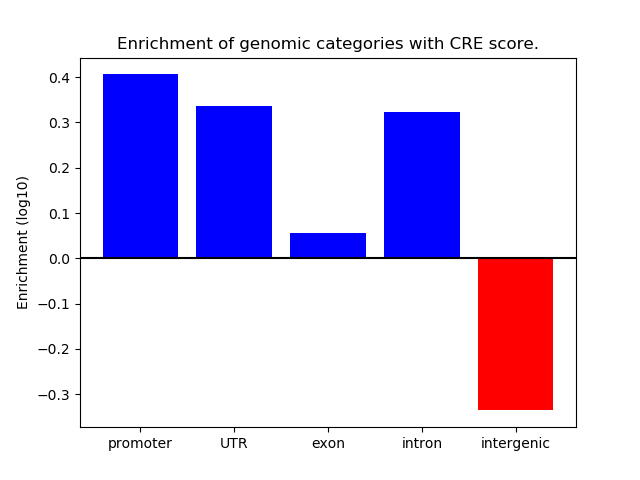
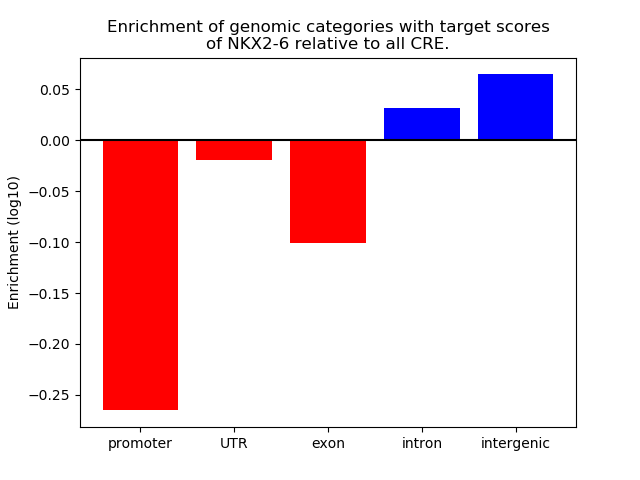
Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
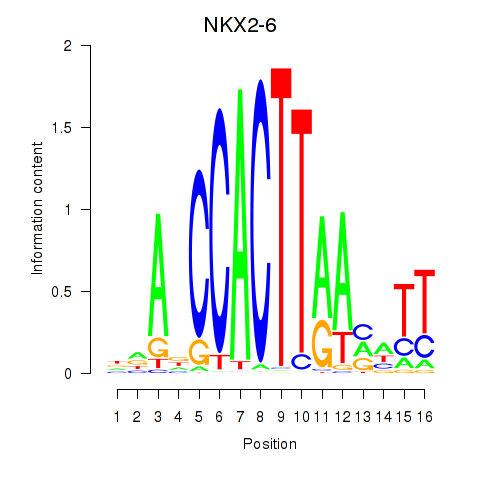
Results for NKX2-6
Z-value: 1.37
Transcription factors associated with NKX2-6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NKX2-6
|
ENSG00000180053.6 | NK2 homeobox 6 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr8_23553705_23553856 | NKX2-6 | 10142 | 0.157480 | 0.53 | 1.4e-01 | Click! |
| chr8_23554046_23554197 | NKX2-6 | 9801 | 0.158195 | 0.49 | 1.8e-01 | Click! |
| chr8_23563753_23563904 | NKX2-6 | 94 | 0.925228 | -0.42 | 2.6e-01 | Click! |
| chr8_23563478_23563629 | NKX2-6 | 369 | 0.766455 | -0.41 | 2.7e-01 | Click! |
| chr8_23560501_23560652 | NKX2-6 | 3346 | 0.200322 | -0.37 | 3.3e-01 | Click! |
Activity of the NKX2-6 motif across conditions
Conditions sorted by the z-value of the NKX2-6 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
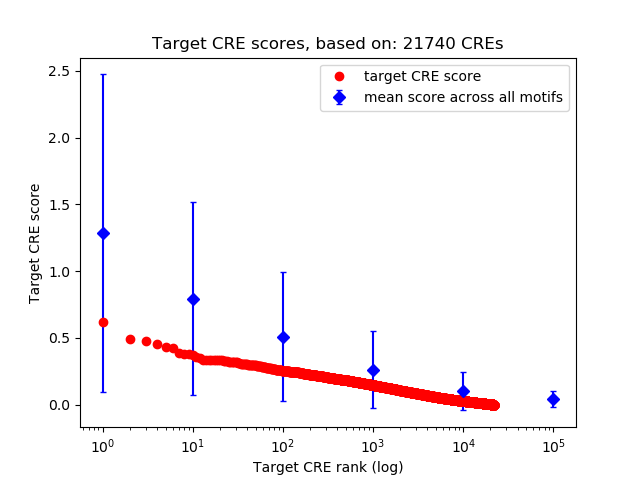
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr9_16540961_16541659 | 0.62 |
RP11-183I6.2 |
|
68124 |
0.12 |
| chr11_73990200_73990857 | 0.49 |
ENSG00000206913 |
. |
26931 |
0.14 |
| chr22_40830866_40831668 | 0.48 |
RP5-1042K10.13 |
|
20260 |
0.14 |
| chr21_35918859_35919211 | 0.46 |
RCAN1 |
regulator of calcineurin 1 |
13442 |
0.2 |
| chr3_45687605_45688015 | 0.44 |
LIMD1-AS1 |
LIMD1 antisense RNA 1 |
42564 |
0.12 |
| chr15_71060497_71060707 | 0.43 |
UACA |
uveal autoantigen with coiled-coil domains and ankyrin repeats |
4670 |
0.23 |
| chr2_37947734_37948100 | 0.39 |
CDC42EP3 |
CDC42 effector protein (Rho GTPase binding) 3 |
17694 |
0.27 |
| chr1_77941867_77942099 | 0.38 |
AK5 |
adenylate kinase 5 |
55809 |
0.12 |
| chr8_125192495_125192646 | 0.38 |
FER1L6-AS2 |
FER1L6 antisense RNA 2 |
8807 |
0.26 |
| chr2_217354341_217354722 | 0.37 |
AC098820.4 |
|
2612 |
0.22 |
| chr12_63122687_63123266 | 0.36 |
ENSG00000238475 |
. |
78021 |
0.09 |
| chr2_172388783_172388934 | 0.35 |
CYBRD1 |
cytochrome b reductase 1 |
9269 |
0.25 |
| chr2_145263610_145263985 | 0.34 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
11318 |
0.24 |
| chr17_32574614_32574944 | 0.34 |
CCL2 |
chemokine (C-C motif) ligand 2 |
7525 |
0.13 |
| chr20_17629504_17629678 | 0.33 |
RRBP1 |
ribosome binding protein 1 |
11561 |
0.2 |
| chr1_86458784_86459076 | 0.33 |
ENSG00000201620 |
. |
109449 |
0.07 |
| chr14_100376642_100376793 | 0.33 |
EML1 |
echinoderm microtubule associated protein like 1 |
12147 |
0.2 |
| chr3_38871334_38871622 | 0.33 |
RP11-134J21.1 |
|
6085 |
0.2 |
| chr7_81396029_81396416 | 0.33 |
HGF |
hepatocyte growth factor (hepapoietin A; scatter factor) |
3065 |
0.4 |
| chr8_134244229_134244380 | 0.33 |
WISP1-OT1 |
WISP1 overlapping transcript 1 (non-protein coding) |
3005 |
0.31 |
| chr20_9063656_9063954 | 0.33 |
PLCB4 |
phospholipase C, beta 4 |
14057 |
0.24 |
| chr13_31294257_31294613 | 0.33 |
ALOX5AP |
arachidonate 5-lipoxygenase-activating protein |
15210 |
0.26 |
| chr10_3856587_3856738 | 0.33 |
KLF6 |
Kruppel-like factor 6 |
29189 |
0.2 |
| chr10_29130225_29130376 | 0.33 |
C10orf126 |
chromosome 10 open reading frame 126 |
5037 |
0.26 |
| chr10_54412447_54412741 | 0.32 |
RP11-556E13.1 |
|
91729 |
0.09 |
| chr16_78719280_78719578 | 0.32 |
RP11-264L1.3 |
|
151158 |
0.04 |
| chr8_129054673_129054878 | 0.32 |
ENSG00000221176 |
. |
6623 |
0.21 |
| chr5_111115219_111115750 | 0.32 |
NREP |
neuronal regeneration related protein |
21569 |
0.21 |
| chr9_86786252_86786403 | 0.32 |
RP11-380F14.2 |
|
106807 |
0.06 |
| chr15_80271357_80271985 | 0.32 |
BCL2A1 |
BCL2-related protein A1 |
7883 |
0.2 |
| chr14_71839341_71839562 | 0.32 |
ENSG00000207444 |
. |
25603 |
0.19 |
| chr6_143281120_143281272 | 0.31 |
HIVEP2 |
human immunodeficiency virus type I enhancer binding protein 2 |
14858 |
0.23 |
| chr7_13959398_13959662 | 0.31 |
ETV1 |
ets variant 1 |
66536 |
0.14 |
| chr1_225942912_225943116 | 0.31 |
ENSG00000223306 |
. |
14037 |
0.14 |
| chr15_99271535_99272053 | 0.31 |
IGF1R |
insulin-like growth factor 1 receptor |
20734 |
0.22 |
| chr1_192988213_192988364 | 0.31 |
UCHL5 |
ubiquitin carboxyl-terminal hydrolase L5 |
4764 |
0.21 |
| chr12_29752319_29752470 | 0.31 |
TMTC1 |
transmembrane and tetratricopeptide repeat containing 1 |
4763 |
0.31 |
| chr17_9249524_9249871 | 0.31 |
RP11-85B7.4 |
|
63206 |
0.12 |
| chr5_158126612_158126763 | 0.30 |
CTD-2363C16.1 |
|
283327 |
0.01 |
| chr5_158448429_158448934 | 0.30 |
CTD-2363C16.2 |
|
35867 |
0.17 |
| chr12_109042760_109042911 | 0.30 |
CORO1C |
coronin, actin binding protein, 1C |
3286 |
0.17 |
| chr10_97047482_97047633 | 0.30 |
PDLIM1 |
PDZ and LIM domain 1 |
3224 |
0.31 |
| chr3_45684381_45684734 | 0.30 |
LIMD1-AS1 |
LIMD1 antisense RNA 1 |
45817 |
0.11 |
| chr8_96715315_96715466 | 0.30 |
ENSG00000223297 |
. |
24247 |
0.26 |
| chr12_2494439_2494710 | 0.30 |
CACNA1C-IT3 |
CACNA1C intronic transcript 3 (non-protein coding) |
115632 |
0.06 |
| chr12_88756954_88757105 | 0.30 |
ENSG00000199245 |
. |
67190 |
0.14 |
| chr2_192116508_192116659 | 0.30 |
MYO1B |
myosin IB |
5574 |
0.29 |
| chr3_45589006_45589723 | 0.30 |
LARS2-AS1 |
LARS2 antisense RNA 1 |
38327 |
0.14 |
| chr1_97004836_97004987 | 0.30 |
ENSG00000241992 |
. |
43854 |
0.2 |
| chr1_68187782_68188175 | 0.29 |
GADD45A |
growth arrest and DNA-damage-inducible, alpha |
37094 |
0.16 |
| chr11_121961832_121961983 | 0.29 |
ENSG00000207971 |
. |
8645 |
0.17 |
| chr15_67065770_67066102 | 0.29 |
SMAD6 |
SMAD family member 6 |
61901 |
0.13 |
| chr20_23132592_23133523 | 0.29 |
ENSG00000201527 |
. |
8551 |
0.22 |
| chr1_200337482_200337689 | 0.29 |
ZNF281 |
zinc finger protein 281 |
41535 |
0.21 |
| chr1_178055842_178055993 | 0.29 |
RASAL2 |
RAS protein activator like 2 |
6947 |
0.32 |
| chr7_76867154_76867700 | 0.29 |
AC073635.5 |
|
20013 |
0.2 |
| chr2_55339476_55339889 | 0.29 |
RTN4 |
reticulon 4 |
75 |
0.97 |
| chr8_122001527_122001678 | 0.29 |
ENSG00000252093 |
. |
80026 |
0.11 |
| chr17_47076838_47077332 | 0.29 |
IGF2BP1 |
insulin-like growth factor 2 mRNA binding protein 1 |
1977 |
0.17 |
| chr16_67274601_67274968 | 0.29 |
SLC9A5 |
solute carrier family 9, subfamily A (NHE5, cation proton antiporter 5), member 5 |
6034 |
0.08 |
| chr8_105689851_105690002 | 0.28 |
ENSG00000263732 |
. |
77477 |
0.1 |
| chr10_77290347_77290581 | 0.28 |
ENSG00000207583 |
. |
21752 |
0.23 |
| chr13_27157989_27158140 | 0.28 |
WASF3 |
WAS protein family, member 3 |
26177 |
0.23 |
| chr12_20543454_20543605 | 0.28 |
RP11-284H19.1 |
|
20333 |
0.23 |
| chr6_52467732_52468424 | 0.28 |
TRAM2 |
translocation associated membrane protein 2 |
26365 |
0.19 |
| chr1_219299907_219300058 | 0.28 |
LYPLAL1 |
lysophospholipase-like 1 |
47204 |
0.17 |
| chr1_211775743_211775938 | 0.28 |
RP11-359E8.5 |
|
18773 |
0.16 |
| chr7_17001293_17001444 | 0.28 |
AGR3 |
anterior gradient 3 |
79757 |
0.1 |
| chr4_157895789_157896050 | 0.27 |
PDGFC |
platelet derived growth factor C |
3373 |
0.27 |
| chr4_157416635_157416786 | 0.27 |
RP11-171N4.2 |
Uncharacterized protein |
146749 |
0.04 |
| chr1_61370549_61370700 | 0.27 |
NFIA |
nuclear factor I/A |
39693 |
0.19 |
| chr20_40138790_40139006 | 0.27 |
CHD6 |
chromodomain helicase DNA binding protein 6 |
10941 |
0.28 |
| chr13_30095383_30095797 | 0.27 |
MTUS2-AS1 |
MTUS2 antisense RNA 1 |
31348 |
0.21 |
| chr1_17448827_17448978 | 0.27 |
PADI2 |
peptidyl arginine deiminase, type II |
2954 |
0.25 |
| chr6_133032333_133032512 | 0.27 |
VNN1 |
vanin 1 |
2766 |
0.21 |
| chr4_14061132_14061426 | 0.27 |
ENSG00000252092 |
. |
401028 |
0.01 |
| chr4_88296347_88296610 | 0.27 |
ENSG00000240966 |
. |
11848 |
0.15 |
| chr17_53504679_53504830 | 0.27 |
MMD |
monocyte to macrophage differentiation-associated |
5401 |
0.32 |
| chr3_67047069_67047341 | 0.27 |
KBTBD8 |
kelch repeat and BTB (POZ) domain containing 8 |
1522 |
0.59 |
| chr11_2324783_2325431 | 0.27 |
TSPAN32 |
tetraspanin 32 |
1009 |
0.38 |
| chr10_44958607_44958758 | 0.27 |
CXCL12 |
chemokine (C-X-C motif) ligand 12 |
78142 |
0.11 |
| chr17_67139065_67139216 | 0.26 |
ABCA6 |
ATP-binding cassette, sub-family A (ABC1), member 6 |
1111 |
0.51 |
| chr5_34524127_34524278 | 0.26 |
RAI14 |
retinoic acid induced 14 |
132140 |
0.05 |
| chr4_157860965_157861230 | 0.26 |
PDGFC |
platelet derived growth factor C |
30958 |
0.19 |
| chr15_40171438_40171589 | 0.26 |
GPR176 |
G protein-coupled receptor 176 |
40913 |
0.11 |
| chr2_153469749_153469919 | 0.26 |
FMNL2 |
formin-like 2 |
6258 |
0.31 |
| chr7_137557971_137558230 | 0.26 |
DGKI |
diacylglycerol kinase, iota |
26262 |
0.2 |
| chr2_40354084_40354235 | 0.26 |
SLC8A1-AS1 |
SLC8A1 antisense RNA 1 |
11990 |
0.31 |
| chr7_137628746_137628897 | 0.26 |
AC022173.2 |
|
9273 |
0.21 |
| chr4_120439477_120439628 | 0.26 |
PDE5A |
phosphodiesterase 5A, cGMP-specific |
702 |
0.73 |
| chr6_123032140_123032424 | 0.26 |
PKIB |
protein kinase (cAMP-dependent, catalytic) inhibitor beta |
6424 |
0.24 |
| chr17_74711629_74712064 | 0.26 |
MXRA7 |
matrix-remodelling associated 7 |
4748 |
0.09 |
| chr3_171283226_171283448 | 0.26 |
ENSG00000207114 |
. |
61197 |
0.13 |
| chr7_120633768_120634071 | 0.26 |
CPED1 |
cadherin-like and PC-esterase domain containing 1 |
4243 |
0.23 |
| chr10_93871030_93871181 | 0.26 |
ENSG00000252993 |
. |
35106 |
0.16 |
| chr1_101093719_101093870 | 0.26 |
GPR88 |
G protein-coupled receptor 88 |
90101 |
0.08 |
| chr1_21451523_21451771 | 0.26 |
EIF4G3 |
eukaryotic translation initiation factor 4 gamma, 3 |
35946 |
0.17 |
| chr2_200603538_200603689 | 0.26 |
FTCDNL1 |
formiminotransferase cyclodeaminase N-terminal like |
111983 |
0.07 |
| chr5_33369882_33370033 | 0.26 |
CTD-2203K17.1 |
|
70768 |
0.12 |
| chr5_138875477_138875767 | 0.26 |
TMEM173 |
transmembrane protein 173 |
13247 |
0.13 |
| chr6_82472333_82472746 | 0.26 |
ENSG00000206886 |
. |
1202 |
0.5 |
| chr12_13270933_13271084 | 0.25 |
GSG1 |
germ cell associated 1 |
14389 |
0.21 |
| chr12_93770029_93770180 | 0.25 |
RP11-486A14.2 |
|
1380 |
0.31 |
| chr4_26143484_26143734 | 0.25 |
RBPJ |
recombination signal binding protein for immunoglobulin kappa J region |
21468 |
0.28 |
| chr6_110417870_110418143 | 0.25 |
WASF1 |
WAS protein family, member 1 |
82846 |
0.09 |
| chr15_71625454_71625605 | 0.25 |
RP11-592N21.2 |
|
9134 |
0.29 |
| chr10_5504056_5504535 | 0.25 |
NET1 |
neuroepithelial cell transforming 1 |
15721 |
0.15 |
| chr6_35185894_35186601 | 0.25 |
SCUBE3 |
signal peptide, CUB domain, EGF-like 3 |
4051 |
0.24 |
| chr2_210337616_210337973 | 0.25 |
MAP2 |
microtubule-associated protein 2 |
49012 |
0.19 |
| chr5_124820297_124820570 | 0.25 |
ENSG00000222107 |
. |
133609 |
0.06 |
| chr10_92681329_92681480 | 0.25 |
ANKRD1 |
ankyrin repeat domain 1 (cardiac muscle) |
371 |
0.79 |
| chr7_29905918_29906329 | 0.25 |
WIPF3 |
WAS/WASL interacting protein family, member 3 |
31782 |
0.17 |
| chr10_31550924_31551075 | 0.25 |
ENSG00000252479 |
. |
2471 |
0.35 |
| chr7_114627811_114627962 | 0.25 |
MDFIC |
MyoD family inhibitor domain containing |
53962 |
0.18 |
| chr1_29528780_29529144 | 0.25 |
SRSF4 |
serine/arginine-rich splicing factor 4 |
20463 |
0.13 |
| chr1_235104740_235105149 | 0.25 |
ENSG00000239690 |
. |
65011 |
0.12 |
| chr11_121965409_121965560 | 0.25 |
ENSG00000207971 |
. |
5068 |
0.18 |
| chr4_186837108_186837259 | 0.25 |
ENSG00000239034 |
. |
30054 |
0.18 |
| chr7_41517588_41517820 | 0.25 |
INHBA-AS1 |
INHBA antisense RNA 1 |
215810 |
0.02 |
| chr8_98983880_98984031 | 0.25 |
MATN2 |
matrilin 2 |
10245 |
0.2 |
| chr2_218550468_218550619 | 0.25 |
DIRC3 |
disrupted in renal carcinoma 3 |
70735 |
0.12 |
| chr15_51225693_51226195 | 0.25 |
AP4E1 |
adaptor-related protein complex 4, epsilon 1 subunit |
24952 |
0.23 |
| chr7_76855540_76855691 | 0.25 |
FGL2 |
fibrinogen-like 2 |
26472 |
0.18 |
| chr10_91921633_91921784 | 0.25 |
ENSG00000222451 |
. |
2009 |
0.5 |
| chr7_80264399_80264550 | 0.25 |
CD36 |
CD36 molecule (thrombospondin receptor) |
3486 |
0.37 |
| chr6_131428128_131428279 | 0.25 |
AKAP7 |
A kinase (PRKA) anchor protein 7 |
28603 |
0.22 |
| chr2_183283239_183283390 | 0.25 |
PDE1A |
phosphodiesterase 1A, calmodulin-dependent |
8435 |
0.3 |
| chr8_59966515_59966743 | 0.25 |
RP11-328K2.1 |
|
61132 |
0.11 |
| chr12_104518100_104518251 | 0.25 |
ENSG00000199415 |
. |
839 |
0.6 |
| chr2_43303903_43304168 | 0.25 |
ENSG00000207087 |
. |
14597 |
0.28 |
| chr2_226982606_226982904 | 0.25 |
ENSG00000263363 |
. |
540754 |
0.0 |
| chr2_26661594_26661750 | 0.25 |
DRC1 |
dynein regulatory complex subunit 1 homolog (Chlamydomonas) |
8315 |
0.21 |
| chr10_3902800_3902951 | 0.25 |
KLF6 |
Kruppel-like factor 6 |
75402 |
0.11 |
| chr20_2839933_2840363 | 0.24 |
VPS16 |
vacuolar protein sorting 16 homolog (S. cerevisiae) |
2955 |
0.18 |
| chr8_104117881_104118032 | 0.24 |
ENSG00000265667 |
. |
13007 |
0.13 |
| chr5_114915172_114915323 | 0.24 |
AC010226.4 |
|
22507 |
0.14 |
| chr7_33948512_33948663 | 0.24 |
BMPER |
BMP binding endothelial regulator |
3442 |
0.38 |
| chr17_30070933_30071094 | 0.24 |
ENSG00000202026 |
. |
30241 |
0.18 |
| chr17_36586871_36587022 | 0.24 |
ARHGAP23 |
Rho GTPase activating protein 23 |
2216 |
0.26 |
| chr1_10441222_10441373 | 0.24 |
ENSG00000265945 |
. |
8762 |
0.13 |
| chr4_169652420_169652571 | 0.24 |
PALLD |
palladin, cytoskeletal associated protein |
19185 |
0.21 |
| chr8_108314695_108314915 | 0.24 |
ANGPT1 |
angiopoietin 1 |
33945 |
0.21 |
| chr2_144711401_144711552 | 0.24 |
AC016910.1 |
|
16836 |
0.25 |
| chr7_48285699_48285984 | 0.24 |
ABCA13 |
ATP-binding cassette, sub-family A (ABC1), member 13 |
74784 |
0.12 |
| chr5_125351586_125351737 | 0.24 |
ENSG00000265637 |
. |
180867 |
0.03 |
| chr18_455946_456097 | 0.24 |
RP11-720L2.2 |
|
31605 |
0.16 |
| chr6_82454549_82454764 | 0.24 |
FAM46A |
family with sequence similarity 46, member A |
6814 |
0.2 |
| chr6_127514210_127514472 | 0.24 |
RNF146 |
ring finger protein 146 |
73414 |
0.11 |
| chr7_134574979_134575413 | 0.24 |
CALD1 |
caldesmon 1 |
955 |
0.69 |
| chr18_46238546_46238697 | 0.24 |
RP11-426J5.2 |
|
23831 |
0.21 |
| chr9_110399429_110399916 | 0.24 |
ENSG00000202308 |
. |
25855 |
0.23 |
| chr12_20061614_20061845 | 0.24 |
ENSG00000221679 |
. |
64070 |
0.15 |
| chr17_29894018_29894169 | 0.24 |
ENSG00000221038 |
. |
3180 |
0.14 |
| chr13_50783531_50783810 | 0.24 |
DLEU1 |
deleted in lymphocytic leukemia 1 (non-protein coding) |
127363 |
0.05 |
| chr11_34367020_34367283 | 0.24 |
ABTB2 |
ankyrin repeat and BTB (POZ) domain containing 2 |
11651 |
0.26 |
| chr1_56162477_56162628 | 0.24 |
ENSG00000272051 |
. |
211907 |
0.02 |
| chr11_122556790_122557092 | 0.24 |
UBASH3B |
ubiquitin associated and SH3 domain containing B |
30558 |
0.18 |
| chr2_69464357_69464627 | 0.24 |
ENSG00000199460 |
. |
54483 |
0.12 |
| chr1_180000870_180001021 | 0.24 |
CEP350 |
centrosomal protein 350kDa |
9393 |
0.26 |
| chr7_41518329_41518480 | 0.24 |
INHBA-AS1 |
INHBA antisense RNA 1 |
215110 |
0.02 |
| chr4_16780959_16781110 | 0.24 |
LDB2 |
LIM domain binding 2 |
105098 |
0.08 |
| chr7_100016771_100016922 | 0.24 |
ZCWPW1 |
zinc finger, CW type with PWWP domain 1 |
654 |
0.47 |
| chr12_109117281_109117726 | 0.24 |
CORO1C |
coronin, actin binding protein, 1C |
6824 |
0.16 |
| chr3_99681966_99682117 | 0.24 |
ENSG00000264897 |
. |
1201 |
0.6 |
| chr3_45100675_45100826 | 0.24 |
ENSG00000252410 |
. |
24670 |
0.16 |
| chr18_18744847_18745057 | 0.24 |
ENSG00000251886 |
. |
44485 |
0.15 |
| chr3_123509920_123510071 | 0.24 |
MYLK |
myosin light chain kinase |
2693 |
0.3 |
| chr3_177649836_177649987 | 0.23 |
ENSG00000199858 |
. |
75656 |
0.13 |
| chr8_104241607_104241758 | 0.23 |
RP11-318M2.3 |
|
140 |
0.96 |
| chr12_127768981_127769132 | 0.23 |
ENSG00000253089 |
. |
36844 |
0.23 |
| chr12_66455744_66455895 | 0.23 |
ENSG00000251857 |
. |
4182 |
0.25 |
| chr8_23381826_23381979 | 0.23 |
SLC25A37 |
solute carrier family 25 (mitochondrial iron transporter), member 37 |
4416 |
0.2 |
| chr6_116733337_116733739 | 0.23 |
DSE |
dermatan sulfate epimerase |
41428 |
0.1 |
| chr11_12841613_12841764 | 0.23 |
RP11-47J17.3 |
|
3526 |
0.26 |
| chr10_3591240_3591463 | 0.23 |
RP11-184A2.3 |
|
201908 |
0.03 |
| chr1_109936676_109936954 | 0.23 |
SORT1 |
sortilin 1 |
836 |
0.57 |
| chr4_106117299_106117679 | 0.23 |
TET2-AS1 |
TET2 antisense RNA 1 |
18269 |
0.22 |
| chr3_179435405_179435556 | 0.23 |
USP13 |
ubiquitin specific peptidase 13 (isopeptidase T-3) |
3872 |
0.28 |
| chr10_99478408_99478800 | 0.23 |
MARVELD1 |
MARVEL domain containing 1 |
5123 |
0.17 |
| chr6_11522959_11523110 | 0.23 |
TMEM170B |
transmembrane protein 170B |
14904 |
0.22 |
| chr9_129135479_129135756 | 0.23 |
MVB12B |
multivesicular body subunit 12B |
3105 |
0.29 |
| chr12_27701552_27701930 | 0.23 |
PPFIBP1 |
PTPRF interacting protein, binding protein 1 (liprin beta 1) |
10685 |
0.23 |
| chr14_30392743_30392894 | 0.23 |
PRKD1 |
protein kinase D1 |
4030 |
0.28 |
| chr16_57142254_57142881 | 0.23 |
CPNE2 |
copine II |
1896 |
0.27 |
| chrX_18979389_18979540 | 0.23 |
PHKA2 |
phosphorylase kinase, alpha 2 (liver) |
23252 |
0.19 |
| chr13_27540158_27540309 | 0.23 |
USP12-AS1 |
USP12 antisense RNA 1 |
196759 |
0.02 |
| chr12_66050364_66050557 | 0.23 |
HMGA2 |
high mobility group AT-hook 2 |
167451 |
0.03 |
| chr11_67781235_67781527 | 0.23 |
ALDH3B1 |
aldehyde dehydrogenase 3 family, member B1 |
3591 |
0.13 |
| chr6_113396081_113396232 | 0.23 |
ENSG00000201386 |
. |
103461 |
0.08 |
| chr7_34078618_34078769 | 0.23 |
BMPER |
BMP binding endothelial regulator |
133548 |
0.05 |
| chr6_132455519_132455670 | 0.23 |
ENSG00000265669 |
. |
19191 |
0.27 |
| chr1_25119704_25119971 | 0.23 |
ENSG00000238482 |
. |
15367 |
0.21 |
| chr4_141031157_141031308 | 0.23 |
RP11-392B6.1 |
|
17937 |
0.23 |
| chr13_60265756_60265907 | 0.23 |
ENSG00000239003 |
. |
211627 |
0.02 |
| chr1_178599347_178599498 | 0.23 |
ENSG00000266417 |
. |
47462 |
0.12 |
| chr10_31888840_31889209 | 0.23 |
ENSG00000222412 |
. |
156040 |
0.04 |
| chr3_12750431_12750810 | 0.23 |
TMEM40 |
transmembrane protein 40 |
33396 |
0.14 |
| chr3_30204653_30204804 | 0.23 |
ENSG00000266546 |
. |
24204 |
0.21 |
| chr4_74698717_74699078 | 0.23 |
CXCL6 |
chemokine (C-X-C motif) ligand 6 |
3317 |
0.2 |
| chr3_113345260_113345597 | 0.23 |
ENSG00000241529 |
. |
6123 |
0.18 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.1 | 0.3 | GO:0021801 | cerebral cortex radial glia guided migration(GO:0021801) telencephalon glial cell migration(GO:0022030) |
| 0.1 | 0.1 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.1 | 0.2 | GO:0060831 | smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:0060831) |
| 0.1 | 0.3 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.1 | 0.3 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.1 | 0.2 | GO:0070874 | negative regulation of glycogen metabolic process(GO:0070874) |
| 0.1 | 0.2 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.1 | 0.2 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.1 | 0.2 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) |
| 0.1 | 0.2 | GO:0030091 | protein repair(GO:0030091) |
| 0.1 | 0.3 | GO:0032277 | negative regulation of gonadotropin secretion(GO:0032277) |
| 0.1 | 0.2 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.1 | 0.2 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 0.2 | GO:1900115 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.1 | 0.1 | GO:0048245 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.1 | 0.1 | GO:0060449 | bud elongation involved in lung branching(GO:0060449) |
| 0.1 | 0.2 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) |
| 0.1 | 0.3 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.1 | 0.2 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.1 | 0.1 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.0 | 0.3 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 0.1 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.0 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.0 | 0.2 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.2 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.0 | 0.1 | GO:0006065 | UDP-glucuronate biosynthetic process(GO:0006065) |
| 0.0 | 0.1 | GO:0048669 | collateral sprouting in absence of injury(GO:0048669) |
| 0.0 | 0.1 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.0 | 0.2 | GO:0018101 | protein citrullination(GO:0018101) |
| 0.0 | 0.1 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.0 | GO:0060242 | contact inhibition(GO:0060242) |
| 0.0 | 0.2 | GO:0097061 | dendritic spine morphogenesis(GO:0060997) regulation of dendritic spine morphogenesis(GO:0061001) dendritic spine organization(GO:0097061) |
| 0.0 | 0.2 | GO:0071731 | response to nitric oxide(GO:0071731) |
| 0.0 | 0.2 | GO:0060753 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 0.3 | GO:0045652 | regulation of megakaryocyte differentiation(GO:0045652) regulation of hematopoietic progenitor cell differentiation(GO:1901532) |
| 0.0 | 0.1 | GO:0060638 | mesenchymal-epithelial cell signaling(GO:0060638) |
| 0.0 | 0.1 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.0 | 0.1 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.0 | 0.2 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.1 | GO:0045343 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) |
| 0.0 | 0.1 | GO:0055089 | fatty acid homeostasis(GO:0055089) |
| 0.0 | 0.2 | GO:0060180 | regulation of female receptivity(GO:0045924) female mating behavior(GO:0060180) |
| 0.0 | 0.2 | GO:0001547 | antral ovarian follicle growth(GO:0001547) |
| 0.0 | 0.1 | GO:0010766 | negative regulation of sodium ion transport(GO:0010766) |
| 0.0 | 0.1 | GO:0072311 | renal filtration cell differentiation(GO:0061318) glomerular epithelium development(GO:0072010) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.0 | 0.1 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 | 0.2 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 0.1 | GO:0045425 | positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) |
| 0.0 | 0.1 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 0.1 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.1 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.0 | 0.1 | GO:1903053 | regulation of extracellular matrix disassembly(GO:0010715) negative regulation of extracellular matrix disassembly(GO:0010716) regulation of extracellular matrix organization(GO:1903053) negative regulation of extracellular matrix organization(GO:1903054) |
| 0.0 | 0.0 | GO:0042661 | regulation of mesodermal cell fate specification(GO:0042661) regulation of mesoderm development(GO:2000380) |
| 0.0 | 0.1 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.1 | GO:0016102 | retinoic acid biosynthetic process(GO:0002138) diterpenoid biosynthetic process(GO:0016102) |
| 0.0 | 0.1 | GO:0051305 | chromosome movement towards spindle pole(GO:0051305) |
| 0.0 | 0.2 | GO:0031017 | exocrine pancreas development(GO:0031017) |
| 0.0 | 0.1 | GO:0034393 | positive regulation of muscle cell apoptotic process(GO:0010661) positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.0 | 0.2 | GO:0032462 | regulation of protein homooligomerization(GO:0032462) |
| 0.0 | 0.1 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.1 | GO:0033962 | cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.0 | 0.1 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) negative regulation of neural precursor cell proliferation(GO:2000178) |
| 0.0 | 0.1 | GO:0032344 | regulation of ketone biosynthetic process(GO:0010566) regulation of aldosterone metabolic process(GO:0032344) regulation of aldosterone biosynthetic process(GO:0032347) |
| 0.0 | 0.1 | GO:0007028 | cytoplasm organization(GO:0007028) |
| 0.0 | 0.2 | GO:0015840 | urea transport(GO:0015840) |
| 0.0 | 0.0 | GO:0060197 | cloaca development(GO:0035844) cloacal septation(GO:0060197) |
| 0.0 | 0.1 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.1 | GO:2000095 | regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.0 | 0.2 | GO:0070875 | positive regulation of glycogen biosynthetic process(GO:0045725) positive regulation of glycogen metabolic process(GO:0070875) |
| 0.0 | 0.1 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.0 | 0.1 | GO:0008300 | isoprenoid catabolic process(GO:0008300) |
| 0.0 | 0.1 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.0 | 0.1 | GO:0070561 | vitamin D receptor signaling pathway(GO:0070561) |
| 0.0 | 0.0 | GO:0060083 | smooth muscle contraction involved in micturition(GO:0060083) |
| 0.0 | 0.1 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.1 | GO:0015911 | plasma membrane long-chain fatty acid transport(GO:0015911) |
| 0.0 | 0.1 | GO:0060129 | thyroid-stimulating hormone-secreting cell differentiation(GO:0060129) |
| 0.0 | 0.1 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.0 | 0.2 | GO:0043097 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine nucleoside salvage(GO:0043097) |
| 0.0 | 0.1 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.0 | 0.2 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.1 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.0 | GO:0003321 | positive regulation of blood pressure by epinephrine-norepinephrine(GO:0003321) |
| 0.0 | 0.1 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.0 | 0.0 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.0 | 0.1 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.0 | 0.1 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.1 | GO:0045992 | negative regulation of embryonic development(GO:0045992) |
| 0.0 | 0.1 | GO:0060841 | venous blood vessel development(GO:0060841) |
| 0.0 | 0.0 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.0 | 0.1 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 0.0 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.0 | GO:0060433 | bronchus development(GO:0060433) |
| 0.0 | 0.1 | GO:0010193 | response to ozone(GO:0010193) |
| 0.0 | 0.1 | GO:0008215 | spermine metabolic process(GO:0008215) |
| 0.0 | 0.0 | GO:0002689 | negative regulation of leukocyte chemotaxis(GO:0002689) |
| 0.0 | 0.0 | GO:0002887 | negative regulation of myeloid leukocyte mediated immunity(GO:0002887) negative regulation of leukocyte degranulation(GO:0043301) |
| 0.0 | 0.0 | GO:0040019 | positive regulation of embryonic development(GO:0040019) |
| 0.0 | 0.0 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.0 | 0.0 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.0 | 0.1 | GO:0051124 | synaptic growth at neuromuscular junction(GO:0051124) |
| 0.0 | 0.1 | GO:0033008 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of leukocyte degranulation(GO:0043302) positive regulation of mast cell degranulation(GO:0043306) |
| 0.0 | 0.1 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.1 | GO:0022601 | menstrual cycle phase(GO:0022601) |
| 0.0 | 0.1 | GO:0031641 | regulation of myelination(GO:0031641) |
| 0.0 | 0.1 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.1 | GO:0007343 | egg activation(GO:0007343) |
| 0.0 | 0.3 | GO:0021846 | cell proliferation in forebrain(GO:0021846) |
| 0.0 | 0.0 | GO:0048680 | positive regulation of axon regeneration(GO:0048680) positive regulation of neuron projection regeneration(GO:0070572) |
| 0.0 | 0.1 | GO:0071360 | cellular response to exogenous dsRNA(GO:0071360) |
| 0.0 | 0.0 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 0.0 | GO:0033860 | regulation of NAD(P)H oxidase activity(GO:0033860) |
| 0.0 | 0.2 | GO:0048844 | artery morphogenesis(GO:0048844) |
| 0.0 | 0.1 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 0.1 | GO:0036465 | synaptic vesicle recycling(GO:0036465) synaptic vesicle endocytosis(GO:0048488) clathrin-mediated endocytosis(GO:0072583) |
| 0.0 | 0.1 | GO:0008614 | pyridoxine metabolic process(GO:0008614) pyridoxine biosynthetic process(GO:0008615) vitamin B6 metabolic process(GO:0042816) vitamin B6 biosynthetic process(GO:0042819) |
| 0.0 | 0.1 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.3 | GO:0048008 | platelet-derived growth factor receptor signaling pathway(GO:0048008) |
| 0.0 | 0.1 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.1 | GO:0046504 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.1 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.1 | GO:0009200 | deoxyribonucleoside triphosphate metabolic process(GO:0009200) |
| 0.0 | 0.1 | GO:0000052 | citrulline metabolic process(GO:0000052) |
| 0.0 | 0.1 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.0 | GO:0042711 | maternal behavior(GO:0042711) parental behavior(GO:0060746) |
| 0.0 | 0.1 | GO:2001235 | positive regulation of mitochondrion organization(GO:0010822) positive regulation of release of cytochrome c from mitochondria(GO:0090200) positive regulation of apoptotic signaling pathway(GO:2001235) |
| 0.0 | 0.1 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) platelet activating factor metabolic process(GO:0046469) |
| 0.0 | 0.0 | GO:0042253 | granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0042253) |
| 0.0 | 0.1 | GO:0009128 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.0 | 0.1 | GO:0032455 | nerve growth factor processing(GO:0032455) |
| 0.0 | 0.0 | GO:0061213 | positive regulation of mesonephros development(GO:0061213) positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.0 | 0.0 | GO:0060839 | endothelial cell fate commitment(GO:0060839) |
| 0.0 | 0.0 | GO:0060435 | bronchiole development(GO:0060435) |
| 0.0 | 0.0 | GO:0043578 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.0 | GO:0034616 | response to laminar fluid shear stress(GO:0034616) |
| 0.0 | 0.0 | GO:0060872 | semicircular canal development(GO:0060872) |
| 0.0 | 0.2 | GO:0007184 | SMAD protein import into nucleus(GO:0007184) |
| 0.0 | 0.0 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.2 | GO:0007128 | meiotic prophase I(GO:0007128) |
| 0.0 | 0.1 | GO:0050872 | white fat cell differentiation(GO:0050872) |
| 0.0 | 0.1 | GO:0009261 | ribonucleotide catabolic process(GO:0009261) |
| 0.0 | 0.1 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.1 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.1 | GO:0009151 | purine deoxyribonucleotide metabolic process(GO:0009151) |
| 0.0 | 0.0 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.0 | 0.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.1 | GO:1902001 | fatty acid transmembrane transport(GO:1902001) |
| 0.0 | 0.0 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.0 | GO:0021591 | ventricular system development(GO:0021591) |
| 0.0 | 0.1 | GO:0006927 | obsolete transformed cell apoptotic process(GO:0006927) |
| 0.0 | 0.0 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.0 | 0.0 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.0 | 0.0 | GO:0060061 | Spemann organizer formation(GO:0060061) |
| 0.0 | 0.1 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.0 | 0.1 | GO:0048712 | negative regulation of astrocyte differentiation(GO:0048712) |
| 0.0 | 0.1 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.0 | GO:0090025 | regulation of monocyte chemotaxis(GO:0090025) positive regulation of monocyte chemotaxis(GO:0090026) |
| 0.0 | 0.0 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 | 0.0 | GO:0042091 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.0 | 0.1 | GO:0010092 | organ induction(GO:0001759) specification of organ identity(GO:0010092) |
| 0.0 | 0.0 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.1 | GO:0051307 | resolution of meiotic recombination intermediates(GO:0000712) meiotic chromosome separation(GO:0051307) |
| 0.0 | 0.0 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.1 | GO:0060259 | regulation of feeding behavior(GO:0060259) |
| 0.0 | 0.0 | GO:0043461 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.0 | 0.0 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.0 | 0.0 | GO:0042659 | regulation of cell fate specification(GO:0042659) |
| 0.0 | 0.0 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.0 | 0.0 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 0.3 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.0 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.0 | 0.0 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.1 | GO:0001516 | prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.0 | 0.0 | GO:0046831 | regulation of RNA export from nucleus(GO:0046831) |
| 0.0 | 0.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.0 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.0 | GO:0006591 | ornithine metabolic process(GO:0006591) |
| 0.0 | 0.0 | GO:1903020 | positive regulation of glycoprotein biosynthetic process(GO:0010560) positive regulation of glycoprotein metabolic process(GO:1903020) |
| 0.0 | 0.1 | GO:0002040 | sprouting angiogenesis(GO:0002040) |
| 0.0 | 0.0 | GO:0010886 | positive regulation of cholesterol storage(GO:0010886) |
| 0.0 | 0.0 | GO:0014912 | negative regulation of smooth muscle cell migration(GO:0014912) |
| 0.0 | 0.0 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.0 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.0 | 0.0 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.0 | 0.0 | GO:0007501 | mesodermal cell fate specification(GO:0007501) |
| 0.0 | 0.0 | GO:0071462 | cellular response to water stimulus(GO:0071462) |
| 0.0 | 0.0 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.0 | 0.0 | GO:0070254 | mucus secretion(GO:0070254) |
| 0.0 | 0.0 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.0 | GO:0033137 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) |
| 0.0 | 0.0 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.0 | 0.0 | GO:0046121 | deoxyribonucleoside metabolic process(GO:0009120) deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.1 | GO:0015824 | proline transport(GO:0015824) |
| 0.0 | 0.2 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.0 | 0.1 | GO:0009629 | response to gravity(GO:0009629) |
| 0.0 | 0.0 | GO:0045628 | regulation of T-helper 2 cell differentiation(GO:0045628) |
| 0.0 | 0.1 | GO:0010225 | response to UV-C(GO:0010225) |
| 0.0 | 0.0 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.2 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 0.1 | GO:0006195 | purine nucleotide catabolic process(GO:0006195) |
| 0.0 | 0.0 | GO:0032239 | regulation of nucleobase-containing compound transport(GO:0032239) |
| 0.0 | 0.1 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 0.0 | GO:0019255 | glucose 1-phosphate metabolic process(GO:0019255) |
| 0.0 | 0.1 | GO:0032964 | collagen biosynthetic process(GO:0032964) |
| 0.0 | 0.1 | GO:0006206 | pyrimidine nucleobase metabolic process(GO:0006206) |
| 0.0 | 0.0 | GO:0010587 | miRNA metabolic process(GO:0010586) miRNA catabolic process(GO:0010587) |
| 0.0 | 0.1 | GO:0042693 | muscle cell fate commitment(GO:0042693) |
| 0.0 | 0.0 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.1 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 0.0 | GO:0043129 | surfactant homeostasis(GO:0043129) chemical homeostasis within a tissue(GO:0048875) |
| 0.0 | 0.0 | GO:0003032 | detection of oxygen(GO:0003032) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.2 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.1 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.0 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.2 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.3 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.2 | GO:0098642 | network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.2 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.1 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.3 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.1 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.0 | GO:0098651 | basement membrane collagen trimer(GO:0098651) |
| 0.0 | 0.1 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.0 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.1 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.4 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.1 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 0.2 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.1 | GO:0036454 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.0 | 0.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.1 | GO:0031512 | motile primary cilium(GO:0031512) |
| 0.0 | 0.1 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.1 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.0 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.1 | GO:0031312 | extrinsic component of organelle membrane(GO:0031312) |
| 0.0 | 0.6 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.0 | GO:0043230 | extracellular organelle(GO:0043230) |
| 0.0 | 0.1 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.1 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 0.0 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.1 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.0 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.0 | 0.0 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.1 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.0 | GO:0001520 | outer dense fiber(GO:0001520) sperm flagellum(GO:0036126) |
| 0.0 | 0.0 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.1 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.1 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.1 | GO:0031430 | M band(GO:0031430) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 0.3 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.1 | 0.2 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.1 | 0.2 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.1 | 0.2 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 0.2 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 0.2 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.1 | 0.2 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.1 | 0.2 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.1 | 0.2 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.1 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.1 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.2 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.2 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.1 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.2 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.2 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.1 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.2 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.1 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.2 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.4 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.0 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.2 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.1 | GO:0016901 | oxidoreductase activity, acting on the CH-OH group of donors, quinone or similar compound as acceptor(GO:0016901) |
| 0.0 | 0.2 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.0 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.1 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.1 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.0 | 0.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.1 | GO:0071553 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.0 | 0.3 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.1 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.0 | 0.2 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.1 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.1 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.2 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.0 | 0.1 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.2 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.3 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.2 | GO:0070696 | transmembrane receptor protein serine/threonine kinase binding(GO:0070696) |
| 0.0 | 0.1 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.1 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.0 | 0.1 | GO:0008486 | diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) |
| 0.0 | 0.1 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.1 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.0 | 0.3 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.1 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.1 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.1 | GO:0004470 | malic enzyme activity(GO:0004470) |
| 0.0 | 0.1 | GO:0051637 | obsolete Gram-positive bacterial cell surface binding(GO:0051637) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.1 | GO:0042808 | obsolete neuronal Cdc2-like kinase binding(GO:0042808) |
| 0.0 | 0.1 | GO:0033765 | steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) |
| 0.0 | 0.2 | GO:0042171 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) lysophosphatidic acid acyltransferase activity(GO:0042171) lysophospholipid acyltransferase activity(GO:0071617) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.1 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.2 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.0 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.1 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.1 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) |
| 0.0 | 0.1 | GO:0052743 | inositol tetrakisphosphate phosphatase activity(GO:0052743) |
| 0.0 | 0.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.1 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.0 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.0 | 0.1 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.1 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.0 | 0.1 | GO:0004075 | biotin carboxylase activity(GO:0004075) |
| 0.0 | 0.1 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.0 | GO:1901567 | icosanoid binding(GO:0050542) fatty acid derivative binding(GO:1901567) |
| 0.0 | 0.1 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.4 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.1 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.1 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 0.8 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 0.0 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.0 | 0.0 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 0.1 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.0 | 0.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.1 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.4 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.0 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.0 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.2 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.0 | 0.1 | GO:0004645 | phosphorylase activity(GO:0004645) |
| 0.0 | 0.2 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.1 | GO:0051635 | obsolete bacterial cell surface binding(GO:0051635) |
| 0.0 | 0.0 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.0 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.0 | 0.0 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.0 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.0 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.0 | 0.2 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.0 | 0.0 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.1 | GO:0001595 | angiotensin receptor activity(GO:0001595) angiotensin type II receptor activity(GO:0004945) |
| 0.0 | 0.3 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.0 | 0.0 | GO:0097493 | extracellular matrix constituent conferring elasticity(GO:0030023) structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.0 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.1 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.2 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) protein tyrosine kinase binding(GO:1990782) |
| 0.0 | 0.0 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.0 | 0.2 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.0 | 0.0 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.0 | 0.1 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.1 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.2 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.0 | 0.1 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.0 | 0.0 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.0 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.0 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.0 | 0.1 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 0.0 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.1 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.1 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.1 | GO:0016248 | channel inhibitor activity(GO:0016248) |
| 0.0 | 0.0 | GO:0008061 | chitin binding(GO:0008061) |
| 0.0 | 0.1 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.0 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.0 | 0.1 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 0.1 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.1 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.0 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.0 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.0 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.0 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.0 | 0.0 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.2 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.0 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.0 | 0.0 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.1 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.1 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 0.0 | 0.0 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.0 | 0.1 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.1 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.1 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 0.2 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.3 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.7 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.0 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.0 | 0.1 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.1 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.0 | 0.3 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.0 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.0 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.0 | 0.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.1 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | REACTOME DAG AND IP3 SIGNALING | Genes involved in DAG and IP3 signaling |
| 0.0 | 0.4 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.9 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.0 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.6 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.1 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.0 | 0.1 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.1 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.0 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.4 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.5 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.2 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.0 | REACTOME IL 3 5 AND GM CSF SIGNALING | Genes involved in Interleukin-3, 5 and GM-CSF signaling |
| 0.0 | 0.3 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.3 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.1 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.3 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.1 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.1 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.7 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.3 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.0 | 0.2 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.0 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.3 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 0.1 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.4 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.0 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.0 | 0.1 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.2 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.1 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |