chr7_37460206_37460357 | 0.14 |
ENSG00000200113 |
. |
1050 |
0.54 |
| chr9_93558751_93558954 | 0.14 |
SYK |
spleen tyrosine kinase |
5217 |
0.35 |
| chr10_7347823_7348170 | 0.14 |
SFMBT2 |
Scm-like with four mbt domains 2 |
102711 |
0.08 |
| chr20_57844647_57845067 | 0.14 |
EDN3 |
endothelin 3 |
30625 |
0.2 |
| chr20_61611257_61611490 | 0.14 |
SLC17A9 |
solute carrier family 17 (vesicular nucleotide transporter), member 9 |
26975 |
0.13 |
| chr1_206225703_206225865 | 0.14 |
AVPR1B |
arginine vasopressin receptor 1B |
1808 |
0.28 |
| chr14_68808631_68808789 | 0.14 |
RAD51B |
RAD51 paralog B |
69517 |
0.12 |
| chr14_71512203_71512354 | 0.13 |
PCNX |
pecanex homolog (Drosophila) |
2399 |
0.43 |
| chr1_117056573_117057331 | 0.13 |
CD58 |
CD58 molecule |
30260 |
0.14 |
| chr2_30557961_30558163 | 0.13 |
ENSG00000221377 |
. |
97252 |
0.07 |
| chr3_15334844_15335113 | 0.13 |
SH3BP5 |
SH3-domain binding protein 5 (BTK-associated) |
17426 |
0.13 |
| chr7_139555811_139556201 | 0.13 |
TBXAS1 |
thromboxane A synthase 1 (platelet) |
26896 |
0.22 |
| chr2_177805314_177805465 | 0.13 |
ENSG00000206866 |
. |
10571 |
0.29 |
| chr11_103747159_103747310 | 0.13 |
ENSG00000264200 |
. |
26600 |
0.23 |
| chr9_36218264_36218537 | 0.13 |
CLTA |
clathrin, light chain A |
27440 |
0.14 |
| chr21_36276778_36277002 | 0.13 |
RUNX1 |
runt-related transcription factor 1 |
14803 |
0.29 |
| chr17_38376031_38376425 | 0.13 |
WIPF2 |
WAS/WASL interacting protein family, member 2 |
124 |
0.95 |
| chr20_11150036_11150187 | 0.13 |
C20orf187 |
chromosome 20 open reading frame 187 |
141300 |
0.05 |
| chr2_38151916_38152120 | 0.13 |
RMDN2 |
regulator of microtubule dynamics 2 |
444 |
0.89 |
| chr2_62433452_62433745 | 0.13 |
ENSG00000266097 |
. |
637 |
0.7 |
| chr20_24936381_24936539 | 0.13 |
CST7 |
cystatin F (leukocystatin) |
6594 |
0.2 |
| chr8_141180516_141180667 | 0.13 |
C8orf17 |
chromosome 8 open reading frame 17 |
237175 |
0.02 |
| chr15_40489916_40490242 | 0.13 |
PAK6 |
p21 protein (Cdc42/Rac)-activated kinase 6 |
19550 |
0.12 |
| chr5_75600048_75600202 | 0.13 |
RP11-466P24.6 |
|
7162 |
0.3 |
| chr17_4615808_4616101 | 0.13 |
ARRB2 |
arrestin, beta 2 |
1960 |
0.15 |
| chr3_193494689_193494840 | 0.13 |
ENSG00000243991 |
. |
36254 |
0.2 |
| chr12_29393294_29393456 | 0.13 |
FAR2 |
fatty acyl CoA reductase 2 |
16697 |
0.24 |
| chr14_65297386_65297537 | 0.12 |
SPTB |
spectrin, beta, erythrocytic |
7595 |
0.2 |
| chr16_73092870_73093114 | 0.12 |
ZFHX3 |
zinc finger homeobox 3 |
605 |
0.79 |
| chr14_70091720_70091880 | 0.12 |
KIAA0247 |
KIAA0247 |
13487 |
0.23 |
| chr4_15471936_15472128 | 0.12 |
CC2D2A |
coiled-coil and C2 domain containing 2A |
395 |
0.86 |
| chr18_9404689_9404840 | 0.12 |
TWSG1 |
twisted gastrulation BMP signaling modulator 1 |
69916 |
0.08 |
| chr1_245344487_245344638 | 0.12 |
KIF26B |
kinesin family member 26B |
26275 |
0.17 |
| chr22_21922789_21923120 | 0.12 |
UBE2L3 |
ubiquitin-conjugating enzyme E2L 3 |
928 |
0.38 |
| chr16_85337462_85337809 | 0.12 |
ENSG00000266307 |
. |
2296 |
0.37 |
| chr5_36842497_36842706 | 0.12 |
NIPBL |
Nipped-B homolog (Drosophila) |
34260 |
0.23 |
| chrX_19795655_19795879 | 0.12 |
SH3KBP1 |
SH3-domain kinase binding protein 1 |
22102 |
0.26 |
| chr5_64332628_64332878 | 0.12 |
ENSG00000207439 |
. |
86443 |
0.1 |
| chr6_53109375_53109667 | 0.12 |
ENSG00000206908 |
. |
25462 |
0.15 |
| chr7_37697149_37697300 | 0.12 |
GPR141 |
G protein-coupled receptor 141 |
26176 |
0.2 |
| chr6_117804732_117805043 | 0.12 |
DCBLD1 |
discoidin, CUB and LCCL domain containing 1 |
1062 |
0.49 |
| chr20_31149162_31149437 | 0.12 |
RP11-410N8.3 |
|
404 |
0.82 |
| chr6_15078179_15078392 | 0.12 |
ENSG00000242989 |
. |
34914 |
0.21 |
| chr3_151963328_151963479 | 0.12 |
MBNL1 |
muscleblind-like splicing regulator 1 |
22426 |
0.21 |
| chr5_61613165_61613316 | 0.12 |
KIF2A |
kinesin heavy chain member 2A |
5600 |
0.28 |
| chr14_101601175_101601448 | 0.12 |
ENSG00000206761 |
. |
10721 |
0.06 |
| chr19_47948776_47948927 | 0.12 |
MEIS3 |
Meis homeobox 3 |
26071 |
0.11 |
| chr14_77873001_77873391 | 0.12 |
FKSG61 |
|
9545 |
0.11 |
| chr1_167458663_167458959 | 0.12 |
CD247 |
CD247 molecule |
28964 |
0.16 |
| chr1_235439552_235439703 | 0.12 |
GGPS1 |
geranylgeranyl diphosphate synthase 1 |
51038 |
0.11 |
| chr20_48955704_48955952 | 0.12 |
ENSG00000244376 |
. |
90192 |
0.08 |
| chr9_92078215_92078535 | 0.12 |
SEMA4D |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D |
7507 |
0.25 |
| chr11_128479669_128479993 | 0.12 |
ETS1 |
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
22378 |
0.2 |
| chr2_207025046_207025348 | 0.11 |
EEF1B2 |
eukaryotic translation elongation factor 1 beta 2 |
866 |
0.28 |
| chr16_17461094_17461491 | 0.11 |
XYLT1 |
xylosyltransferase I |
103446 |
0.08 |
| chr1_117269856_117270128 | 0.11 |
CD2 |
CD2 molecule |
27015 |
0.15 |
| chr16_1013502_1013938 | 0.11 |
LMF1 |
lipase maturation factor 1 |
402 |
0.77 |
| chr3_58292293_58292444 | 0.11 |
RPP14 |
ribonuclease P/MRP 14kDa subunit |
331 |
0.89 |
| chr20_61613987_61614138 | 0.11 |
BHLHE23 |
basic helix-loop-helix family, member e23 |
24325 |
0.13 |
| chr1_27941254_27941405 | 0.11 |
FGR |
feline Gardner-Rasheed sarcoma viral oncogene homolog |
9244 |
0.14 |
| chr5_111109748_111109899 | 0.11 |
NREP |
neuronal regeneration related protein |
15908 |
0.22 |
| chr7_142966484_142967056 | 0.11 |
GSTK1 |
glutathione S-transferase kappa 1 |
6221 |
0.1 |
| chr5_156695529_156695722 | 0.11 |
CYFIP2 |
cytoplasmic FMR1 interacting protein 2 |
737 |
0.55 |
| chr14_91855068_91855898 | 0.11 |
CCDC88C |
coiled-coil domain containing 88C |
28207 |
0.19 |
| chr2_114189862_114190032 | 0.11 |
CBWD2 |
COBW domain containing 2 |
5321 |
0.16 |
| chr20_8349732_8349927 | 0.11 |
PLCB1-IT1 |
PLCB1 intronic transcript 1 (non-protein coding) |
120457 |
0.06 |
| chr6_147204276_147204427 | 0.11 |
STXBP5-AS1 |
STXBP5 antisense RNA 1 |
28391 |
0.24 |
| chr3_16971345_16971496 | 0.11 |
PLCL2 |
phospholipase C-like 2 |
3162 |
0.28 |
| chr9_133711275_133711489 | 0.11 |
ABL1 |
c-abl oncogene 1, non-receptor tyrosine kinase |
929 |
0.63 |
| chr7_123387796_123388107 | 0.11 |
WASL |
Wiskott-Aldrich syndrome-like |
1170 |
0.35 |
| chr3_4951013_4951164 | 0.11 |
BHLHE40-AS1 |
BHLHE40 antisense RNA 1 |
2902 |
0.27 |
| chr9_135382524_135382903 | 0.11 |
BARHL1 |
BarH-like homeobox 1 |
74859 |
0.09 |
| chr20_47334105_47334366 | 0.11 |
ENSG00000251876 |
. |
21750 |
0.26 |
| chr15_91912605_91912789 | 0.11 |
SV2B |
synaptic vesicle glycoprotein 2B |
143597 |
0.05 |
| chr12_54145980_54146199 | 0.10 |
CALCOCO1 |
calcium binding and coiled-coil domain 1 |
24560 |
0.14 |
| chr18_46654785_46654936 | 0.10 |
RP11-15F12.3 |
|
12189 |
0.2 |
| chr2_28848726_28848877 | 0.10 |
PLB1 |
phospholipase B1 |
4652 |
0.23 |
| chr20_3389002_3389295 | 0.10 |
C20orf194 |
chromosome 20 open reading frame 194 |
876 |
0.52 |
| chr5_150595265_150595488 | 0.10 |
GM2A |
GM2 ganglioside activator |
3665 |
0.22 |
| chr3_142347554_142347705 | 0.10 |
PLS1 |
plastin 1 |
5380 |
0.18 |
| chr16_75102873_75103024 | 0.10 |
ZNRF1 |
zinc and ring finger 1, E3 ubiquitin protein ligase |
35734 |
0.13 |
| chr14_69366794_69366948 | 0.10 |
ACTN1 |
actinin, alpha 1 |
10012 |
0.23 |
| chr1_58790005_58790188 | 0.10 |
DAB1 |
Dab, reelin signal transducer, homolog 1 (Drosophila) |
73885 |
0.12 |
| chr2_175533335_175533616 | 0.10 |
WIPF1 |
WAS/WASL interacting protein family, member 1 |
14124 |
0.23 |
| chr13_108979091_108979372 | 0.10 |
ENSG00000223177 |
. |
25552 |
0.22 |
| chr3_18411245_18411586 | 0.10 |
RP11-158G18.1 |
|
39712 |
0.18 |
| chr16_80690009_80690168 | 0.10 |
ENSG00000265341 |
. |
9372 |
0.18 |
| chr16_17376292_17376629 | 0.10 |
CTD-2576D5.4 |
|
148099 |
0.05 |
| chr16_23921554_23921705 | 0.10 |
PRKCB |
protein kinase C, beta |
73085 |
0.11 |
| chr22_40322800_40322970 | 0.10 |
GRAP2 |
GRB2-related adaptor protein 2 |
244 |
0.92 |
| chr5_164987815_164987966 | 0.10 |
ENSG00000252794 |
. |
48556 |
0.2 |
| chr5_49726182_49726490 | 0.10 |
EMB |
embigin |
1775 |
0.54 |
| chr5_59892399_59892550 | 0.10 |
DEPDC1B |
DEP domain containing 1B |
103487 |
0.07 |
| chr17_1462563_1462714 | 0.10 |
PITPNA |
phosphatidylinositol transfer protein, alpha |
457 |
0.74 |
| chr14_95929937_95930315 | 0.10 |
SYNE3 |
spectrin repeat containing, nuclear envelope family member 3 |
7600 |
0.21 |
| chr16_81668859_81669010 | 0.10 |
CMIP |
c-Maf inducing protein |
10029 |
0.29 |
| chr10_50012073_50012379 | 0.10 |
WDFY4 |
WDFY family member 4 |
1443 |
0.49 |
| chr20_49001914_49002090 | 0.10 |
ENSG00000244376 |
. |
44018 |
0.16 |
| chr1_156737883_156738034 | 0.10 |
PRCC |
papillary renal cell carcinoma (translocation-associated) |
612 |
0.57 |
| chr13_96421703_96421854 | 0.10 |
ENSG00000251901 |
. |
54746 |
0.15 |
| chr9_70851374_70851561 | 0.10 |
CBWD3 |
COBW domain containing 3 |
4930 |
0.19 |
| chr16_46798396_46798547 | 0.10 |
MYLK3 |
myosin light chain kinase 3 |
1313 |
0.49 |
| chr3_168335883_168336034 | 0.10 |
ENSG00000207717 |
. |
66316 |
0.14 |
| chr9_97089685_97089867 | 0.10 |
NUTM2F |
NUT family member 2F |
1150 |
0.51 |
| chr1_9685978_9686129 | 0.10 |
PIK3CD |
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta |
25737 |
0.14 |
| chr15_69106923_69107111 | 0.10 |
ANP32A |
acidic (leucine-rich) nuclear phosphoprotein 32 family, member A |
6208 |
0.23 |
| chr4_143624938_143625093 | 0.10 |
INPP4B |
inositol polyphosphate-4-phosphatase, type II, 105kDa |
142428 |
0.05 |
| chr3_187717370_187717716 | 0.10 |
LPP |
LIM domain containing preferred translocation partner in lipoma |
153529 |
0.04 |
| chr2_102239938_102240253 | 0.10 |
MAP4K4 |
mitogen-activated protein kinase kinase kinase kinase 4 |
73217 |
0.12 |
| chr1_68697619_68697806 | 0.10 |
WLS |
wntless Wnt ligand secretion mediator |
251 |
0.94 |
| chr20_56482524_56482675 | 0.10 |
ENSG00000221007 |
. |
11341 |
0.21 |
| chr17_4403592_4403861 | 0.10 |
AC118754.4 |
|
757 |
0.49 |
| chr3_186730308_186730463 | 0.10 |
ST6GAL1 |
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
9258 |
0.24 |
| chr3_128402800_128402979 | 0.10 |
RPN1 |
ribophorin I |
2971 |
0.26 |
| chr14_81423199_81423350 | 0.10 |
TSHR |
thyroid stimulating hormone receptor |
1275 |
0.49 |
| chr11_111317717_111317915 | 0.10 |
RP11-794P6.3 |
|
2777 |
0.15 |
| chr19_55008845_55009031 | 0.10 |
LAIR2 |
leukocyte-associated immunoglobulin-like receptor 2 |
162 |
0.91 |
| chr10_29612226_29612377 | 0.10 |
LYZL1 |
lysozyme-like 1 |
34311 |
0.21 |
| chr7_142024322_142024543 | 0.10 |
PRSS3P3 |
protease, serine, 3 pseudogene 3 |
34823 |
0.18 |
| chr2_8615200_8615353 | 0.10 |
AC011747.7 |
|
200620 |
0.03 |
| chr7_28631139_28631290 | 0.09 |
CREB5 |
cAMP responsive element binding protein 5 |
17464 |
0.3 |
| chr5_79432109_79432283 | 0.09 |
CTC-458I2.2 |
|
8012 |
0.23 |
| chr6_16713160_16713355 | 0.09 |
RP1-151F17.1 |
|
48112 |
0.16 |
| chr12_1613825_1614060 | 0.09 |
WNT5B |
wingless-type MMTV integration site family, member 5B |
25115 |
0.21 |
| chr1_33115546_33115708 | 0.09 |
ZBTB8OS |
zinc finger and BTB domain containing 8 opposite strand |
534 |
0.62 |
| chr13_27867259_27867592 | 0.09 |
RASL11A |
RAS-like, family 11, member A |
22961 |
0.11 |
| chr2_69859573_69859724 | 0.09 |
AAK1 |
AP2 associated kinase 1 |
11138 |
0.21 |
| chr17_40574094_40574415 | 0.09 |
PTRF |
polymerase I and transcript release factor |
1281 |
0.3 |
| chr3_72357618_72357769 | 0.09 |
ENSG00000212070 |
. |
46114 |
0.19 |
| chr14_59950418_59950569 | 0.09 |
L3HYPDH |
L-3-hydroxyproline dehydratase (trans-) |
90 |
0.91 |
| chr16_16193929_16194080 | 0.09 |
ABCC1 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 1 |
90291 |
0.07 |
| chr16_29630080_29630315 | 0.09 |
ENSG00000266758 |
. |
19611 |
0.13 |
| chr2_158768231_158768382 | 0.09 |
ENSG00000251980 |
. |
978 |
0.56 |
| chr1_186268373_186268524 | 0.09 |
PRG4 |
proteoglycan 4 |
3037 |
0.25 |
| chr3_186745073_186745228 | 0.09 |
ST6GAL1 |
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
1879 |
0.42 |
| chr3_153535873_153536024 | 0.09 |
ENSG00000238755 |
. |
55633 |
0.16 |
| chr17_76235442_76235593 | 0.09 |
TMEM235 |
transmembrane protein 235 |
7395 |
0.14 |
| chr8_133770079_133770329 | 0.09 |
TMEM71 |
transmembrane protein 71 |
2590 |
0.28 |
| chr12_68028301_68028534 | 0.09 |
DYRK2 |
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 2 |
13701 |
0.26 |
| chr5_145583999_145584318 | 0.09 |
RBM27 |
RNA binding motif protein 27 |
995 |
0.56 |
| chr1_200120948_200121380 | 0.09 |
ENSG00000221403 |
. |
7202 |
0.28 |
| chr18_74182005_74182224 | 0.09 |
ZNF516 |
zinc finger protein 516 |
20621 |
0.16 |
| chr3_187840204_187840389 | 0.09 |
LPP |
LIM domain containing preferred translocation partner in lipoma |
30776 |
0.18 |
| chr7_139315604_139315856 | 0.09 |
HIPK2 |
homeodomain interacting protein kinase 2 |
106111 |
0.06 |
| chr2_225806667_225806962 | 0.09 |
DOCK10 |
dedicator of cytokinesis 10 |
4968 |
0.33 |
| chr8_49862673_49862887 | 0.09 |
SNAI2 |
snail family zinc finger 2 |
28481 |
0.25 |
| chr20_40221307_40221497 | 0.09 |
CHD6 |
chromodomain helicase DNA binding protein 6 |
22010 |
0.23 |
| chr18_5709587_5709738 | 0.09 |
ENSG00000252432 |
. |
47613 |
0.15 |
| chr1_236284257_236284560 | 0.09 |
GPR137B |
G protein-coupled receptor 137B |
21424 |
0.19 |
| chr4_185201552_185201820 | 0.09 |
ENSG00000244512 |
. |
1597 |
0.41 |
| chr10_98411356_98411597 | 0.09 |
PIK3AP1 |
phosphoinositide-3-kinase adaptor protein 1 |
17892 |
0.19 |
| chr8_80733314_80733465 | 0.09 |
ENSG00000238884 |
. |
20959 |
0.23 |
| chr18_13260507_13260658 | 0.09 |
LDLRAD4 |
low density lipoprotein receptor class A domain containing 4 |
17520 |
0.17 |
| chr20_31261981_31262132 | 0.09 |
COMMD7 |
COMM domain containing 7 |
69156 |
0.08 |
| chr9_33154550_33154783 | 0.09 |
RP11-326F20.5 |
|
12307 |
0.14 |
| chr12_65108821_65108972 | 0.09 |
AC025262.1 |
Mesenchymal stem cell protein DSC96; Uncharacterized protein |
18567 |
0.13 |
| chr5_98103966_98104240 | 0.09 |
RGMB |
repulsive guidance molecule family member b |
896 |
0.64 |
| chr2_89167006_89167157 | 0.09 |
IGKJ1 |
immunoglobulin kappa joining 1 |
5646 |
0.09 |
| chr1_174929832_174929983 | 0.09 |
RABGAP1L |
RAB GTPase activating protein 1-like |
3998 |
0.18 |
| chr7_135656817_135657003 | 0.09 |
MTPN |
myotrophin |
5154 |
0.19 |
| chr17_73381904_73382055 | 0.09 |
GRB2 |
growth factor receptor-bound protein 2 |
7758 |
0.1 |
| chr3_16384687_16384897 | 0.09 |
RP11-415F23.4 |
|
1841 |
0.32 |
| chr1_179801518_179801669 | 0.09 |
RP11-12M5.3 |
|
14267 |
0.16 |
| chr3_39330707_39330989 | 0.09 |
CX3CR1 |
chemokine (C-X3-C motif) receptor 1 |
7622 |
0.2 |
| chr10_64515756_64515907 | 0.09 |
ADO |
2-aminoethanethiol (cysteamine) dioxygenase |
48685 |
0.13 |
| chr11_110349538_110349689 | 0.09 |
FDX1 |
ferredoxin 1 |
49006 |
0.18 |
| chr21_26782848_26782999 | 0.09 |
ENSG00000238314 |
. |
59590 |
0.13 |
| chr20_57828749_57828900 | 0.09 |
EDN3 |
endothelin 3 |
46658 |
0.15 |
| chr8_57126457_57126608 | 0.08 |
CHCHD7 |
coiled-coil-helix-coiled-coil-helix domain containing 7 |
455 |
0.81 |
| chr5_76696412_76696563 | 0.08 |
PDE8B |
phosphodiesterase 8B |
9762 |
0.26 |
| chr3_71862606_71862965 | 0.08 |
ENSG00000239250 |
. |
13543 |
0.2 |
| chr16_54033588_54033739 | 0.08 |
RP11-357N13.2 |
|
218 |
0.93 |
| chr2_29593489_29593640 | 0.08 |
ENSG00000266310 |
. |
133295 |
0.05 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0032632 | interleukin-3 production(GO:0032632) |
| 0.0 | 0.2 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.1 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.0 | 0.2 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.1 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.0 | 0.1 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.1 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.1 | GO:0031946 | regulation of glucocorticoid biosynthetic process(GO:0031946) |
| 0.0 | 0.1 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.0 | 0.1 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.0 | GO:0042109 | lymphotoxin A production(GO:0032641) lymphotoxin A biosynthetic process(GO:0042109) |
| 0.0 | 0.0 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.0 | GO:1900120 | regulation of receptor binding(GO:1900120) negative regulation of receptor binding(GO:1900121) |
| 0.0 | 0.1 | GO:0055064 | cellular chloride ion homeostasis(GO:0030644) chloride ion homeostasis(GO:0055064) |
| 0.0 | 0.1 | GO:0090400 | stress-induced premature senescence(GO:0090400) |
| 0.0 | 0.1 | GO:0034086 | maintenance of sister chromatid cohesion(GO:0034086) maintenance of mitotic sister chromatid cohesion(GO:0034088) |
| 0.0 | 0.0 | GO:0071224 | cellular response to peptidoglycan(GO:0071224) |
| 0.0 | 0.0 | GO:0002328 | pro-B cell differentiation(GO:0002328) |
| 0.0 | 0.0 | GO:0001865 | NK T cell differentiation(GO:0001865) regulation of NK T cell differentiation(GO:0051136) positive regulation of NK T cell differentiation(GO:0051138) |
| 0.0 | 0.1 | GO:0001911 | negative regulation of leukocyte mediated cytotoxicity(GO:0001911) |
| 0.0 | 0.0 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 0.0 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.0 | 0.0 | GO:0009296 | obsolete flagellum assembly(GO:0009296) |
| 0.0 | 0.0 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.0 | GO:0021542 | dentate gyrus development(GO:0021542) |
| 0.0 | 0.1 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.0 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.0 | 0.1 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 | 0.1 | GO:0051299 | centrosome separation(GO:0051299) |
| 0.0 | 0.0 | GO:0002693 | positive regulation of cellular extravasation(GO:0002693) |
| 0.0 | 0.0 | GO:0010748 | negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) negative regulation of anion transmembrane transport(GO:1903960) negative regulation of fatty acid transport(GO:2000192) |
| 0.0 | 0.1 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.0 | 0.0 | GO:0006551 | leucine metabolic process(GO:0006551) |
| 0.0 | 0.0 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.1 | GO:1901503 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.0 | GO:0046010 | positive regulation of circadian sleep/wake cycle, non-REM sleep(GO:0046010) |
| 0.0 | 0.1 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.0 | GO:0006924 | activation-induced cell death of T cells(GO:0006924) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.0 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.1 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.1 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.0 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.1 | GO:0071778 | obsolete WINAC complex(GO:0071778) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0016149 | translation release factor activity, codon specific(GO:0016149) |
| 0.0 | 0.1 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.1 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.1 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.0 | 0.1 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.2 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.0 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.1 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.2 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.0 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0004904 | interferon receptor activity(GO:0004904) |
| 0.0 | 0.1 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.1 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.0 | GO:0031705 | bombesin receptor binding(GO:0031705) endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.0 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.0 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.0 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.0 | GO:0042608 | T cell receptor binding(GO:0042608) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.2 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.0 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.4 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.0 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |