Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
Results for NKX6-3
Z-value: 1.12
Transcription factors associated with NKX6-3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NKX6-3
|
ENSG00000165066.11 | NK6 homeobox 3 |
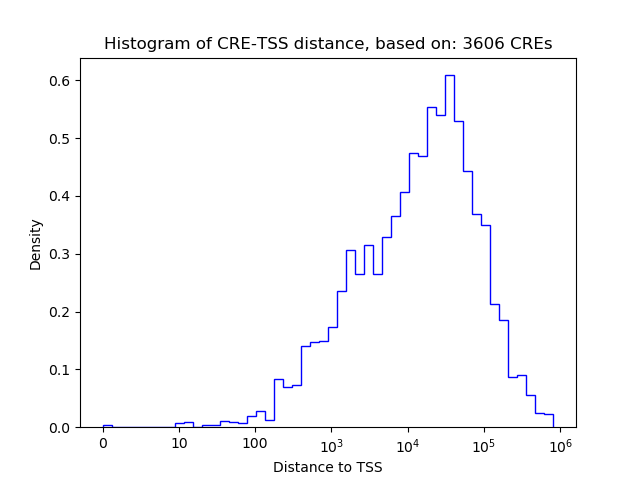
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr8_41503976_41504252 | NKX6-3 | 764 | 0.539993 | 0.52 | 1.6e-01 | Click! |
| chr8_41512290_41512441 | NKX6-3 | 3510 | 0.156058 | -0.51 | 1.6e-01 | Click! |
| chr8_41506141_41506292 | NKX6-3 | 1338 | 0.326383 | -0.42 | 2.6e-01 | Click! |
| chr8_41503640_41503791 | NKX6-3 | 1163 | 0.373407 | 0.39 | 3.0e-01 | Click! |
| chr8_41503291_41503442 | NKX6-3 | 1512 | 0.291593 | 0.36 | 3.4e-01 | Click! |
Activity of the NKX6-3 motif across conditions
Conditions sorted by the z-value of the NKX6-3 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
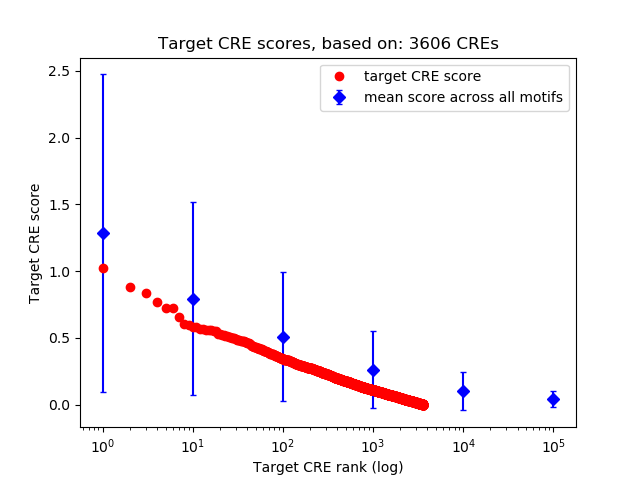
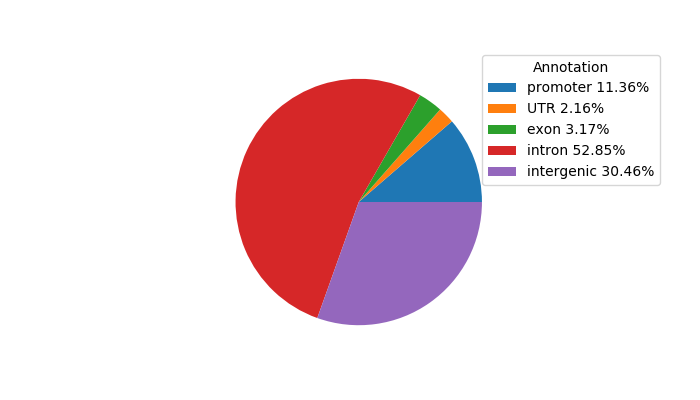
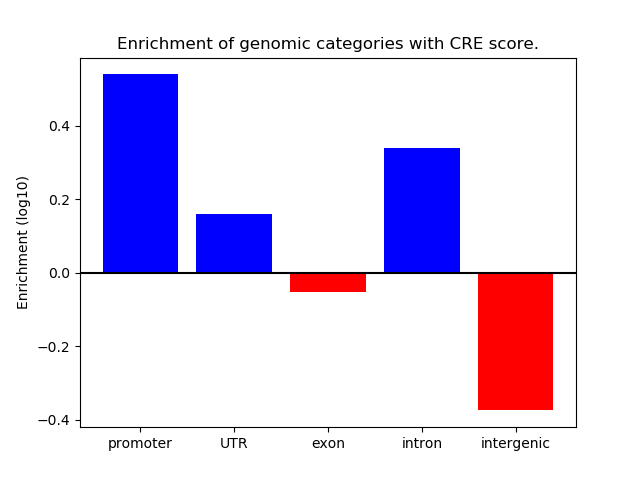
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr11_33712841_33713004 | 1.02 |
RP4-541C22.5 |
|
5325 |
0.18 |
| chr22_18539461_18539695 | 0.88 |
XXbac-B476C20.9 |
|
20985 |
0.13 |
| chr11_102722401_102722552 | 0.83 |
MMP3 |
matrix metallopeptidase 3 (stromelysin 1, progelatinase) |
7942 |
0.19 |
| chr4_145504640_145504791 | 0.77 |
HHIP-AS1 |
HHIP antisense RNA 1 |
62349 |
0.14 |
| chr9_14251921_14252072 | 0.73 |
NFIB |
nuclear factor I/B |
56016 |
0.15 |
| chr6_114196551_114196945 | 0.73 |
MARCKS |
myristoylated alanine-rich protein kinase C substrate |
18207 |
0.17 |
| chr6_52425530_52425681 | 0.66 |
TRAM2 |
translocation associated membrane protein 2 |
16108 |
0.24 |
| chr1_206603221_206603372 | 0.61 |
SRGAP2 |
SLIT-ROBO Rho GTPase activating protein 2 |
20488 |
0.16 |
| chr12_10545690_10545841 | 0.60 |
KLRK1 |
killer cell lectin-like receptor subfamily K, member 1 |
3148 |
0.16 |
| chr2_225681896_225682047 | 0.58 |
DOCK10 |
dedicator of cytokinesis 10 |
602 |
0.85 |
| chr10_65460709_65460902 | 0.58 |
REEP3 |
receptor accessory protein 3 |
179682 |
0.03 |
| chr5_139626504_139626769 | 0.57 |
CTB-131B5.2 |
|
46888 |
0.09 |
| chr17_61357894_61358045 | 0.57 |
RP11-269G24.2 |
|
32913 |
0.16 |
| chr3_177651530_177651798 | 0.56 |
ENSG00000199858 |
. |
73903 |
0.13 |
| chr2_48571922_48572463 | 0.56 |
ENSG00000251889 |
. |
4227 |
0.27 |
| chr16_27590290_27590441 | 0.56 |
KIAA0556 |
KIAA0556 |
5137 |
0.22 |
| chr5_16890159_16890310 | 0.55 |
MYO10 |
myosin X |
26402 |
0.18 |
| chr4_47215322_47215473 | 0.55 |
ENSG00000238301 |
. |
92657 |
0.09 |
| chr4_14728667_14728818 | 0.53 |
ENSG00000202449 |
. |
36408 |
0.24 |
| chr5_158370461_158370612 | 0.53 |
CTD-2363C16.1 |
|
39478 |
0.18 |
| chr3_188995375_188995558 | 0.52 |
TPRG1-AS2 |
TPRG1 antisense RNA 2 |
37083 |
0.21 |
| chr14_54405199_54405350 | 0.52 |
ENSG00000266431 |
. |
9928 |
0.25 |
| chr4_187608141_187608292 | 0.52 |
FAT1 |
FAT atypical cadherin 1 |
36793 |
0.2 |
| chr5_64504974_64505125 | 0.51 |
ENSG00000207439 |
. |
85853 |
0.1 |
| chr7_72619380_72619686 | 0.51 |
NCF1B |
neutrophil cytosolic factor 1B pseudogene |
15078 |
0.17 |
| chr4_125335710_125335861 | 0.51 |
ANKRD50 |
ankyrin repeat domain 50 |
298102 |
0.01 |
| chr17_454384_454535 | 0.50 |
RP5-1029F21.2 |
|
1347 |
0.44 |
| chr16_84765282_84765475 | 0.50 |
USP10 |
ubiquitin specific peptidase 10 |
31732 |
0.17 |
| chr3_149298844_149298995 | 0.50 |
WWTR1 |
WW domain containing transcription regulator 1 |
4880 |
0.23 |
| chr6_56591218_56591369 | 0.49 |
DST |
dystonin |
59384 |
0.15 |
| chr6_44414403_44414554 | 0.49 |
ENSG00000266619 |
. |
11100 |
0.18 |
| chr6_158591128_158591427 | 0.48 |
GTF2H5 |
general transcription factor IIH, polypeptide 5 |
1893 |
0.27 |
| chr2_150601107_150601258 | 0.48 |
ENSG00000207270 |
. |
133894 |
0.05 |
| chr22_33108393_33108637 | 0.48 |
LL22NC01-116C6.1 |
|
70648 |
0.1 |
| chr11_111685692_111685843 | 0.48 |
ALG9-IT1 |
ALG9 intronic transcript 1 (non-protein coding) |
14169 |
0.14 |
| chr12_10344371_10344522 | 0.48 |
TMEM52B |
transmembrane protein 52B |
12333 |
0.12 |
| chr1_246132469_246132620 | 0.47 |
RP11-83A16.1 |
|
64309 |
0.14 |
| chr2_182941626_182941777 | 0.47 |
ENSG00000265129 |
. |
11642 |
0.22 |
| chr8_131156068_131156219 | 0.47 |
ENSG00000212342 |
. |
19075 |
0.2 |
| chr1_67397953_67399117 | 0.46 |
MIER1 |
mesoderm induction early response 1, transcriptional regulator |
2609 |
0.29 |
| chr8_126231993_126232161 | 0.46 |
ENSG00000242170 |
. |
50769 |
0.15 |
| chr2_217461850_217462001 | 0.46 |
AC073321.3 |
|
9736 |
0.19 |
| chr10_28601668_28601819 | 0.46 |
ENSG00000222374 |
. |
1961 |
0.37 |
| chr4_170165351_170165677 | 0.44 |
SH3RF1 |
SH3 domain containing ring finger 1 |
25594 |
0.22 |
| chr6_161590013_161590164 | 0.44 |
AGPAT4 |
1-acylglycerol-3-phosphate O-acyltransferase 4 |
19767 |
0.28 |
| chr18_46056441_46056592 | 0.44 |
CTIF |
CBP80/20-dependent translation initiation factor |
8901 |
0.25 |
| chr2_153441109_153441260 | 0.44 |
FMNL2 |
formin-like 2 |
34908 |
0.22 |
| chr1_222341096_222341247 | 0.44 |
ENSG00000212094 |
. |
154739 |
0.04 |
| chr7_112636182_112636333 | 0.43 |
C7orf60 |
chromosome 7 open reading frame 60 |
56286 |
0.14 |
| chr7_42233492_42233928 | 0.43 |
GLI3 |
GLI family zinc finger 3 |
33610 |
0.25 |
| chr20_11120768_11120919 | 0.43 |
C20orf187 |
chromosome 20 open reading frame 187 |
112032 |
0.07 |
| chr4_41150165_41150316 | 0.43 |
ENSG00000207198 |
. |
34281 |
0.16 |
| chr3_188067044_188067254 | 0.42 |
LPP |
LIM domain containing preferred translocation partner in lipoma |
109497 |
0.07 |
| chr14_101459833_101459984 | 0.42 |
ENSG00000200089 |
. |
335 |
0.31 |
| chr4_141866091_141866242 | 0.42 |
RNF150 |
ring finger protein 150 |
52024 |
0.17 |
| chr8_128755138_128755289 | 0.42 |
MYC |
v-myc avian myelocytomatosis viral oncogene homolog |
6736 |
0.28 |
| chr4_186196562_186196713 | 0.42 |
SNX25 |
sorting nexin 25 |
65404 |
0.09 |
| chr17_17640963_17641296 | 0.41 |
RAI1-AS1 |
RAI1 antisense RNA 1 |
33006 |
0.11 |
| chr9_75764547_75765336 | 0.41 |
ANXA1 |
annexin A1 |
1732 |
0.5 |
| chr13_43580173_43580324 | 0.41 |
EPSTI1 |
epithelial stromal interaction 1 (breast) |
13863 |
0.24 |
| chr12_27763041_27763300 | 0.40 |
PPFIBP1 |
PTPRF interacting protein, binding protein 1 (liprin beta 1) |
10889 |
0.22 |
| chr15_72045393_72045544 | 0.40 |
CTD-2524L6.3 |
|
65269 |
0.12 |
| chr12_125160668_125160889 | 0.40 |
NCOR2 |
nuclear receptor corepressor 2 |
108768 |
0.07 |
| chr2_46325558_46325709 | 0.40 |
AC017006.2 |
|
19666 |
0.24 |
| chr10_71734773_71734924 | 0.40 |
COL13A1 |
collagen, type XIII, alpha 1 |
44639 |
0.15 |
| chr8_58980536_58980687 | 0.40 |
FAM110B |
family with sequence similarity 110, member B |
73498 |
0.13 |
| chr11_111848937_111849153 | 0.39 |
DIXDC1 |
DIX domain containing 1 |
1012 |
0.41 |
| chr3_188104571_188104722 | 0.39 |
LPP |
LIM domain containing preferred translocation partner in lipoma |
146994 |
0.04 |
| chr2_196547314_196547465 | 0.39 |
ENSG00000201813 |
. |
12191 |
0.23 |
| chr11_110228298_110228580 | 0.39 |
RDX |
radixin |
60992 |
0.14 |
| chr6_79614930_79615081 | 0.39 |
IRAK1BP1 |
interleukin-1 receptor-associated kinase 1 binding protein 1 |
7096 |
0.29 |
| chr2_55328902_55329053 | 0.38 |
ENSG00000266376 |
. |
6869 |
0.2 |
| chr12_89461977_89462128 | 0.38 |
ENSG00000238302 |
. |
214010 |
0.02 |
| chr12_94181628_94181779 | 0.38 |
ENSG00000264978 |
. |
43149 |
0.13 |
| chr6_139293869_139294337 | 0.38 |
REPS1 |
RALBP1 associated Eps domain containing 1 |
14674 |
0.24 |
| chr11_60686848_60686999 | 0.38 |
TMEM109 |
transmembrane protein 109 |
227 |
0.84 |
| chr4_7881276_7881455 | 0.38 |
AFAP1 |
actin filament associated protein 1 |
7325 |
0.25 |
| chr7_116211652_116211947 | 0.38 |
AC006159.4 |
|
511 |
0.78 |
| chr16_74514100_74514392 | 0.37 |
ENSG00000207525 |
. |
16360 |
0.16 |
| chr2_161608908_161609059 | 0.37 |
ENSG00000244372 |
. |
125075 |
0.06 |
| chr11_65047025_65047288 | 0.37 |
POLA2 |
polymerase (DNA directed), alpha 2, accessory subunit |
2822 |
0.18 |
| chr2_197264610_197264832 | 0.37 |
HECW2 |
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2 |
37775 |
0.2 |
| chr6_108975764_108975915 | 0.37 |
FOXO3 |
forkhead box O3 |
1710 |
0.51 |
| chr4_55408709_55408988 | 0.37 |
KIT |
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog |
115237 |
0.07 |
| chr7_129933263_129933414 | 0.36 |
CPA4 |
carboxypeptidase A4 |
219 |
0.91 |
| chrX_39832066_39832217 | 0.36 |
BCOR |
BCL6 corepressor |
90049 |
0.09 |
| chr11_126788305_126788456 | 0.36 |
KIRREL3-AS2 |
KIRREL3 antisense RNA 2 |
22262 |
0.2 |
| chr8_143757168_143757384 | 0.36 |
PSCA |
prostate stem cell antigen |
4598 |
0.13 |
| chr1_8138703_8138854 | 0.36 |
ERRFI1 |
ERBB receptor feedback inhibitor 1 |
52410 |
0.13 |
| chr8_90739175_90739326 | 0.36 |
RIPK2 |
receptor-interacting serine-threonine kinase 2 |
30725 |
0.24 |
| chr8_91783068_91783219 | 0.36 |
NECAB1 |
N-terminal EF-hand calcium binding protein 1 |
20635 |
0.19 |
| chr3_182992678_182993398 | 0.35 |
B3GNT5 |
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5 |
9906 |
0.2 |
| chr4_154388689_154389189 | 0.35 |
KIAA0922 |
KIAA0922 |
1438 |
0.51 |
| chr3_197098695_197098846 | 0.35 |
ENSG00000238491 |
. |
32348 |
0.17 |
| chr2_47101079_47101246 | 0.35 |
AC016722.3 |
|
20527 |
0.13 |
| chr5_95064137_95064401 | 0.35 |
RHOBTB3 |
Rho-related BTB domain containing 3 |
2307 |
0.25 |
| chr2_36581626_36581777 | 0.35 |
CRIM1 |
cysteine rich transmembrane BMP regulator 1 (chordin-like) |
1368 |
0.59 |
| chr5_72327163_72327314 | 0.35 |
ENSG00000200833 |
. |
68662 |
0.09 |
| chr10_31604223_31604374 | 0.35 |
ENSG00000237036 |
. |
3563 |
0.23 |
| chr2_161232105_161232912 | 0.34 |
ENSG00000252465 |
. |
20971 |
0.19 |
| chr3_145882013_145882164 | 0.34 |
PLOD2 |
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2 |
648 |
0.77 |
| chr10_89536127_89536278 | 0.34 |
ATAD1 |
ATPase family, AAA domain containing 1 |
16312 |
0.18 |
| chr6_15095366_15095517 | 0.34 |
ENSG00000242989 |
. |
17758 |
0.25 |
| chr7_106873690_106873841 | 0.34 |
HBP1 |
HMG-box transcription factor 1 |
37472 |
0.15 |
| chr8_53031421_53031572 | 0.34 |
RP11-26M5.3 |
|
31884 |
0.21 |
| chr10_80832109_80832421 | 0.34 |
ZMIZ1 |
zinc finger, MIZ-type containing 1 |
3473 |
0.29 |
| chr8_107951495_107951646 | 0.34 |
ABRA |
actin-binding Rho activating protein |
169097 |
0.04 |
| chr7_116350835_116350986 | 0.34 |
MET |
met proto-oncogene |
11771 |
0.25 |
| chr12_14990014_14990290 | 0.33 |
RP11-233G1.4 |
|
3874 |
0.14 |
| chr7_36487948_36488099 | 0.33 |
AC006960.7 |
|
14609 |
0.19 |
| chr8_38259724_38259888 | 0.33 |
RP11-350N15.3 |
|
605 |
0.59 |
| chr18_73120073_73120224 | 0.33 |
RP11-321M21.3 |
HCG1996301; Uncharacterized protein |
1181 |
0.58 |
| chr11_131765892_131766043 | 0.33 |
AP004372.1 |
|
1035 |
0.61 |
| chr5_142682227_142682378 | 0.33 |
ARHGAP26 |
Rho GTPase activating protein 26 |
95537 |
0.08 |
| chr16_24742094_24743166 | 0.33 |
TNRC6A |
trinucleotide repeat containing 6A |
1596 |
0.5 |
| chr1_198874035_198874197 | 0.33 |
ENSG00000207759 |
. |
45834 |
0.17 |
| chr4_26344038_26344485 | 0.33 |
RBPJ |
recombination signal binding protein for immunoglobulin kappa J region |
501 |
0.88 |
| chr1_113666672_113666823 | 0.33 |
LRIG2 |
leucine-rich repeats and immunoglobulin-like domains 2 |
50916 |
0.17 |
| chr9_80078507_80078658 | 0.33 |
RP11-466A17.1 |
|
7371 |
0.28 |
| chr20_45905361_45905512 | 0.33 |
ZMYND8 |
zinc finger, MYND-type containing 8 |
22208 |
0.14 |
| chr2_218258772_218258923 | 0.33 |
ENSG00000251982 |
. |
142568 |
0.05 |
| chr1_172855064_172855215 | 0.33 |
TNFSF18 |
tumor necrosis factor (ligand) superfamily, member 18 |
164917 |
0.04 |
| chr6_117769723_117769874 | 0.32 |
ENSG00000221434 |
. |
3245 |
0.19 |
| chr4_170081579_170081730 | 0.32 |
RP11-327O17.2 |
|
41291 |
0.18 |
| chr6_21004450_21004601 | 0.32 |
ENSG00000252046 |
. |
10726 |
0.32 |
| chr14_52795656_52796058 | 0.32 |
PTGER2 |
prostaglandin E receptor 2 (subtype EP2), 53kDa |
14744 |
0.25 |
| chr3_150422131_150422452 | 0.32 |
RP11-103G8.2 |
|
461 |
0.51 |
| chr10_29772721_29772872 | 0.32 |
SVIL |
supervillin |
12651 |
0.23 |
| chr11_28855936_28856087 | 0.32 |
ENSG00000211499 |
. |
442225 |
0.01 |
| chr2_142315504_142315655 | 0.32 |
ENSG00000265224 |
. |
80834 |
0.12 |
| chr16_81638343_81638641 | 0.32 |
CMIP |
c-Maf inducing protein |
40471 |
0.2 |
| chr10_62187776_62187927 | 0.31 |
ANK3 |
ankyrin 3, node of Ranvier (ankyrin G) |
38363 |
0.23 |
| chr2_56181551_56181702 | 0.31 |
ENSG00000202344 |
. |
2477 |
0.26 |
| chr6_38228109_38228260 | 0.31 |
ENSG00000200706 |
. |
53134 |
0.16 |
| chr4_184558434_184558585 | 0.31 |
ENSG00000252157 |
. |
16320 |
0.14 |
| chr3_45521464_45521615 | 0.31 |
LARS2-AS1 |
LARS2 antisense RNA 1 |
29498 |
0.2 |
| chr3_136505593_136506225 | 0.31 |
SLC35G2 |
solute carrier family 35, member G2 |
31952 |
0.12 |
| chr17_39218871_39219022 | 0.31 |
KRTAP2-3 |
keratin associated protein 2-3 |
2602 |
0.09 |
| chr18_67711328_67711479 | 0.30 |
RTTN |
rotatin |
23449 |
0.25 |
| chr8_77711424_77711575 | 0.30 |
ZFHX4 |
zinc finger homeobox 4 |
95219 |
0.08 |
| chr12_66345138_66345289 | 0.30 |
RP11-366L20.3 |
|
2954 |
0.27 |
| chr2_43986548_43986990 | 0.30 |
ENSG00000252490 |
. |
12720 |
0.19 |
| chr14_90557642_90557793 | 0.30 |
KCNK13 |
potassium channel, subfamily K, member 13 |
29608 |
0.2 |
| chr11_20505330_20505481 | 0.30 |
PRMT3 |
protein arginine methyltransferase 3 |
96143 |
0.08 |
| chr4_15013014_15013165 | 0.30 |
CPEB2 |
cytoplasmic polyadenylation element binding protein 2 |
7529 |
0.3 |
| chr1_25666704_25667414 | 0.30 |
ENSG00000252515 |
. |
403 |
0.75 |
| chr5_57485225_57485376 | 0.30 |
ENSG00000238899 |
. |
119285 |
0.06 |
| chr12_65288524_65288675 | 0.30 |
ENSG00000221564 |
. |
44204 |
0.15 |
| chr1_61989098_61989249 | 0.30 |
ENSG00000264551 |
. |
105530 |
0.07 |
| chr4_183161080_183161231 | 0.30 |
TENM3 |
teneurin transmembrane protein 3 |
3427 |
0.32 |
| chrX_41135797_41136128 | 0.30 |
ENSG00000199564 |
. |
16089 |
0.18 |
| chr9_117877775_117877926 | 0.30 |
TNC |
tenascin C |
2628 |
0.36 |
| chr11_74659216_74659741 | 0.30 |
XRRA1 |
X-ray radiation resistance associated 1 |
587 |
0.48 |
| chr10_62570411_62570562 | 0.30 |
CDK1 |
cyclin-dependent kinase 1 |
30575 |
0.22 |
| chr1_198226866_198227037 | 0.30 |
NEK7 |
NIMA-related kinase 7 |
37022 |
0.23 |
| chr2_145088605_145089481 | 0.30 |
GTDC1 |
glycosyltransferase-like domain containing 1 |
1040 |
0.68 |
| chr1_198238756_198238907 | 0.30 |
NEK7 |
NIMA-related kinase 7 |
48902 |
0.19 |
| chr13_51104174_51104472 | 0.29 |
ENSG00000221198 |
. |
166950 |
0.04 |
| chr15_45881879_45882313 | 0.29 |
BLOC1S6 |
biogenesis of lysosomal organelles complex-1, subunit 6, pallidin |
2073 |
0.2 |
| chr1_165739030_165739243 | 0.29 |
TMCO1 |
transmembrane and coiled-coil domains 1 |
719 |
0.5 |
| chr7_13985521_13985672 | 0.29 |
ETV1 |
ets variant 1 |
40470 |
0.2 |
| chr15_38690005_38690267 | 0.29 |
FAM98B |
family with sequence similarity 98, member B |
56192 |
0.14 |
| chr2_9171149_9171380 | 0.29 |
MBOAT2 |
membrane bound O-acyltransferase domain containing 2 |
27322 |
0.22 |
| chr3_149189968_149190119 | 0.29 |
TM4SF4 |
transmembrane 4 L six family member 4 |
1718 |
0.4 |
| chr12_67665547_67665828 | 0.29 |
CAND1 |
cullin-associated and neddylation-dissociated 1 |
2006 |
0.5 |
| chr18_43819137_43819353 | 0.29 |
C18orf25 |
chromosome 18 open reading frame 25 |
65245 |
0.11 |
| chr7_137534903_137535054 | 0.29 |
DGKI |
diacylglycerol kinase, iota |
3140 |
0.34 |
| chr6_111273369_111273520 | 0.29 |
GTF3C6 |
general transcription factor IIIC, polypeptide 6, alpha 35kDa |
6319 |
0.15 |
| chr4_24069141_24069414 | 0.29 |
PPARGC1A |
peroxisome proliferator-activated receptor gamma, coactivator 1 alpha |
177577 |
0.03 |
| chr4_138974616_138974767 | 0.29 |
ENSG00000250033 |
. |
3927 |
0.38 |
| chr3_151985241_151985971 | 0.29 |
MBNL1 |
muscleblind-like splicing regulator 1 |
223 |
0.93 |
| chr2_225027538_225027689 | 0.29 |
ENSG00000211987 |
. |
76660 |
0.11 |
| chr14_80052151_80052302 | 0.29 |
RP11-242P2.2 |
|
47428 |
0.2 |
| chr1_179148045_179148196 | 0.29 |
ENSG00000212338 |
. |
17488 |
0.17 |
| chr4_138975505_138975656 | 0.28 |
ENSG00000250033 |
. |
3038 |
0.41 |
| chr21_28941972_28942234 | 0.28 |
ADAMTS5 |
ADAM metallopeptidase with thrombospondin type 1 motif, 5 |
603271 |
0.0 |
| chr2_179070454_179070605 | 0.28 |
OSBPL6 |
oxysterol binding protein-like 6 |
11153 |
0.22 |
| chr9_89952372_89952738 | 0.28 |
ENSG00000212421 |
. |
77190 |
0.11 |
| chr10_15368183_15368334 | 0.28 |
FAM171A1 |
family with sequence similarity 171, member A1 |
44800 |
0.17 |
| chr20_37614000_37614151 | 0.28 |
DHX35 |
DEAH (Asp-Glu-Ala-His) box polypeptide 35 |
23088 |
0.18 |
| chr8_29109260_29109557 | 0.28 |
KIF13B |
kinesin family member 13B |
11182 |
0.17 |
| chr8_25032357_25032508 | 0.28 |
ENSG00000241811 |
. |
6803 |
0.25 |
| chr5_64459127_64459278 | 0.28 |
ENSG00000207439 |
. |
40006 |
0.22 |
| chr10_95235571_95235788 | 0.28 |
MYOF |
myoferlin |
6272 |
0.2 |
| chr2_100218822_100218973 | 0.28 |
AFF3 |
AF4/FMR2 family, member 3 |
24065 |
0.23 |
| chr21_40683413_40684326 | 0.28 |
BRWD1 |
bromodomain and WD repeat domain containing 1 |
1627 |
0.24 |
| chr5_92694884_92695035 | 0.28 |
ENSG00000237187 |
. |
63160 |
0.15 |
| chr8_128937773_128937924 | 0.28 |
TMEM75 |
transmembrane protein 75 |
22743 |
0.19 |
| chr2_1657675_1657826 | 0.28 |
AC144450.1 |
|
33865 |
0.19 |
| chr5_154158547_154158698 | 0.28 |
LARP1 |
La ribonucleoprotein domain family, member 1 |
13671 |
0.17 |
| chr3_142719040_142719191 | 0.28 |
U2SURP |
U2 snRNP-associated SURP domain containing |
1251 |
0.46 |
| chr1_150450551_150450702 | 0.28 |
TARS2 |
threonyl-tRNA synthetase 2, mitochondrial (putative) |
9261 |
0.13 |
| chr10_59788519_59788670 | 0.28 |
ENSG00000238970 |
. |
209927 |
0.02 |
| chr1_61918584_61919140 | 0.28 |
NFIA |
nuclear factor I/A |
46518 |
0.19 |
| chr5_92353866_92354017 | 0.28 |
ENSG00000221810 |
. |
300721 |
0.01 |
| chr5_76134379_76134541 | 0.28 |
ENSG00000264391 |
. |
1440 |
0.37 |
| chr16_58597342_58597493 | 0.28 |
ENSG00000206952 |
. |
3582 |
0.17 |
| chr18_30492153_30492304 | 0.27 |
RP11-680N20.1 |
|
41811 |
0.18 |
| chr1_76231815_76231966 | 0.27 |
RABGGTB |
Rab geranylgeranyltransferase, beta subunit |
19989 |
0.11 |
| chr12_10544948_10545233 | 0.27 |
KLRK1 |
killer cell lectin-like receptor subfamily K, member 1 |
2473 |
0.19 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 0.2 | GO:0060831 | smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:0060831) |
| 0.1 | 0.2 | GO:1901722 | regulation of cell proliferation involved in kidney development(GO:1901722) |
| 0.0 | 0.2 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.1 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.0 | 0.2 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 | 0.1 | GO:0010837 | regulation of keratinocyte proliferation(GO:0010837) |
| 0.0 | 0.2 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.1 | GO:0045046 | protein import into peroxisome membrane(GO:0045046) |
| 0.0 | 0.1 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.0 | 0.2 | GO:0001547 | antral ovarian follicle growth(GO:0001547) |
| 0.0 | 0.1 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.0 | 0.1 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.1 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.1 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.0 | 0.2 | GO:0060390 | regulation of SMAD protein import into nucleus(GO:0060390) |
| 0.0 | 0.1 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 0.1 | GO:0006166 | purine ribonucleoside salvage(GO:0006166) |
| 0.0 | 0.1 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.0 | 0.1 | GO:0007028 | cytoplasm organization(GO:0007028) |
| 0.0 | 0.1 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.0 | 0.0 | GO:0060686 | negative regulation of prostatic bud formation(GO:0060686) |
| 0.0 | 0.0 | GO:0071504 | cellular response to heparin(GO:0071504) |
| 0.0 | 0.1 | GO:0042797 | 5S class rRNA transcription from RNA polymerase III type 1 promoter(GO:0042791) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
| 0.0 | 0.2 | GO:0009886 | post-embryonic morphogenesis(GO:0009886) |
| 0.0 | 0.0 | GO:0090081 | regulation of heart induction by regulation of canonical Wnt signaling pathway(GO:0090081) |
| 0.0 | 0.2 | GO:0097354 | protein prenylation(GO:0018342) prenylation(GO:0097354) |
| 0.0 | 0.1 | GO:0046719 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) regulation by virus of viral protein levels in host cell(GO:0046719) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.0 | 0.2 | GO:0006896 | Golgi to endosome transport(GO:0006895) Golgi to vacuole transport(GO:0006896) |
| 0.0 | 0.0 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 | 0.1 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.0 | GO:0072112 | renal filtration cell differentiation(GO:0061318) glomerular epithelium development(GO:0072010) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.0 | 0.1 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.1 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.0 | 0.1 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.0 | 0.0 | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.0 | 0.1 | GO:0010566 | regulation of ketone biosynthetic process(GO:0010566) regulation of aldosterone metabolic process(GO:0032344) regulation of aldosterone biosynthetic process(GO:0032347) |
| 0.0 | 0.1 | GO:0034695 | response to prostaglandin E(GO:0034695) |
| 0.0 | 0.0 | GO:0035385 | Roundabout signaling pathway(GO:0035385) |
| 0.0 | 0.1 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.1 | GO:0051305 | chromosome movement towards spindle pole(GO:0051305) |
| 0.0 | 0.4 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 0.0 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.1 | GO:0031657 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G1/S transition of mitotic cell cycle(GO:0031657) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G1/S transition of mitotic cell cycle(GO:0031659) |
| 0.0 | 0.1 | GO:0070254 | mucus secretion(GO:0070254) |
| 0.0 | 0.1 | GO:0060638 | mesenchymal-epithelial cell signaling(GO:0060638) |
| 0.0 | 0.0 | GO:0021834 | chemorepulsion involved in embryonic olfactory bulb interneuron precursor migration(GO:0021834) |
| 0.0 | 0.0 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.3 | GO:1905037 | autophagosome assembly(GO:0000045) autophagosome organization(GO:1905037) |
| 0.0 | 0.1 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.0 | 0.1 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.0 | 0.1 | GO:0009649 | entrainment of circadian clock(GO:0009649) |
| 0.0 | 0.1 | GO:0060039 | pericardium development(GO:0060039) |
| 0.0 | 0.1 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.1 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.0 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
| 0.0 | 0.4 | GO:0048009 | insulin-like growth factor receptor signaling pathway(GO:0048009) |
| 0.0 | 0.1 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.0 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.0 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.0 | GO:0060789 | hair follicle placode formation(GO:0060789) |
| 0.0 | 0.0 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.0 | 0.0 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.0 | 0.0 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.0 | 0.1 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.1 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 0.1 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.0 | GO:0032928 | regulation of superoxide anion generation(GO:0032928) |
| 0.0 | 0.0 | GO:0015864 | pyrimidine nucleoside transport(GO:0015864) |
| 0.0 | 0.0 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.0 | 0.0 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.0 | 0.1 | GO:0001945 | lymph vessel development(GO:0001945) |
| 0.0 | 0.1 | GO:0033261 | obsolete regulation of S phase(GO:0033261) |
| 0.0 | 0.0 | GO:0016999 | antibiotic metabolic process(GO:0016999) antibiotic biosynthetic process(GO:0017000) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.1 | GO:1901339 | activation of store-operated calcium channel activity(GO:0032237) regulation of store-operated calcium channel activity(GO:1901339) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.0 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.1 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.0 | 0.0 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.0 | 0.0 | GO:0002220 | innate immune response activating cell surface receptor signaling pathway(GO:0002220) |
| 0.0 | 0.2 | GO:0032964 | collagen biosynthetic process(GO:0032964) |
| 0.0 | 0.0 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.0 | 0.0 | GO:0070561 | vitamin D receptor signaling pathway(GO:0070561) |
| 0.0 | 0.0 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.0 | 0.0 | GO:0033600 | negative regulation of mammary gland epithelial cell proliferation(GO:0033600) |
| 0.0 | 0.0 | GO:0035511 | oxidative DNA demethylation(GO:0035511) |
| 0.0 | 0.0 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.0 | 0.1 | GO:0010700 | negative regulation of norepinephrine secretion(GO:0010700) |
| 0.0 | 0.0 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.1 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.2 | GO:0043631 | RNA polyadenylation(GO:0043631) |
| 0.0 | 0.1 | GO:0006853 | carnitine shuttle(GO:0006853) carnitine transmembrane transport(GO:1902603) |
| 0.0 | 0.2 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.1 | GO:0072665 | protein targeting to lysosome(GO:0006622) protein targeting to vacuole(GO:0006623) protein localization to lysosome(GO:0061462) protein localization to vacuole(GO:0072665) establishment of protein localization to vacuole(GO:0072666) |
| 0.0 | 0.0 | GO:0060242 | contact inhibition(GO:0060242) |
| 0.0 | 0.0 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.0 | 0.0 | GO:0060457 | negative regulation of digestive system process(GO:0060457) |
| 0.0 | 0.0 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.0 | 0.2 | GO:0035195 | gene silencing by miRNA(GO:0035195) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.1 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.1 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.1 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.1 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.1 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.1 | GO:0005954 | calcium- and calmodulin-dependent protein kinase complex(GO:0005954) |
| 0.0 | 0.2 | GO:0030990 | intraciliary transport particle(GO:0030990) |
| 0.0 | 0.1 | GO:0043656 | host(GO:0018995) host cell part(GO:0033643) host intracellular part(GO:0033646) intracellular region of host(GO:0043656) host cell(GO:0043657) other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) |
| 0.0 | 0.1 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.1 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.1 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.1 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.1 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.2 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.1 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.0 | GO:0071942 | XPC complex(GO:0071942) |
| 0.0 | 0.0 | GO:0071778 | obsolete WINAC complex(GO:0071778) |
| 0.0 | 0.1 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.1 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.1 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.3 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.1 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.3 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.0 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.1 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 0.0 | GO:0001740 | Barr body(GO:0001740) |
| 0.0 | 0.1 | GO:0030122 | AP-2 adaptor complex(GO:0030122) clathrin coat of endocytic vesicle(GO:0030128) |
| 0.0 | 0.0 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.1 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.1 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.1 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.0 | 0.0 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.0 | 0.4 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.0 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.1 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.1 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.0 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.1 | 0.3 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.1 | 0.2 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.1 | 0.2 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.1 | GO:0016901 | oxidoreductase activity, acting on the CH-OH group of donors, quinone or similar compound as acceptor(GO:0016901) |
| 0.0 | 0.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.2 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.1 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.1 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.1 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.1 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.3 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.1 | GO:0004904 | interferon receptor activity(GO:0004904) |
| 0.0 | 0.1 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.1 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.2 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.0 | 0.1 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.5 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.1 | GO:0047115 | trans-1,2-dihydrobenzene-1,2-diol dehydrogenase activity(GO:0047115) |
| 0.0 | 0.8 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 0.1 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.1 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.1 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.1 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.1 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) |
| 0.0 | 0.1 | GO:0008318 | protein prenyltransferase activity(GO:0008318) |
| 0.0 | 0.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.1 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.0 | 0.1 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.1 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.0 | 0.2 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.2 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.1 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.0 | 0.1 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.0 | 0.1 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.1 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.1 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 0.1 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.0 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.0 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.1 | GO:0003709 | obsolete RNA polymerase III transcription factor activity(GO:0003709) |
| 0.0 | 0.1 | GO:0004470 | malic enzyme activity(GO:0004470) |
| 0.0 | 0.1 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.1 | GO:0071617 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) lysophosphatidic acid acyltransferase activity(GO:0042171) lysophospholipid acyltransferase activity(GO:0071617) |
| 0.0 | 0.0 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.0 | 0.1 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.1 | GO:0016565 | obsolete general transcriptional repressor activity(GO:0016565) |
| 0.0 | 0.1 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.1 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 0.0 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.0 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 0.1 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.1 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.0 | GO:0031705 | bombesin receptor binding(GO:0031705) endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.0 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.0 | 0.0 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.1 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 0.0 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.0 | 0.0 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.0 | GO:0043734 | DNA demethylase activity(GO:0035514) DNA-N1-methyladenine dioxygenase activity(GO:0043734) |
| 0.0 | 0.0 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.0 | GO:0032296 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.0 | 0.1 | GO:0070717 | poly-purine tract binding(GO:0070717) |
| 0.0 | 0.0 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.0 | 0.0 | GO:0016215 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.0 | 0.0 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.1 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.2 | GO:0004532 | exoribonuclease activity(GO:0004532) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.1 | GO:0003706 | obsolete ligand-regulated transcription factor activity(GO:0003706) |
| 0.0 | 0.0 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.0 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 0.0 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.0 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 0.6 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.2 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.1 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.5 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.0 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.4 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.7 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.2 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.0 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF RAS | Genes involved in CREB phosphorylation through the activation of Ras |
| 0.0 | 0.1 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.3 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.1 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.0 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.1 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.1 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.1 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.1 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |