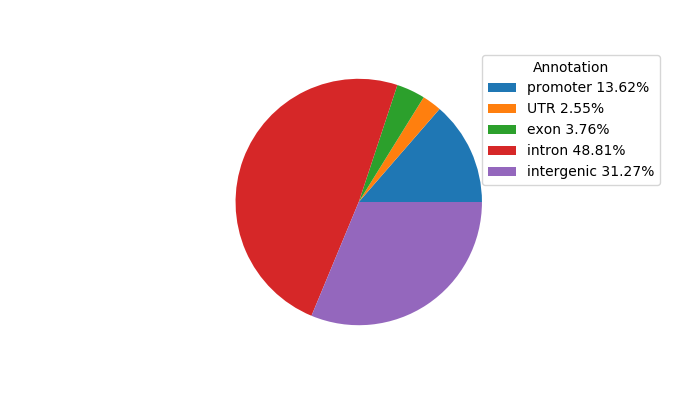
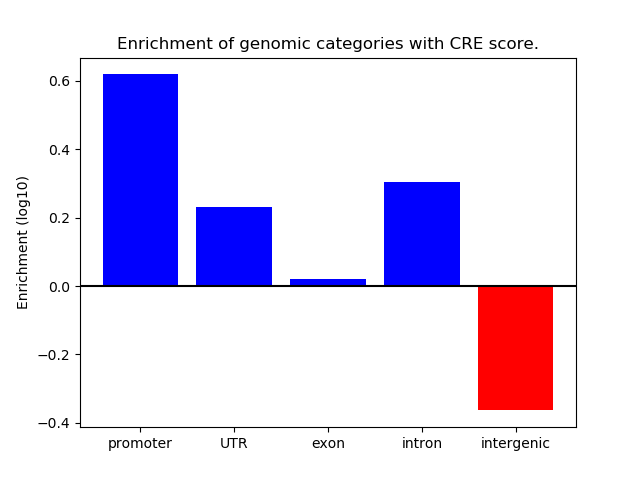
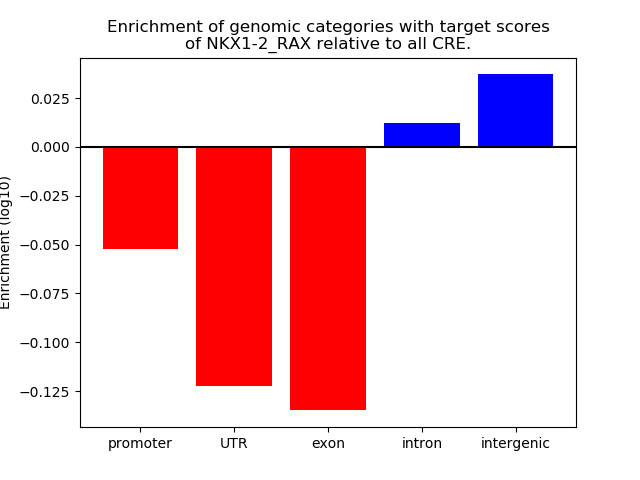
Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
Results for NKX1-2_RAX
Z-value: 0.72
Transcription factors associated with NKX1-2_RAX
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
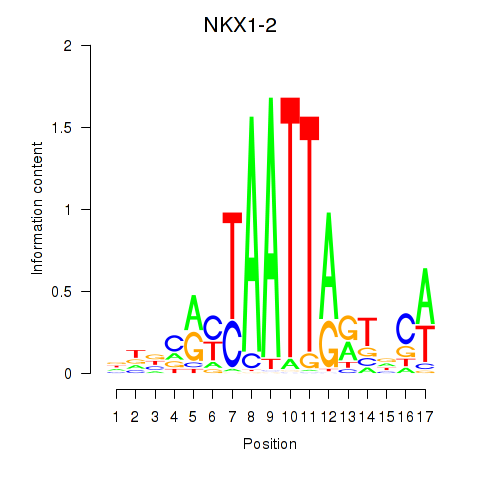
NKX1-2
|
ENSG00000229544.6 | NK1 homeobox 2 |
|
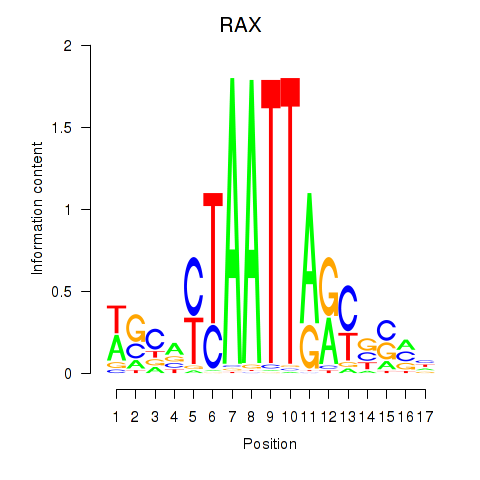
RAX
|
ENSG00000134438.9 | retina and anterior neural fold homeobox |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr10_126138239_126138552 | NKX1-2 | 155 | 0.942068 | 0.43 | 2.5e-01 | Click! |
| chr10_126137738_126137889 | NKX1-2 | 737 | 0.564633 | 0.33 | 3.8e-01 | Click! |
| chr10_126137330_126137616 | NKX1-2 | 1077 | 0.371726 | 0.31 | 4.2e-01 | Click! |
| chr10_126138898_126139049 | NKX1-2 | 220 | 0.919742 | 0.21 | 5.9e-01 | Click! |
| chr10_126137966_126138119 | NKX1-2 | 508 | 0.725484 | 0.01 | 9.8e-01 | Click! |
| chr18_56938469_56938620 | RAX | 2073 | 0.349346 | -0.61 | 8.0e-02 | Click! |
| chr18_56935922_56936202 | RAX | 4555 | 0.241904 | 0.52 | 1.5e-01 | Click! |
| chr18_56936662_56936813 | RAX | 3880 | 0.254334 | 0.42 | 2.6e-01 | Click! |
| chr18_56939397_56939548 | RAX | 1145 | 0.534613 | 0.40 | 2.9e-01 | Click! |
| chr18_56936206_56936357 | RAX | 4336 | 0.245372 | 0.39 | 3.0e-01 | Click! |
Activity of the NKX1-2_RAX motif across conditions
Conditions sorted by the z-value of the NKX1-2_RAX motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
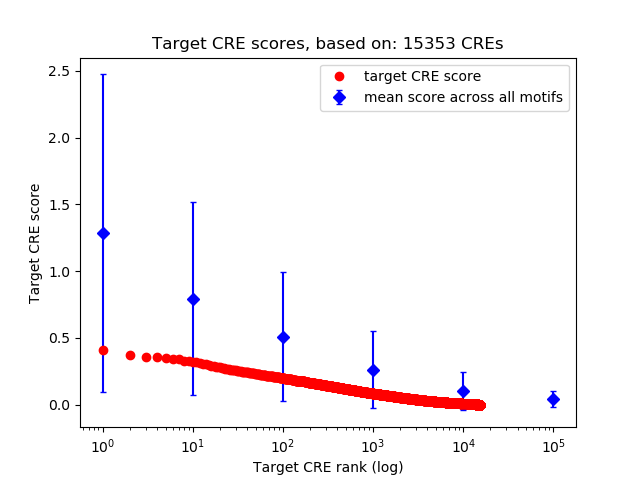
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr6_117804732_117805043 | 0.41 |
DCBLD1 |
discoidin, CUB and LCCL domain containing 1 |
1062 |
0.49 |
| chr2_202077121_202077490 | 0.37 |
CASP8 |
caspase 8, apoptosis-related cysteine peptidase |
20861 |
0.15 |
| chr6_25005285_25005494 | 0.36 |
ENSG00000244618 |
. |
26121 |
0.17 |
| chr5_124214579_124214730 | 0.36 |
ZNF608 |
zinc finger protein 608 |
130154 |
0.05 |
| chr9_134852338_134852532 | 0.35 |
MED27 |
mediator complex subunit 27 |
102818 |
0.07 |
| chr17_72457353_72457898 | 0.34 |
CD300A |
CD300a molecule |
4930 |
0.16 |
| chr18_74872232_74872572 | 0.34 |
MBP |
myelin basic protein |
27602 |
0.24 |
| chr5_73534753_73534915 | 0.33 |
ENSG00000222551 |
. |
32756 |
0.22 |
| chr9_123424941_123425186 | 0.33 |
MEGF9 |
multiple EGF-like-domains 9 |
51549 |
0.15 |
| chr1_87082986_87083335 | 0.32 |
ENSG00000221702 |
. |
11120 |
0.18 |
| chr14_75795544_75795840 | 0.32 |
FOS |
FBJ murine osteosarcoma viral oncogene homolog |
48796 |
0.11 |
| chr2_160789612_160789827 | 0.31 |
LY75-CD302 |
LY75-CD302 readthrough |
28498 |
0.19 |
| chr4_140724600_140724880 | 0.31 |
ENSG00000252233 |
. |
9153 |
0.26 |
| chr12_1725180_1725331 | 0.31 |
WNT5B |
wingless-type MMTV integration site family, member 5B |
967 |
0.58 |
| chr3_187491464_187491692 | 0.30 |
BCL6 |
B-cell CLL/lymphoma 6 |
28063 |
0.2 |
| chr2_64814973_64815124 | 0.29 |
ENSG00000252414 |
. |
9022 |
0.22 |
| chr1_175157741_175158025 | 0.29 |
KIAA0040 |
KIAA0040 |
4007 |
0.31 |
| chr3_112193935_112194086 | 0.28 |
BTLA |
B and T lymphocyte associated |
24195 |
0.21 |
| chr13_41548373_41548524 | 0.28 |
ELF1 |
E74-like factor 1 (ets domain transcription factor) |
7970 |
0.2 |
| chr2_68650725_68651008 | 0.28 |
FBXO48 |
F-box protein 48 |
43524 |
0.12 |
| chr18_13124723_13124874 | 0.28 |
RP11-794M8.1 |
|
91526 |
0.07 |
| chr10_11155205_11156000 | 0.28 |
CELF2-AS2 |
CELF2 antisense RNA 2 |
15166 |
0.21 |
| chr14_92973076_92973375 | 0.27 |
RIN3 |
Ras and Rab interactor 3 |
6893 |
0.29 |
| chr4_14996539_14996690 | 0.27 |
CPEB2 |
cytoplasmic polyadenylation element binding protein 2 |
7684 |
0.31 |
| chr8_142160611_142160762 | 0.27 |
DENND3 |
DENN/MADD domain containing 3 |
9384 |
0.2 |
| chr7_139555811_139556201 | 0.26 |
TBXAS1 |
thromboxane A synthase 1 (platelet) |
26896 |
0.22 |
| chr2_202861641_202861792 | 0.26 |
FZD7 |
frizzled family receptor 7 |
37594 |
0.13 |
| chr1_89949288_89949439 | 0.26 |
LRRC8B |
leucine rich repeat containing 8 family, member B |
41032 |
0.16 |
| chr4_153608932_153609083 | 0.26 |
TMEM154 |
transmembrane protein 154 |
7690 |
0.22 |
| chr10_21807059_21807447 | 0.26 |
SKIDA1 |
SKI/DACH domain containing 1 |
405 |
0.78 |
| chr15_92455349_92455602 | 0.25 |
SLCO3A1 |
solute carrier organic anion transporter family, member 3A1 |
58124 |
0.15 |
| chr8_27223358_27224128 | 0.25 |
PTK2B |
protein tyrosine kinase 2 beta |
14425 |
0.22 |
| chr14_103586056_103586207 | 0.25 |
TNFAIP2 |
tumor necrosis factor, alpha-induced protein 2 |
3667 |
0.2 |
| chr2_153267355_153267720 | 0.25 |
FMNL2 |
formin-like 2 |
75786 |
0.12 |
| chr18_61561537_61561693 | 0.25 |
SERPINB10 |
serpin peptidase inhibitor, clade B (ovalbumin), member 10 |
2793 |
0.24 |
| chr5_55443907_55444286 | 0.25 |
ENSG00000223003 |
. |
1423 |
0.42 |
| chr5_59527719_59528015 | 0.25 |
PDE4D |
phosphodiesterase 4D, cAMP-specific |
46442 |
0.18 |
| chr4_108721534_108721685 | 0.25 |
SGMS2 |
sphingomyelin synthase 2 |
24110 |
0.21 |
| chr14_70110903_70111120 | 0.25 |
KIAA0247 |
KIAA0247 |
32698 |
0.18 |
| chr1_200120948_200121380 | 0.25 |
ENSG00000221403 |
. |
7202 |
0.28 |
| chr2_225451534_225451729 | 0.24 |
CUL3 |
cullin 3 |
1521 |
0.57 |
| chr5_17413551_17413702 | 0.24 |
ENSG00000201715 |
. |
67901 |
0.13 |
| chr7_102072492_102072712 | 0.24 |
ORAI2 |
ORAI calcium release-activated calcium modulator 2 |
951 |
0.34 |
| chr1_246233834_246234131 | 0.24 |
RP11-83A16.1 |
|
37129 |
0.17 |
| chr22_27836384_27836535 | 0.24 |
RP11-375H17.1 |
|
276009 |
0.01 |
| chr7_80742262_80742413 | 0.24 |
AC005008.2 |
Uncharacterized protein |
62487 |
0.16 |
| chr1_90290727_90290878 | 0.24 |
LRRC8D |
leucine rich repeat containing 8 family, member D |
3322 |
0.28 |
| chr17_40207202_40207353 | 0.24 |
ZNF385C |
zinc finger protein 385C |
7798 |
0.1 |
| chr3_142347554_142347705 | 0.23 |
PLS1 |
plastin 1 |
5380 |
0.18 |
| chr10_74579021_74579203 | 0.23 |
RP11-354E23.5 |
|
52794 |
0.12 |
| chr8_48296668_48296819 | 0.23 |
SPIDR |
scaffolding protein involved in DNA repair |
56220 |
0.16 |
| chrX_39003232_39003473 | 0.23 |
ENSG00000207122 |
. |
62814 |
0.16 |
| chr17_8311463_8311614 | 0.23 |
NDEL1 |
nudE neurodevelopment protein 1-like 1 |
4911 |
0.12 |
| chr4_130543160_130543402 | 0.23 |
ENSG00000222861 |
. |
238374 |
0.02 |
| chr8_130559417_130559631 | 0.23 |
ENSG00000266387 |
. |
49611 |
0.09 |
| chr16_73092870_73093114 | 0.23 |
ZFHX3 |
zinc finger homeobox 3 |
605 |
0.79 |
| chr5_36842497_36842706 | 0.22 |
NIPBL |
Nipped-B homolog (Drosophila) |
34260 |
0.23 |
| chr1_245120739_245120890 | 0.22 |
ENSG00000252073 |
. |
2654 |
0.2 |
| chr20_1550565_1550716 | 0.22 |
SIRPB1 |
signal-regulatory protein beta 1 |
1918 |
0.25 |
| chr4_26729613_26729764 | 0.22 |
ENSG00000200999 |
. |
25757 |
0.23 |
| chr3_121990121_121990409 | 0.22 |
ENSG00000221474 |
. |
10435 |
0.16 |
| chr20_18487226_18487377 | 0.22 |
SEC23B |
Sec23 homolog B (S. cerevisiae) |
836 |
0.5 |
| chr21_48025165_48025458 | 0.22 |
S100B |
S100 calcium binding protein B |
190 |
0.95 |
| chr2_45490582_45490733 | 0.22 |
SIX2 |
SIX homeobox 2 |
254088 |
0.02 |
| chr12_10282940_10283100 | 0.22 |
CLEC7A |
C-type lectin domain family 7, member A |
164 |
0.93 |
| chr16_4674083_4674234 | 0.22 |
MGRN1 |
mahogunin ring finger 1, E3 ubiquitin protein ligase |
633 |
0.59 |
| chr10_45559099_45559250 | 0.22 |
RSU1P2 |
Ras suppressor protein 1 pseudogene 2 |
52375 |
0.1 |
| chr12_55435891_55436042 | 0.22 |
NEUROD4 |
neuronal differentiation 4 |
22237 |
0.2 |
| chr16_50720309_50720716 | 0.22 |
SNX20 |
sorting nexin 20 |
5248 |
0.13 |
| chr12_25208694_25208845 | 0.22 |
LRMP |
lymphoid-restricted membrane protein |
3095 |
0.27 |
| chr15_93372987_93373270 | 0.22 |
FAM174B |
family with sequence similarity 174, member B |
20100 |
0.15 |
| chr2_8038740_8038891 | 0.22 |
ENSG00000221255 |
. |
321843 |
0.01 |
| chr18_19178052_19178356 | 0.21 |
ESCO1 |
establishment of sister chromatid cohesion N-acetyltransferase 1 |
2493 |
0.23 |
| chr16_17376292_17376629 | 0.21 |
CTD-2576D5.4 |
|
148099 |
0.05 |
| chr5_75600048_75600202 | 0.21 |
RP11-466P24.6 |
|
7162 |
0.3 |
| chr8_56885039_56885250 | 0.21 |
ENSG00000240905 |
. |
7952 |
0.16 |
| chr1_169663386_169663580 | 0.21 |
SELL |
selectin L |
17356 |
0.18 |
| chr1_169571123_169571274 | 0.21 |
F5 |
coagulation factor V (proaccelerin, labile factor) |
15372 |
0.19 |
| chr5_75688534_75688866 | 0.21 |
IQGAP2 |
IQ motif containing GTPase activating protein 2 |
10374 |
0.28 |
| chr7_28712663_28712814 | 0.21 |
CREB5 |
cAMP responsive element binding protein 5 |
12860 |
0.31 |
| chr7_69119865_69120016 | 0.21 |
AUTS2 |
autism susceptibility candidate 2 |
55343 |
0.16 |
| chr4_101813891_101814042 | 0.21 |
EMCN-IT1 |
EMCN intronic transcript 1 (non-protein coding) |
78529 |
0.11 |
| chr11_60250640_60250791 | 0.21 |
MS4A12 |
membrane-spanning 4-domains, subfamily A, member 12 |
9536 |
0.16 |
| chr2_172008488_172008813 | 0.21 |
ENSG00000238567 |
. |
5810 |
0.25 |
| chr10_7299593_7299834 | 0.21 |
SFMBT2 |
Scm-like with four mbt domains 2 |
150994 |
0.04 |
| chr9_92032244_92032395 | 0.21 |
SEMA4D |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D |
1429 |
0.51 |
| chr15_94796190_94796879 | 0.21 |
MCTP2 |
multiple C2 domains, transmembrane 2 |
21767 |
0.29 |
| chr20_61070311_61070462 | 0.21 |
GATA5 |
GATA binding protein 5 |
19360 |
0.15 |
| chr4_157891332_157891565 | 0.21 |
PDGFC |
platelet derived growth factor C |
607 |
0.78 |
| chr21_16064096_16064532 | 0.20 |
AF165138.7 |
Protein LOC388813 |
33172 |
0.21 |
| chr11_9715214_9715365 | 0.20 |
ENSG00000201564 |
. |
12818 |
0.18 |
| chr15_101790476_101790819 | 0.20 |
CHSY1 |
chondroitin sulfate synthase 1 |
1490 |
0.37 |
| chr4_36305674_36305831 | 0.20 |
DTHD1 |
death domain containing 1 |
20108 |
0.21 |
| chr15_48251538_48251689 | 0.20 |
SLC24A5 |
solute carrier family 24 (sodium/potassium/calcium exchanger), member 5 |
161556 |
0.03 |
| chr3_128402800_128402979 | 0.20 |
RPN1 |
ribophorin I |
2971 |
0.26 |
| chr2_222385882_222386088 | 0.20 |
ENSG00000221432 |
. |
1608 |
0.44 |
| chr7_133563283_133563434 | 0.20 |
EXOC4 |
exocyst complex component 4 |
51864 |
0.19 |
| chr6_91584945_91585096 | 0.20 |
MAP3K7 |
mitogen-activated protein kinase kinase kinase 7 |
288256 |
0.01 |
| chr1_9741200_9741351 | 0.20 |
RP11-558F24.4 |
|
6338 |
0.18 |
| chr9_137251018_137251277 | 0.20 |
ENSG00000263897 |
. |
20110 |
0.22 |
| chr3_14467854_14468179 | 0.20 |
SLC6A6 |
solute carrier family 6 (neurotransmitter transporter), member 6 |
6162 |
0.25 |
| chr10_14584774_14585305 | 0.20 |
FAM107B |
family with sequence similarity 107, member B |
5630 |
0.27 |
| chr8_96160918_96161069 | 0.20 |
PLEKHF2 |
pleckstrin homology domain containing, family F (with FYVE domain) member 2 |
14689 |
0.21 |
| chr14_70168130_70168306 | 0.20 |
SRSF5 |
serine/arginine-rich splicing factor 5 |
25399 |
0.21 |
| chr2_175207474_175207756 | 0.20 |
AC018470.1 |
Uncharacterized protein FLJ46347 |
5464 |
0.18 |
| chr3_194601465_194601616 | 0.20 |
FAM43A |
family with sequence similarity 43, member A |
194918 |
0.02 |
| chr7_80219632_80219783 | 0.20 |
CD36 |
CD36 molecule (thrombospondin receptor) |
11759 |
0.29 |
| chr5_59558878_59559029 | 0.20 |
ENSG00000202017 |
. |
41025 |
0.2 |
| chr17_63043618_63043821 | 0.20 |
RP11-583F2.5 |
|
4308 |
0.21 |
| chr2_169927871_169928022 | 0.20 |
DHRS9 |
dehydrogenase/reductase (SDR family) member 9 |
1064 |
0.56 |
| chr7_41527568_41527719 | 0.20 |
INHBA-AS1 |
INHBA antisense RNA 1 |
205871 |
0.03 |
| chr2_176418395_176418546 | 0.20 |
ENSG00000221347 |
. |
223369 |
0.02 |
| chr2_163263039_163263190 | 0.20 |
GCA |
grancalcin, EF-hand calcium binding protein |
50166 |
0.15 |
| chr7_16818603_16818754 | 0.20 |
AC073333.1 |
Uncharacterized protein |
10188 |
0.17 |
| chr17_72033410_72033561 | 0.19 |
RPL38 |
ribosomal protein L38 |
166236 |
0.03 |
| chr14_72064329_72064928 | 0.19 |
SIPA1L1 |
signal-induced proliferation-associated 1 like 1 |
323 |
0.94 |
| chr16_28942567_28942718 | 0.19 |
CD19 |
CD19 molecule |
618 |
0.49 |
| chr2_8007405_8007668 | 0.19 |
ENSG00000221255 |
. |
290564 |
0.01 |
| chr8_134075895_134076274 | 0.19 |
SLA |
Src-like-adaptor |
3481 |
0.31 |
| chr8_80873858_80874009 | 0.19 |
RP11-26J3.3 |
|
4317 |
0.28 |
| chr6_54888900_54889051 | 0.19 |
RP3-523K23.2 |
|
81010 |
0.11 |
| chr2_26634709_26634860 | 0.19 |
DRC1 |
dynein regulatory complex subunit 1 homolog (Chlamydomonas) |
10000 |
0.21 |
| chr6_17700032_17700183 | 0.19 |
RP11-500C11.3 |
|
6381 |
0.17 |
| chrX_147584575_147584726 | 0.19 |
AFF2 |
AF4/FMR2 family, member 2 |
2121 |
0.31 |
| chr3_37010622_37010773 | 0.19 |
ENSG00000210181 |
. |
890 |
0.56 |
| chr12_104940888_104941108 | 0.19 |
CHST11 |
carbohydrate (chondroitin 4) sulfotransferase 11 |
41628 |
0.17 |
| chr8_19552777_19552928 | 0.19 |
CSGALNACT1 |
chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
12580 |
0.3 |
| chr4_38663643_38663794 | 0.19 |
KLF3 |
Kruppel-like factor 3 (basic) |
2099 |
0.25 |
| chr18_3790975_3791126 | 0.19 |
RP11-874J12.3 |
|
19620 |
0.17 |
| chr6_144171782_144171933 | 0.19 |
LTV1 |
LTV1 homolog (S. cerevisiae) |
7376 |
0.24 |
| chr12_121548159_121548376 | 0.19 |
ENSG00000201945 |
. |
3421 |
0.22 |
| chr8_56446230_56446381 | 0.18 |
RP11-628E19.2 |
|
12769 |
0.28 |
| chr10_94452668_94452819 | 0.18 |
HHEX |
hematopoietically expressed homeobox |
1131 |
0.51 |
| chr4_124341560_124341711 | 0.18 |
SPRY1 |
sprouty homolog 1, antagonist of FGF signaling (Drosophila) |
20512 |
0.29 |
| chr22_17956676_17956827 | 0.18 |
CECR2 |
cat eye syndrome chromosome region, candidate 2 |
124 |
0.97 |
| chr1_117056573_117057331 | 0.18 |
CD58 |
CD58 molecule |
30260 |
0.14 |
| chr3_177443249_177443545 | 0.18 |
ENSG00000200288 |
. |
101200 |
0.08 |
| chr11_127376331_127376497 | 0.18 |
ENSG00000223315 |
. |
98668 |
0.08 |
| chr1_233218809_233218980 | 0.18 |
PCNXL2 |
pecanex-like 2 (Drosophila) |
24630 |
0.26 |
| chr13_32523758_32523999 | 0.18 |
EEF1DP3 |
eukaryotic translation elongation factor 1 delta pseudogene 3 |
2839 |
0.36 |
| chr5_140934914_140935065 | 0.18 |
CTD-2024I7.13 |
|
2889 |
0.17 |
| chr10_131583616_131583767 | 0.18 |
EBF3 |
early B-cell factor 3 |
56744 |
0.14 |
| chr1_172442879_172443090 | 0.18 |
C1orf105 |
chromosome 1 open reading frame 105 |
20951 |
0.18 |
| chr6_24523019_24523170 | 0.18 |
ALDH5A1 |
aldehyde dehydrogenase 5 family, member A1 |
27897 |
0.17 |
| chr2_122458404_122458555 | 0.18 |
ENSG00000238341 |
. |
5154 |
0.2 |
| chr4_74945123_74945274 | 0.18 |
CXCL2 |
chemokine (C-X-C motif) ligand 2 |
19812 |
0.13 |
| chr6_35002148_35002432 | 0.18 |
TCP11 |
t-complex 11, testis-specific |
86532 |
0.08 |
| chr5_64919668_64919819 | 0.18 |
TRIM23 |
tripartite motif containing 23 |
380 |
0.59 |
| chr12_82343405_82343568 | 0.18 |
PPFIA2 |
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
190154 |
0.03 |
| chr10_76625035_76625298 | 0.18 |
KAT6B |
K(lysine) acetyltransferase 6B |
26708 |
0.24 |
| chr1_212325573_212325724 | 0.18 |
ENSG00000252879 |
. |
52630 |
0.12 |
| chr12_115941638_115941789 | 0.18 |
RP11-116D17.1 |
HCG2038717; Uncharacterized protein |
140896 |
0.05 |
| chr4_89526439_89526704 | 0.18 |
HERC3 |
HECT and RLD domain containing E3 ubiquitin protein ligase 3 |
374 |
0.86 |
| chr17_55827874_55828025 | 0.18 |
CCDC182 |
coiled-coil domain containing 182 |
5276 |
0.22 |
| chr16_3146077_3146254 | 0.18 |
ZSCAN10 |
zinc finger and SCAN domain containing 10 |
3064 |
0.09 |
| chr12_54150663_54151040 | 0.18 |
CALCOCO1 |
calcium binding and coiled-coil domain 1 |
29322 |
0.13 |
| chr6_47207680_47207905 | 0.18 |
TNFRSF21 |
tumor necrosis factor receptor superfamily, member 21 |
69849 |
0.13 |
| chr4_75466339_75466608 | 0.18 |
RP11-727M10.2 |
|
9499 |
0.2 |
| chr19_37288631_37289102 | 0.18 |
CTD-2162K18.5 |
|
401 |
0.8 |
| chr15_78462261_78462412 | 0.18 |
IDH3A |
isocitrate dehydrogenase 3 (NAD+) alpha |
6278 |
0.16 |
| chr1_226084453_226084634 | 0.18 |
LEFTY1 |
left-right determination factor 1 |
7697 |
0.12 |
| chr4_75296529_75296796 | 0.18 |
RP11-727M10.1 |
|
9512 |
0.2 |
| chr1_99337480_99337815 | 0.18 |
RP5-896L10.1 |
|
132185 |
0.05 |
| chr10_121459914_121460065 | 0.18 |
INPP5F |
inositol polyphosphate-5-phosphatase F |
25620 |
0.18 |
| chr14_70111444_70111741 | 0.18 |
KIAA0247 |
KIAA0247 |
33279 |
0.18 |
| chr12_64798932_64799366 | 0.18 |
XPOT |
exportin, tRNA |
323 |
0.88 |
| chrX_20456293_20456479 | 0.18 |
ENSG00000252978 |
. |
13840 |
0.3 |
| chr10_77695413_77695615 | 0.18 |
ENSG00000215921 |
. |
111978 |
0.06 |
| chr15_83733229_83733380 | 0.17 |
BTBD1 |
BTB (POZ) domain containing 1 |
2599 |
0.17 |
| chr10_131555094_131555311 | 0.17 |
RP11-109A6.2 |
|
55381 |
0.15 |
| chr13_99718141_99718440 | 0.17 |
DOCK9 |
dedicator of cytokinesis 9 |
20370 |
0.18 |
| chrX_119739145_119739532 | 0.17 |
MCTS1 |
malignant T cell amplified sequence 1 |
1162 |
0.48 |
| chr14_78120000_78120151 | 0.17 |
SPTLC2 |
serine palmitoyltransferase, long chain base subunit 2 |
36959 |
0.13 |
| chr2_112368076_112368227 | 0.17 |
ENSG00000266063 |
. |
160560 |
0.04 |
| chr6_51167339_51167725 | 0.17 |
ENSG00000212532 |
. |
161956 |
0.04 |
| chr16_10847383_10847602 | 0.17 |
NUBP1 |
nucleotide binding protein 1 |
4238 |
0.21 |
| chr11_77223581_77223900 | 0.17 |
PAK1 |
p21 protein (Cdc42/Rac)-activated kinase 1 |
38060 |
0.13 |
| chr10_72746069_72746220 | 0.17 |
PCBD1 |
pterin-4 alpha-carbinolamine dehydratase/dimerization cofactor of hepatocyte nuclear factor 1 alpha |
97603 |
0.07 |
| chr17_47825562_47826034 | 0.17 |
FAM117A |
family with sequence similarity 117, member A |
15695 |
0.14 |
| chr9_92078215_92078535 | 0.17 |
SEMA4D |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D |
7507 |
0.25 |
| chr4_38602670_38602821 | 0.17 |
RP11-617D20.1 |
|
23451 |
0.2 |
| chr16_79651353_79651504 | 0.17 |
MAF |
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog |
16817 |
0.26 |
| chr12_65369994_65370192 | 0.17 |
ENSG00000221564 |
. |
37290 |
0.18 |
| chr3_169897682_169897967 | 0.17 |
PHC3 |
polyhomeotic homolog 3 (Drosophila) |
1692 |
0.35 |
| chr2_8469788_8469939 | 0.17 |
AC011747.7 |
|
346033 |
0.01 |
| chr2_27273137_27273288 | 0.17 |
AGBL5-AS1 |
AGBL5 antisense RNA 1 |
80 |
0.87 |
| chr14_69196211_69196400 | 0.17 |
ENSG00000207089 |
. |
6440 |
0.23 |
| chr12_4326122_4326423 | 0.17 |
CCND2 |
cyclin D2 |
56666 |
0.11 |
| chr14_81769226_81769438 | 0.17 |
STON2 |
stonin 2 |
26055 |
0.23 |
| chr1_9301737_9301925 | 0.17 |
H6PD |
hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) |
1928 |
0.34 |
| chr5_57841537_57841688 | 0.17 |
CTD-2117L12.1 |
Uncharacterized protein |
12458 |
0.18 |
| chr1_27156953_27157104 | 0.17 |
ZDHHC18 |
zinc finger, DHHC-type containing 18 |
1467 |
0.28 |
| chr6_37241298_37241494 | 0.17 |
TBC1D22B |
TBC1 domain family, member 22B |
15848 |
0.15 |
| chr2_43373091_43373242 | 0.17 |
ENSG00000207087 |
. |
54534 |
0.14 |
| chr11_118097693_118098499 | 0.17 |
AMICA1 |
adhesion molecule, interacts with CXADR antigen 1 |
2287 |
0.23 |
| chr12_52096666_52096923 | 0.17 |
SCN8A |
sodium channel, voltage gated, type VIII, alpha subunit |
16587 |
0.24 |
| chr9_71643128_71643290 | 0.17 |
FXN |
frataxin |
6966 |
0.23 |
| chr5_148127360_148127525 | 0.16 |
ADRB2 |
adrenoceptor beta 2, surface |
78714 |
0.09 |
| chr2_64863847_64864192 | 0.16 |
SERTAD2 |
SERTA domain containing 2 |
17028 |
0.22 |
| chr18_10787891_10788042 | 0.16 |
PIEZO2 |
piezo-type mechanosensitive ion channel component 2 |
826 |
0.73 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) |
| 0.1 | 0.1 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.1 | 0.2 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.1 | 0.2 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.0 | 0.1 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.2 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 0.1 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.0 | 0.2 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.2 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.1 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.0 | 0.1 | GO:0002513 | tolerance induction to self antigen(GO:0002513) |
| 0.0 | 0.1 | GO:0001821 | histamine secretion(GO:0001821) |
| 0.0 | 0.1 | GO:0016102 | retinoic acid biosynthetic process(GO:0002138) diterpenoid biosynthetic process(GO:0016102) |
| 0.0 | 0.1 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.0 | 0.1 | GO:0045425 | positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) |
| 0.0 | 0.2 | GO:0045714 | regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045714) |
| 0.0 | 0.1 | GO:0071569 | protein ufmylation(GO:0071569) |
| 0.0 | 0.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.0 | GO:0034135 | regulation of toll-like receptor 2 signaling pathway(GO:0034135) |
| 0.0 | 0.1 | GO:0032230 | positive regulation of synaptic transmission, GABAergic(GO:0032230) |
| 0.0 | 0.0 | GO:0033689 | negative regulation of osteoblast proliferation(GO:0033689) |
| 0.0 | 0.1 | GO:0046607 | positive regulation of centrosome cycle(GO:0046607) |
| 0.0 | 0.1 | GO:0019987 | obsolete negative regulation of anti-apoptosis(GO:0019987) |
| 0.0 | 0.1 | GO:0046398 | UDP-glucuronate metabolic process(GO:0046398) |
| 0.0 | 0.0 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.0 | 0.1 | GO:0002634 | regulation of germinal center formation(GO:0002634) |
| 0.0 | 0.1 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 0.0 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.0 | 0.0 | GO:0034139 | regulation of toll-like receptor 3 signaling pathway(GO:0034139) |
| 0.0 | 0.1 | GO:0010956 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.0 | 0.4 | GO:0046834 | lipid phosphorylation(GO:0046834) |
| 0.0 | 0.2 | GO:0070373 | negative regulation of ERK1 and ERK2 cascade(GO:0070373) |
| 0.0 | 0.1 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.0 | 0.1 | GO:0014826 | vein smooth muscle contraction(GO:0014826) |
| 0.0 | 0.1 | GO:0090281 | negative regulation of calcium ion import(GO:0090281) |
| 0.0 | 0.0 | GO:0072216 | positive regulation of metanephros development(GO:0072216) |
| 0.0 | 0.1 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.0 | 0.0 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.0 | GO:0034086 | maintenance of sister chromatid cohesion(GO:0034086) maintenance of mitotic sister chromatid cohesion(GO:0034088) |
| 0.0 | 0.1 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.1 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.0 | 0.1 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.0 | 0.2 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.2 | GO:0021527 | spinal cord association neuron differentiation(GO:0021527) |
| 0.0 | 0.1 | GO:0051770 | positive regulation of nitric-oxide synthase biosynthetic process(GO:0051770) |
| 0.0 | 0.1 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.1 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.0 | GO:0002691 | regulation of cellular extravasation(GO:0002691) positive regulation of cellular extravasation(GO:0002693) |
| 0.0 | 0.1 | GO:0032493 | response to bacterial lipoprotein(GO:0032493) response to bacterial lipopeptide(GO:0070339) cellular response to bacterial lipoprotein(GO:0071220) cellular response to bacterial lipopeptide(GO:0071221) |
| 0.0 | 0.2 | GO:0030889 | negative regulation of B cell proliferation(GO:0030889) |
| 0.0 | 0.1 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.1 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.2 | GO:0001556 | oocyte maturation(GO:0001556) |
| 0.0 | 0.0 | GO:0010193 | response to ozone(GO:0010193) |
| 0.0 | 0.1 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.1 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.0 | 0.0 | GO:0043374 | CD8-positive, alpha-beta T cell activation(GO:0036037) CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.1 | GO:0015917 | aminophospholipid transport(GO:0015917) |
| 0.0 | 0.1 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.0 | 0.1 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.0 | 0.1 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.0 | 0.1 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.0 | 0.0 | GO:0071043 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.0 | 0.1 | GO:0045354 | interferon-alpha biosynthetic process(GO:0045349) regulation of interferon-alpha biosynthetic process(GO:0045354) positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.0 | 0.0 | GO:2000095 | regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.0 | 0.1 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.0 | 0.3 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.0 | 0.0 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.1 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 0.1 | GO:0071360 | cellular response to exogenous dsRNA(GO:0071360) |
| 0.0 | 0.0 | GO:0046931 | pore complex assembly(GO:0046931) |
| 0.0 | 0.1 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.1 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 0.1 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.0 | GO:2000542 | negative regulation of gastrulation(GO:2000542) |
| 0.0 | 0.1 | GO:0072531 | pyrimidine-containing compound transmembrane transport(GO:0072531) |
| 0.0 | 0.1 | GO:0050858 | negative regulation of antigen receptor-mediated signaling pathway(GO:0050858) negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.0 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.0 | 0.0 | GO:0009448 | gamma-aminobutyric acid metabolic process(GO:0009448) |
| 0.0 | 0.1 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.1 | GO:0046835 | carbohydrate phosphorylation(GO:0046835) |
| 0.0 | 0.0 | GO:0071436 | sodium ion export(GO:0071436) |
| 0.0 | 0.0 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.0 | 0.0 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 0.0 | 0.1 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.0 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.0 | GO:0061030 | epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.0 | 0.0 | GO:0001878 | response to yeast(GO:0001878) |
| 0.0 | 0.0 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.1 | GO:0003351 | epithelial cilium movement(GO:0003351) epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.1 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.0 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.0 | 0.1 | GO:2001259 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of calcium ion transmembrane transporter activity(GO:1901021) regulation of store-operated calcium channel activity(GO:1901339) positive regulation of store-operated calcium channel activity(GO:1901341) positive regulation of cation channel activity(GO:2001259) |
| 0.0 | 0.0 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.0 | GO:0002669 | lymphocyte anergy(GO:0002249) T cell tolerance induction(GO:0002517) regulation of T cell tolerance induction(GO:0002664) positive regulation of T cell tolerance induction(GO:0002666) regulation of T cell anergy(GO:0002667) positive regulation of T cell anergy(GO:0002669) T cell anergy(GO:0002870) regulation of lymphocyte anergy(GO:0002911) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.0 | 0.1 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 | 0.0 | GO:1901419 | regulation of response to alcohol(GO:1901419) |
| 0.0 | 0.1 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.0 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.0 | 0.1 | GO:0048488 | synaptic vesicle recycling(GO:0036465) synaptic vesicle endocytosis(GO:0048488) clathrin-mediated endocytosis(GO:0072583) |
| 0.0 | 0.0 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 | 0.0 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.0 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.0 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.0 | 0.0 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.0 | 0.0 | GO:0042723 | thiamine metabolic process(GO:0006772) thiamine-containing compound metabolic process(GO:0042723) |
| 0.0 | 0.2 | GO:0090659 | adult walking behavior(GO:0007628) walking behavior(GO:0090659) |
| 0.0 | 0.1 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.0 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.0 | 0.0 | GO:0090195 | chemokine secretion(GO:0090195) regulation of chemokine secretion(GO:0090196) positive regulation of chemokine secretion(GO:0090197) |
| 0.0 | 0.0 | GO:0060535 | trachea cartilage morphogenesis(GO:0060535) cartilage morphogenesis(GO:0060536) |
| 0.0 | 0.0 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.0 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.1 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 0.1 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.1 | GO:0010823 | negative regulation of mitochondrion organization(GO:0010823) negative regulation of release of cytochrome c from mitochondria(GO:0090201) negative regulation of apoptotic signaling pathway(GO:2001234) |
| 0.0 | 0.0 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.0 | 0.0 | GO:0002643 | regulation of tolerance induction(GO:0002643) positive regulation of tolerance induction(GO:0002645) |
| 0.0 | 0.0 | GO:0034625 | fatty acid elongation, monounsaturated fatty acid(GO:0034625) |
| 0.0 | 0.0 | GO:0032898 | neurotrophin production(GO:0032898) |
| 0.0 | 0.1 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.0 | 0.1 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 0.2 | GO:0034199 | activation of protein kinase A activity(GO:0034199) |
| 0.0 | 0.0 | GO:0008215 | spermine metabolic process(GO:0008215) |
| 0.0 | 0.1 | GO:0008653 | lipopolysaccharide metabolic process(GO:0008653) |
| 0.0 | 0.0 | GO:0009296 | obsolete flagellum assembly(GO:0009296) |
| 0.0 | 0.0 | GO:1903053 | regulation of extracellular matrix disassembly(GO:0010715) negative regulation of extracellular matrix disassembly(GO:0010716) regulation of extracellular matrix organization(GO:1903053) negative regulation of extracellular matrix organization(GO:1903054) |
| 0.0 | 0.1 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.0 | 0.0 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.0 | 0.0 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) regulation of dendritic spine morphogenesis(GO:0061001) dendritic spine organization(GO:0097061) |
| 0.0 | 0.0 | GO:0010586 | miRNA metabolic process(GO:0010586) miRNA catabolic process(GO:0010587) |
| 0.0 | 0.1 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.0 | 0.1 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.0 | 0.0 | GO:0002407 | dendritic cell chemotaxis(GO:0002407) dendritic cell migration(GO:0036336) |
| 0.0 | 0.1 | GO:0007128 | meiotic prophase I(GO:0007128) |
| 0.0 | 0.1 | GO:1901661 | quinone metabolic process(GO:1901661) |
| 0.0 | 0.0 | GO:0071072 | negative regulation of phospholipid biosynthetic process(GO:0071072) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.1 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.1 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.1 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.1 | GO:0031313 | extrinsic component of vacuolar membrane(GO:0000306) extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.1 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.2 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.1 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.1 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.1 | GO:0017059 | palmitoyltransferase complex(GO:0002178) serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.1 | GO:0071778 | obsolete WINAC complex(GO:0071778) |
| 0.0 | 0.1 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.3 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.2 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.1 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.1 | GO:0031264 | death-inducing signaling complex(GO:0031264) |
| 0.0 | 0.1 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.1 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.1 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.1 | GO:0000800 | lateral element(GO:0000800) |
| 0.0 | 0.1 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.1 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.0 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 0.1 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 0.1 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.1 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.0 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.0 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.1 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.0 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.0 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.1 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.1 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.1 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.1 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.0 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.2 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.1 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.0 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.2 | GO:0001846 | opsonin binding(GO:0001846) |
| 0.0 | 0.1 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.2 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.3 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 0.1 | GO:0000987 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.0 | 0.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.2 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.0 | 0.1 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.1 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.1 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.1 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.0 | 0.2 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.1 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.1 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 0.1 | GO:0031708 | bombesin receptor binding(GO:0031705) endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.2 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.2 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.1 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.1 | GO:0004904 | interferon receptor activity(GO:0004904) |
| 0.0 | 0.1 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.1 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.1 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.1 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.0 | 0.1 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.1 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.1 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.0 | 0.1 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.3 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.1 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.1 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.1 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.0 | 0.1 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.0 | 0.1 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.0 | 0.1 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.0 | 0.1 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.0 | 0.1 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.1 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.0 | 0.1 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.1 | GO:1901474 | azole transporter activity(GO:0045118) azole transmembrane transporter activity(GO:1901474) |
| 0.0 | 0.1 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.0 | 0.0 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.0 | 0.1 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.1 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.1 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.1 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.1 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.2 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.0 | 0.1 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.0 | 0.0 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.0 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.0 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.0 | GO:0016362 | activin receptor activity, type II(GO:0016362) |
| 0.0 | 0.1 | GO:0016778 | diphosphotransferase activity(GO:0016778) |
| 0.0 | 0.3 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.0 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.0 | 0.0 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.1 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.0 | GO:0004477 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) |
| 0.0 | 0.1 | GO:0016986 | obsolete transcription initiation factor activity(GO:0016986) |
| 0.0 | 0.0 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.0 | 0.0 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.0 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.0 | 0.0 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.2 | GO:0008373 | sialyltransferase activity(GO:0008373) |
| 0.0 | 0.0 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.0 | 0.1 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.1 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.0 | GO:0001848 | complement binding(GO:0001848) |
| 0.0 | 0.2 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.0 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.1 | GO:0050656 | 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.0 | 0.0 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.1 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.0 | 0.1 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.2 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.0 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.1 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.1 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.1 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.0 | GO:0016149 | translation release factor activity, codon specific(GO:0016149) |
| 0.0 | 0.0 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.1 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.2 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.0 | GO:0031701 | angiotensin receptor binding(GO:0031701) |
| 0.0 | 0.1 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.0 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.0 | 0.0 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.1 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.1 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.0 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.1 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.4 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.3 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.6 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.0 | 0.1 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.2 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.3 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.1 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.2 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.0 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.0 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.3 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.3 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 0.1 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 0.1 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 0.1 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.0 | 0.1 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 0.1 | REACTOME PROCESSING OF CAPPED INTRON CONTAINING PRE MRNA | Genes involved in Processing of Capped Intron-Containing Pre-mRNA |
| 0.0 | 0.3 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.0 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 0.2 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.2 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.1 | REACTOME ADP SIGNALLING THROUGH P2RY1 | Genes involved in ADP signalling through P2Y purinoceptor 1 |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.0 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.0 | 0.2 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.2 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.0 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.1 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 0.2 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.2 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.1 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.0 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.0 | 0.1 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.1 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.2 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.3 | REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |
| 0.0 | 0.2 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.0 | REACTOME CYCLIN E ASSOCIATED EVENTS DURING G1 S TRANSITION | Genes involved in Cyclin E associated events during G1/S transition |
| 0.0 | 0.1 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.0 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.2 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.1 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.1 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 0.3 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.1 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.2 | REACTOME IL 2 SIGNALING | Genes involved in Interleukin-2 signaling |