Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
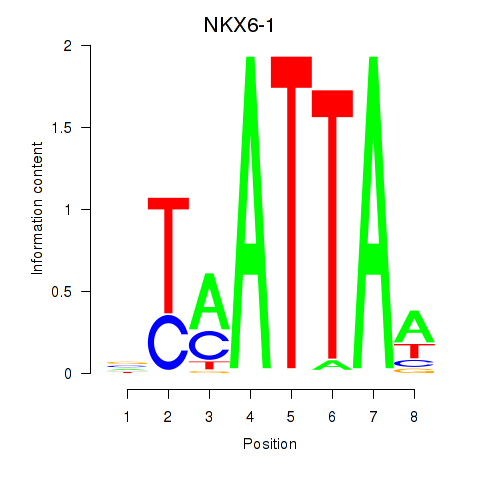
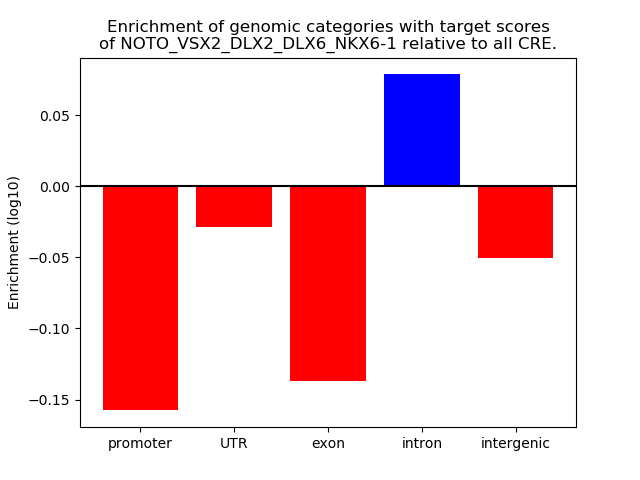
Results for NOTO_VSX2_DLX2_DLX6_NKX6-1
Z-value: 0.36
Transcription factors associated with NOTO_VSX2_DLX2_DLX6_NKX6-1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
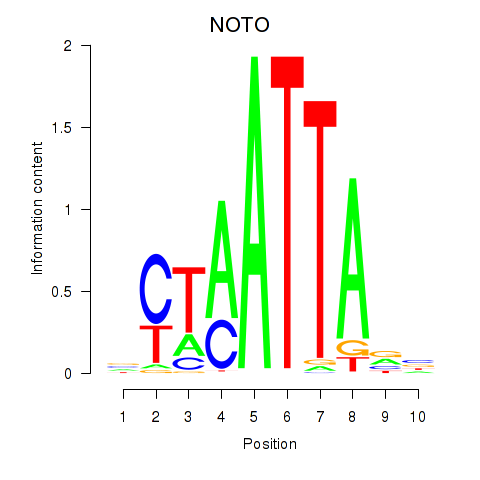
NOTO
|
ENSG00000214513.3 | notochord homeobox |
|
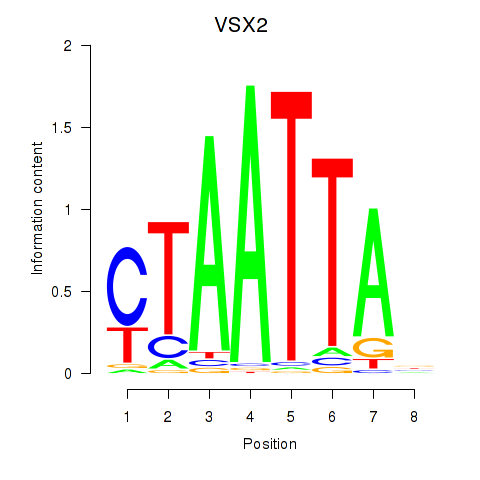
VSX2
|
ENSG00000119614.2 | visual system homeobox 2 |
|
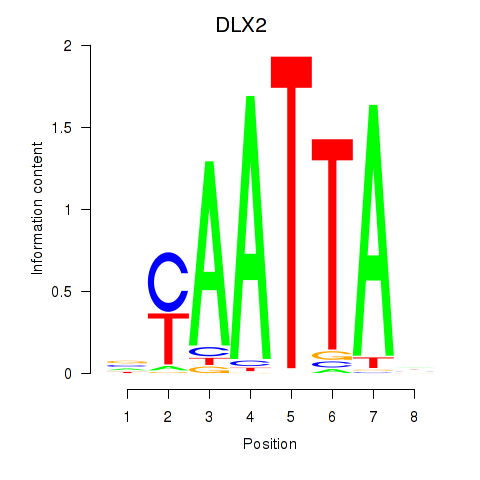
DLX2
|
ENSG00000115844.6 | distal-less homeobox 2 |
|
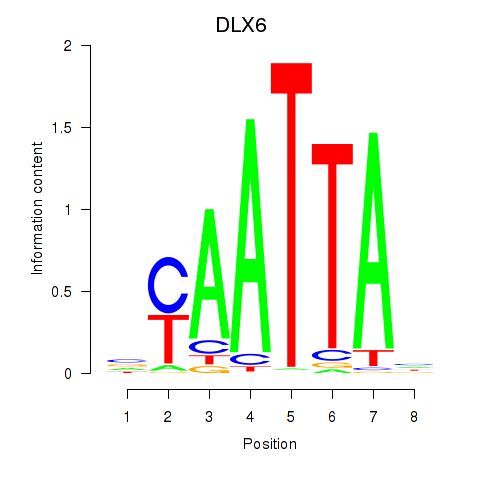
DLX6
|
ENSG00000006377.9 | distal-less homeobox 6 |
|
NKX6-1
|
ENSG00000163623.5 | NK6 homeobox 1 |
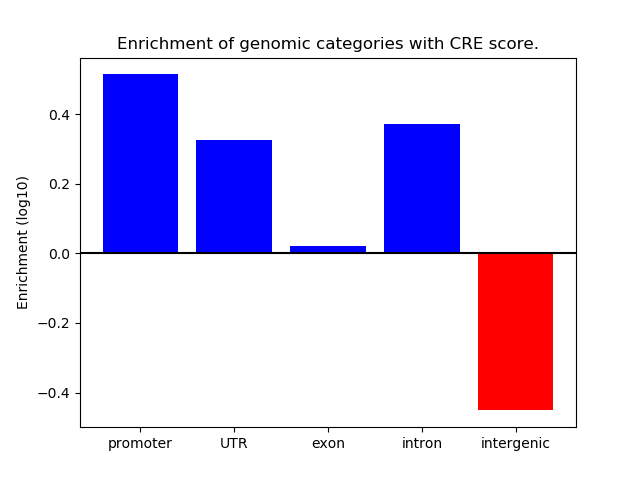
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr2_172963100_172963251 | DLX2 | 4453 | 0.235017 | -0.81 | 8.6e-03 | Click! |
| chr2_172963272_172963426 | DLX2 | 4279 | 0.237927 | -0.81 | 8.6e-03 | Click! |
| chr2_172962678_172962829 | DLX2 | 4875 | 0.229059 | -0.78 | 1.4e-02 | Click! |
| chr2_172970558_172970785 | DLX2 | 3043 | 0.274625 | -0.73 | 2.7e-02 | Click! |
| chr2_172983664_172983815 | DLX2 | 16111 | 0.193934 | -0.70 | 3.6e-02 | Click! |
| chr7_96634629_96634816 | DLX6 | 138 | 0.916168 | -0.76 | 1.6e-02 | Click! |
| chr7_96634448_96634599 | DLX6 | 337 | 0.746776 | -0.43 | 2.5e-01 | Click! |
| chr7_96635544_96635719 | DLX6 | 341 | 0.788171 | -0.31 | 4.2e-01 | Click! |
| chr7_96635897_96636514 | DLX6 | 532 | 0.551753 | 0.21 | 5.9e-01 | Click! |
| chr7_96635193_96635344 | DLX6 | 22 | 0.957836 | -0.11 | 7.8e-01 | Click! |
| chr4_85414076_85414325 | NKX6-1 | 3903 | 0.344154 | 0.88 | 1.5e-03 | Click! |
| chr4_85414774_85414925 | NKX6-1 | 3254 | 0.365933 | -0.73 | 2.7e-02 | Click! |
| chr4_85414514_85414716 | NKX6-1 | 3488 | 0.356869 | -0.69 | 3.9e-02 | Click! |
| chr4_85414978_85415129 | NKX6-1 | 3050 | 0.375302 | -0.64 | 6.3e-02 | Click! |
| chr4_85418461_85418631 | NKX6-1 | 443 | 0.896615 | -0.57 | 1.1e-01 | Click! |
| chr2_73404020_73404496 | NOTO | 25128 | 0.131991 | -0.63 | 7.1e-02 | Click! |
| chr2_73428847_73428998 | NOTO | 464 | 0.764129 | -0.61 | 8.4e-02 | Click! |
| chr2_73402978_73403574 | NOTO | 26110 | 0.130543 | -0.45 | 2.2e-01 | Click! |
| chr2_73403809_73403998 | NOTO | 25483 | 0.131472 | -0.43 | 2.5e-01 | Click! |
| chr2_73402351_73402559 | NOTO | 26931 | 0.129301 | -0.41 | 2.7e-01 | Click! |
| chr14_74707717_74707868 | VSX2 | 1617 | 0.345319 | -0.62 | 7.6e-02 | Click! |
| chr14_74709486_74709637 | VSX2 | 3386 | 0.211647 | 0.60 | 9.0e-02 | Click! |
| chr14_74707166_74707367 | VSX2 | 1091 | 0.479166 | 0.55 | 1.2e-01 | Click! |
| chr14_74684580_74684796 | VSX2 | 21487 | 0.151649 | -0.50 | 1.7e-01 | Click! |
| chr14_74724538_74725207 | VSX2 | 18697 | 0.135256 | -0.47 | 2.1e-01 | Click! |
Activity of the NOTO_VSX2_DLX2_DLX6_NKX6-1 motif across conditions
Conditions sorted by the z-value of the NOTO_VSX2_DLX2_DLX6_NKX6-1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
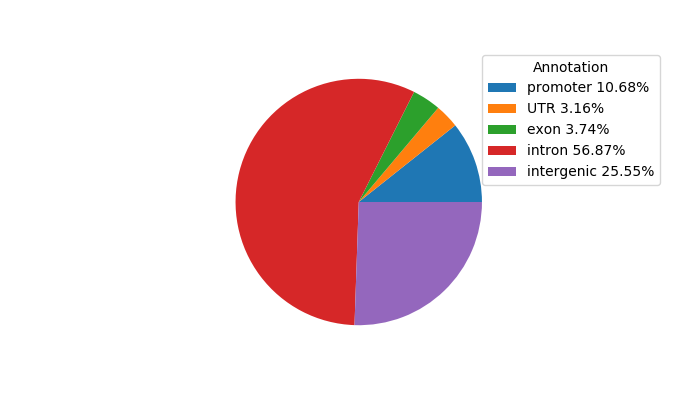
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr14_75982573_75982735 | 0.11 |
BATF |
basic leucine zipper transcription factor, ATF-like |
6114 |
0.21 |
| chrX_11784442_11784722 | 0.10 |
MSL3 |
male-specific lethal 3 homolog (Drosophila) |
6835 |
0.32 |
| chrX_154356045_154356312 | 0.09 |
MTCP1 |
mature T-cell proliferation 1 |
19872 |
0.16 |
| chr16_73092870_73093114 | 0.09 |
ZFHX3 |
zinc finger homeobox 3 |
605 |
0.79 |
| chr13_46893880_46894070 | 0.09 |
ENSG00000223336 |
. |
54750 |
0.1 |
| chr13_32616590_32616779 | 0.09 |
FRY-AS1 |
FRY antisense RNA 1 |
10908 |
0.22 |
| chr2_114633518_114633816 | 0.08 |
ACTR3 |
ARP3 actin-related protein 3 homolog (yeast) |
13870 |
0.21 |
| chr10_4170376_4170710 | 0.08 |
KLF6 |
Kruppel-like factor 6 |
343070 |
0.01 |
| chr3_107697281_107697722 | 0.08 |
CD47 |
CD47 molecule |
79707 |
0.11 |
| chr19_47948776_47948927 | 0.08 |
MEIS3 |
Meis homeobox 3 |
26071 |
0.11 |
| chr17_38376031_38376425 | 0.08 |
WIPF2 |
WAS/WASL interacting protein family, member 2 |
124 |
0.95 |
| chr2_204726088_204726239 | 0.08 |
CTLA4 |
cytotoxic T-lymphocyte-associated protein 4 |
6346 |
0.27 |
| chr5_156695529_156695722 | 0.07 |
CYFIP2 |
cytoplasmic FMR1 interacting protein 2 |
737 |
0.55 |
| chr2_30557961_30558163 | 0.07 |
ENSG00000221377 |
. |
97252 |
0.07 |
| chr7_6298731_6299016 | 0.07 |
CYTH3 |
cytohesin 3 |
13402 |
0.18 |
| chr3_71862606_71862965 | 0.07 |
ENSG00000239250 |
. |
13543 |
0.2 |
| chr14_60761646_60761856 | 0.07 |
PPM1A |
protein phosphatase, Mg2+/Mn2+ dependent, 1A |
38271 |
0.17 |
| chr7_106512561_106512750 | 0.07 |
PIK3CG |
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit gamma |
6731 |
0.28 |
| chr8_19331197_19331348 | 0.07 |
CSGALNACT1 |
chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
128104 |
0.06 |
| chr10_73513150_73513301 | 0.07 |
C10orf54 |
chromosome 10 open reading frame 54 |
4162 |
0.23 |
| chr2_204587544_204587695 | 0.07 |
CD28 |
CD28 molecule |
16203 |
0.22 |
| chr19_55008845_55009031 | 0.07 |
LAIR2 |
leukocyte-associated immunoglobulin-like receptor 2 |
162 |
0.91 |
| chr1_172666866_172667065 | 0.07 |
FASLG |
Fas ligand (TNF superfamily, member 6) |
38807 |
0.19 |
| chr11_20179253_20179404 | 0.07 |
DBX1 |
developing brain homeobox 1 |
2542 |
0.34 |
| chrX_118818778_118818993 | 0.07 |
SEPT6 |
septin 6 |
7907 |
0.18 |
| chr13_111837384_111838319 | 0.07 |
ARHGEF7 |
Rho guanine nucleotide exchange factor (GEF) 7 |
566 |
0.79 |
| chr1_208047000_208047151 | 0.07 |
CD34 |
CD34 molecule |
16762 |
0.25 |
| chr22_36802881_36803061 | 0.07 |
MYH9 |
myosin, heavy chain 9, non-muscle |
18908 |
0.15 |
| chr2_136815081_136815280 | 0.06 |
AC093391.2 |
|
45145 |
0.15 |
| chr4_78721201_78721381 | 0.06 |
CNOT6L |
CCR4-NOT transcription complex, subunit 6-like |
18926 |
0.25 |
| chr4_68440895_68441046 | 0.06 |
STAP1 |
signal transducing adaptor family member 1 |
16524 |
0.2 |
| chrX_41117068_41117219 | 0.06 |
RP5-1172N10.4 |
|
24257 |
0.17 |
| chr6_112069279_112069430 | 0.06 |
FYN |
FYN oncogene related to SRC, FGR, YES |
10963 |
0.26 |
| chr2_8631121_8631315 | 0.06 |
AC011747.7 |
|
184678 |
0.03 |
| chr13_49484698_49484849 | 0.06 |
ENSG00000265585 |
. |
19575 |
0.22 |
| chr4_153738270_153738485 | 0.06 |
ARFIP1 |
ADP-ribosylation factor interacting protein 1 |
37240 |
0.16 |
| chr2_8615200_8615353 | 0.06 |
AC011747.7 |
|
200620 |
0.03 |
| chr7_50461192_50461505 | 0.06 |
ENSG00000200815 |
. |
41730 |
0.16 |
| chr8_82065263_82065697 | 0.06 |
PAG1 |
phosphoprotein associated with glycosphingolipid microdomains 1 |
41177 |
0.19 |
| chr20_57961202_57961353 | 0.06 |
ENSG00000263903 |
. |
39742 |
0.19 |
| chr12_104916695_104916846 | 0.06 |
CHST11 |
carbohydrate (chondroitin 4) sulfotransferase 11 |
65856 |
0.12 |
| chr12_27107136_27107312 | 0.06 |
FGFR1OP2 |
FGFR1 oncogene partner 2 |
15791 |
0.13 |
| chr11_114065637_114066010 | 0.06 |
NNMT |
nicotinamide N-methyltransferase |
62730 |
0.12 |
| chr1_206753706_206754386 | 0.06 |
RASSF5 |
Ras association (RalGDS/AF-6) domain family member 5 |
23553 |
0.13 |
| chr3_16883717_16883868 | 0.06 |
PLCL2 |
phospholipase C-like 2 |
42660 |
0.19 |
| chr15_76578114_76578265 | 0.06 |
ETFA |
electron-transfer-flavoprotein, alpha polypeptide |
6643 |
0.24 |
| chr5_96345721_96346249 | 0.06 |
ENSG00000200884 |
. |
47524 |
0.12 |
| chr11_128603513_128603832 | 0.06 |
FLI1 |
Fli-1 proto-oncogene, ETS transcription factor |
31013 |
0.16 |
| chr2_143889620_143889771 | 0.06 |
ARHGAP15 |
Rho GTPase activating protein 15 |
2812 |
0.35 |
| chr1_192536616_192536790 | 0.06 |
RP5-1011O1.2 |
|
356 |
0.89 |
| chr2_30489326_30489587 | 0.06 |
LBH |
limb bud and heart development |
34410 |
0.19 |
| chr1_108327675_108328746 | 0.06 |
ENSG00000265536 |
. |
9332 |
0.26 |
| chr3_151932265_151932416 | 0.06 |
MBNL1 |
muscleblind-like splicing regulator 1 |
53489 |
0.14 |
| chr19_24222899_24223050 | 0.06 |
ZNF254 |
zinc finger protein 254 |
6698 |
0.24 |
| chr17_72489409_72489599 | 0.06 |
CD300A |
CD300a molecule |
26503 |
0.12 |
| chr16_384366_384517 | 0.06 |
AXIN1 |
axin 1 |
18008 |
0.1 |
| chr10_44277312_44277574 | 0.06 |
ZNF32 |
zinc finger protein 32 |
133139 |
0.05 |
| chr3_18470958_18471238 | 0.06 |
SATB1 |
SATB homeobox 1 |
4237 |
0.24 |
| chr9_33976084_33976235 | 0.06 |
UBAP2 |
ubiquitin associated protein 2 |
12882 |
0.13 |
| chr14_90187580_90187731 | 0.06 |
ENSG00000200312 |
. |
8800 |
0.23 |
| chr16_74620197_74620348 | 0.06 |
GLG1 |
golgi glycoprotein 1 |
20720 |
0.2 |
| chr2_174756889_174757040 | 0.06 |
SP3 |
Sp3 transcription factor |
71983 |
0.13 |
| chr3_196150607_196150758 | 0.06 |
UBXN7-AS1 |
UBXN7 antisense RNA 1 |
7553 |
0.11 |
| chr9_111350242_111350393 | 0.06 |
ENSG00000222512 |
. |
229108 |
0.02 |
| chr3_157824510_157824697 | 0.06 |
SHOX2 |
short stature homeobox 2 |
311 |
0.83 |
| chr4_25953219_25953370 | 0.06 |
SMIM20 |
small integral membrane protein 20 |
37363 |
0.21 |
| chrX_13023409_13023615 | 0.06 |
TMSB4X |
thymosin beta 4, X-linked |
29735 |
0.18 |
| chr22_24908185_24908422 | 0.06 |
AP000355.2 |
|
4173 |
0.16 |
| chr2_99302058_99302209 | 0.06 |
MGAT4A |
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A |
22197 |
0.2 |
| chr16_68502814_68502979 | 0.06 |
ENSG00000199263 |
. |
5671 |
0.17 |
| chr2_86332208_86332925 | 0.06 |
POLR1A |
polymerase (RNA) I polypeptide A, 194kDa |
712 |
0.41 |
| chr1_45100216_45100404 | 0.05 |
RNF220 |
ring finger protein 220 |
2906 |
0.21 |
| chr1_65408789_65408940 | 0.05 |
JAK1 |
Janus kinase 1 |
23323 |
0.22 |
| chr18_67686310_67686461 | 0.05 |
RTTN |
rotatin |
1569 |
0.52 |
| chr11_128600475_128600823 | 0.05 |
FLI1 |
Fli-1 proto-oncogene, ETS transcription factor |
34036 |
0.15 |
| chr1_117311065_117311639 | 0.05 |
CD2 |
CD2 molecule |
14263 |
0.21 |
| chr6_35838270_35838421 | 0.05 |
SRPK1 |
SRSF protein kinase 1 |
50479 |
0.1 |
| chr14_39893718_39893961 | 0.05 |
FBXO33 |
F-box protein 33 |
7240 |
0.26 |
| chrX_15779492_15779655 | 0.05 |
CA5B |
carbonic anhydrase VB, mitochondrial |
11480 |
0.18 |
| chr7_55627006_55627245 | 0.05 |
VOPP1 |
vesicular, overexpressed in cancer, prosurvival protein 1 |
5545 |
0.24 |
| chrX_48533482_48533772 | 0.05 |
WAS |
Wiskott-Aldrich syndrome |
1358 |
0.32 |
| chr10_82288993_82289258 | 0.05 |
RP11-137H2.4 |
|
6573 |
0.22 |
| chr10_17548800_17549076 | 0.05 |
ST8SIA6 |
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 6 |
52609 |
0.13 |
| chr19_9976591_9976870 | 0.05 |
OLFM2 |
olfactomedin 2 |
7886 |
0.11 |
| chr2_215664954_215665105 | 0.05 |
BARD1 |
BRCA1 associated RING domain 1 |
9368 |
0.21 |
| chr16_304069_304251 | 0.05 |
ITFG3 |
integrin alpha FG-GAP repeat containing 3 |
467 |
0.67 |
| chr17_76707598_76707858 | 0.05 |
CYTH1 |
cytohesin 1 |
5379 |
0.21 |
| chr22_36843971_36844162 | 0.05 |
ENSG00000252225 |
. |
7051 |
0.15 |
| chr15_40489916_40490242 | 0.05 |
PAK6 |
p21 protein (Cdc42/Rac)-activated kinase 6 |
19550 |
0.12 |
| chr13_50706871_50707077 | 0.05 |
DLEU1 |
deleted in lymphocytic leukemia 1 (non-protein coding) |
50667 |
0.13 |
| chr14_65547049_65547200 | 0.05 |
RP11-840I19.3 |
|
1628 |
0.32 |
| chr18_12421274_12421492 | 0.05 |
SLMO1 |
slowmo homolog 1 (Drosophila) |
98 |
0.96 |
| chr11_128173540_128173801 | 0.05 |
ETS1 |
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
201619 |
0.03 |
| chr6_117922468_117923064 | 0.05 |
GOPC |
golgi-associated PDZ and coiled-coil motif containing |
761 |
0.74 |
| chr2_204583285_204583561 | 0.05 |
CD28 |
CD28 molecule |
12007 |
0.23 |
| chr12_19573889_19574040 | 0.05 |
AEBP2 |
AE binding protein 2 |
16964 |
0.22 |
| chr5_95150906_95151226 | 0.05 |
GLRX |
glutaredoxin (thioltransferase) |
7349 |
0.16 |
| chr4_123133902_123134053 | 0.05 |
KIAA1109 |
KIAA1109 |
6647 |
0.31 |
| chr9_37340676_37340827 | 0.05 |
ENSG00000206784 |
. |
18495 |
0.19 |
| chr1_93445938_93446179 | 0.05 |
ENSG00000238787 |
. |
1807 |
0.23 |
| chr5_169731037_169731197 | 0.05 |
LCP2 |
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa) |
5886 |
0.24 |
| chr16_30188682_30188851 | 0.05 |
CORO1A |
coronin, actin binding protein, 1A |
5382 |
0.09 |
| chr3_44345343_44345494 | 0.05 |
TCAIM |
T cell activation inhibitor, mitochondrial |
34193 |
0.16 |
| chr20_10652876_10653323 | 0.05 |
RP11-103J8.1 |
|
477 |
0.79 |
| chr11_121198510_121198941 | 0.05 |
SC5D |
sterol-C5-desaturase |
35164 |
0.22 |
| chr17_71199413_71199564 | 0.05 |
COG1 |
component of oligomeric golgi complex 1 |
2266 |
0.24 |
| chr14_71512203_71512354 | 0.05 |
PCNX |
pecanex homolog (Drosophila) |
2399 |
0.43 |
| chrX_96290122_96290273 | 0.05 |
RPA4 |
replication protein A4, 30kDa |
151290 |
0.05 |
| chr3_71543419_71543780 | 0.05 |
ENSG00000221264 |
. |
47641 |
0.15 |
| chr15_86235842_86236056 | 0.05 |
RP11-815J21.1 |
|
8391 |
0.12 |
| chr11_48040089_48040240 | 0.05 |
AC103828.1 |
|
2757 |
0.3 |
| chr2_54763445_54763596 | 0.05 |
AC092839.3 |
|
4314 |
0.22 |
| chr16_3863779_3863930 | 0.05 |
CREBBP |
CREB binding protein |
32986 |
0.17 |
| chr1_25969522_25969744 | 0.05 |
RP1-187B23.1 |
|
15969 |
0.18 |
| chr5_61613165_61613316 | 0.05 |
KIF2A |
kinesin heavy chain member 2A |
5600 |
0.28 |
| chr10_11224652_11224918 | 0.05 |
RP3-323N1.2 |
|
11446 |
0.22 |
| chr2_68470770_68470921 | 0.05 |
PPP3R1 |
protein phosphatase 3, regulatory subunit B, alpha |
7435 |
0.14 |
| chr17_29146107_29146385 | 0.05 |
CRLF3 |
cytokine receptor-like factor 3 |
5443 |
0.13 |
| chrX_24207530_24207774 | 0.05 |
ZFX |
zinc finger protein, X-linked |
16820 |
0.19 |
| chr6_13686993_13687323 | 0.05 |
RANBP9 |
RAN binding protein 9 |
22 |
0.98 |
| chr10_4121743_4121894 | 0.05 |
KLF6 |
Kruppel-like factor 6 |
294345 |
0.01 |
| chr2_29593489_29593640 | 0.05 |
ENSG00000266310 |
. |
133295 |
0.05 |
| chr7_143007723_143007933 | 0.05 |
CLCN1 |
chloride channel, voltage-sensitive 1 |
5391 |
0.1 |
| chr14_73918520_73918671 | 0.05 |
NUMB |
numb homolog (Drosophila) |
6394 |
0.17 |
| chr13_46745351_46745718 | 0.05 |
ENSG00000240767 |
. |
1651 |
0.31 |
| chrX_38757954_38758105 | 0.05 |
MID1IP1-AS1 |
MID1IP1 antisense RNA 1 |
94893 |
0.08 |
| chr14_66052416_66052567 | 0.05 |
FUT8 |
fucosyltransferase 8 (alpha (1,6) fucosyltransferase) |
83642 |
0.1 |
| chr9_132708644_132709361 | 0.05 |
RP11-409K20.6 |
|
13187 |
0.19 |
| chr12_56429367_56429565 | 0.05 |
RP11-603J24.4 |
|
6124 |
0.09 |
| chr11_48078680_48078903 | 0.05 |
ENSG00000263693 |
. |
39543 |
0.14 |
| chr5_156969848_156970007 | 0.05 |
AC106801.1 |
|
23045 |
0.14 |
| chr16_29673426_29673713 | 0.05 |
SPN |
sialophorin |
731 |
0.45 |
| chr1_185564303_185564454 | 0.05 |
ENSG00000207108 |
. |
35282 |
0.18 |
| chr10_63813717_63813907 | 0.05 |
ARID5B |
AT rich interactive domain 5B (MRF1-like) |
4842 |
0.3 |
| chr1_206817821_206817972 | 0.05 |
DYRK3 |
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 3 |
8722 |
0.14 |
| chr5_126093902_126094474 | 0.05 |
ENSG00000252185 |
. |
3079 |
0.3 |
| chr2_63280572_63281076 | 0.05 |
OTX1 |
orthodenticle homeobox 1 |
2887 |
0.3 |
| chr8_19552777_19552928 | 0.05 |
CSGALNACT1 |
chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
12580 |
0.3 |
| chr14_96978213_96978375 | 0.05 |
PAPOLA |
poly(A) polymerase alpha |
9106 |
0.18 |
| chr19_8628819_8628970 | 0.05 |
MYO1F |
myosin IF |
13428 |
0.11 |
| chr5_156711613_156711855 | 0.05 |
CYFIP2 |
cytoplasmic FMR1 interacting protein 2 |
638 |
0.67 |
| chr7_37011204_37011355 | 0.05 |
ELMO1 |
engulfment and cell motility 1 |
13386 |
0.2 |
| chr4_86636333_86636484 | 0.05 |
ENSG00000266421 |
. |
7213 |
0.29 |
| chr9_123882798_123883340 | 0.05 |
CNTRL |
centriolin |
1010 |
0.59 |
| chr8_42607900_42608051 | 0.05 |
CHRNA6 |
cholinergic receptor, nicotinic, alpha 6 (neuronal) |
15778 |
0.17 |
| chr2_214096607_214096758 | 0.05 |
SPAG16 |
sperm associated antigen 16 |
52431 |
0.17 |
| chr5_92921754_92921905 | 0.05 |
ENSG00000237187 |
. |
475 |
0.81 |
| chr17_34906652_34907000 | 0.05 |
GGNBP2 |
gametogenetin binding protein 2 |
5473 |
0.14 |
| chr7_140171728_140171879 | 0.05 |
MKRN1 |
makorin ring finger protein 1 |
6500 |
0.2 |
| chr6_88403463_88403724 | 0.05 |
AKIRIN2 |
akirin 2 |
8334 |
0.2 |
| chr5_110428605_110428905 | 0.05 |
WDR36 |
WD repeat domain 36 |
736 |
0.49 |
| chr11_117863729_117863895 | 0.05 |
IL10RA |
interleukin 10 receptor, alpha |
6703 |
0.2 |
| chr19_13951569_13952139 | 0.05 |
ENSG00000207980 |
. |
4381 |
0.09 |
| chr7_105088467_105088805 | 0.05 |
SRPK2 |
SRSF protein kinase 2 |
58798 |
0.11 |
| chr1_174929832_174929983 | 0.05 |
RABGAP1L |
RAB GTPase activating protein 1-like |
3998 |
0.18 |
| chr8_17788613_17788764 | 0.05 |
PCM1 |
pericentriolar material 1 |
6435 |
0.21 |
| chr11_57480467_57480618 | 0.05 |
TMX2 |
thioredoxin-related transmembrane protein 2 |
463 |
0.58 |
| chr18_2976679_2976953 | 0.05 |
LPIN2 |
lipin 2 |
6055 |
0.16 |
| chr11_118935786_118935937 | 0.05 |
RP11-110I1.13 |
|
2548 |
0.1 |
| chr2_43402387_43402951 | 0.05 |
ZFP36L2 |
ZFP36 ring finger protein-like 2 |
51079 |
0.14 |
| chr6_159453780_159453931 | 0.05 |
RP1-111C20.4 |
|
6877 |
0.19 |
| chr5_98269675_98269959 | 0.05 |
ENSG00000200351 |
. |
2634 |
0.3 |
| chr15_60850340_60850622 | 0.05 |
CTD-2501E16.2 |
|
28309 |
0.16 |
| chr3_195121741_195121910 | 0.05 |
ENSG00000207368 |
. |
18450 |
0.19 |
| chr9_129252809_129252960 | 0.05 |
ENSG00000221768 |
. |
40386 |
0.14 |
| chr15_63503072_63503368 | 0.05 |
RAB8B |
RAB8B, member RAS oncogene family |
21415 |
0.19 |
| chr2_64416040_64416191 | 0.05 |
AC074289.1 |
|
17994 |
0.23 |
| chr17_75456251_75456589 | 0.05 |
SEPT9 |
septin 9 |
3972 |
0.18 |
| chr6_109381834_109382031 | 0.05 |
ENSG00000199944 |
. |
1115 |
0.52 |
| chr5_100212754_100212905 | 0.05 |
ST8SIA4 |
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 4 |
26089 |
0.24 |
| chr17_57866912_57867150 | 0.05 |
RP11-619I22.1 |
|
4857 |
0.18 |
| chr2_210850912_210851168 | 0.05 |
RPE |
ribulose-5-phosphate-3-epimerase |
16249 |
0.17 |
| chr1_100905257_100905795 | 0.05 |
RP5-837M10.4 |
|
46027 |
0.13 |
| chr4_81061096_81061271 | 0.05 |
PRDM8 |
PR domain containing 8 |
44256 |
0.15 |
| chr12_94600310_94600885 | 0.05 |
RP11-74K11.2 |
|
20777 |
0.19 |
| chr21_16057852_16058003 | 0.05 |
AF165138.7 |
Protein LOC388813 |
26785 |
0.23 |
| chr1_198627688_198627997 | 0.05 |
RP11-553K8.5 |
|
8348 |
0.25 |
| chr3_53380504_53380708 | 0.05 |
DCP1A |
decapping mRNA 1A |
946 |
0.5 |
| chr11_82885812_82886113 | 0.05 |
PCF11 |
PCF11 cleavage and polyadenylation factor subunit |
3669 |
0.19 |
| chr15_75414732_75414883 | 0.05 |
ENSG00000252722 |
. |
800 |
0.61 |
| chr20_6758219_6758370 | 0.05 |
BMP2 |
bone morphogenetic protein 2 |
9983 |
0.3 |
| chr12_25432989_25433140 | 0.04 |
KRAS |
Kirsten rat sarcoma viral oncogene homolog |
29194 |
0.18 |
| chr13_28726745_28726896 | 0.04 |
PAN3 |
PAN3 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) |
13650 |
0.18 |
| chr14_98641364_98641616 | 0.04 |
ENSG00000222066 |
. |
156597 |
0.04 |
| chr3_88112020_88112171 | 0.04 |
RP11-159G9.5 |
Uncharacterized protein |
3671 |
0.23 |
| chr12_68530648_68530810 | 0.04 |
IFNG |
interferon, gamma |
22798 |
0.24 |
| chrX_109266234_109266489 | 0.04 |
TMEM164 |
transmembrane protein 164 |
20018 |
0.2 |
| chr15_91880318_91880572 | 0.04 |
SV2B |
synaptic vesicle glycoprotein 2B |
111345 |
0.07 |
| chr13_99892072_99892468 | 0.04 |
GPR18 |
G protein-coupled receptor 18 |
18358 |
0.17 |
| chr5_156636522_156636760 | 0.04 |
CTB-4E7.1 |
|
14749 |
0.12 |
| chr10_26740284_26740599 | 0.04 |
APBB1IP |
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein |
13087 |
0.26 |
| chr11_129956983_129957182 | 0.04 |
APLP2 |
amyloid beta (A4) precursor-like protein 2 |
16619 |
0.2 |
| chr19_42318679_42318830 | 0.04 |
CEACAM3 |
carcinoembryonic antigen-related cell adhesion molecule 3 |
17669 |
0.1 |
| chr7_157674477_157674628 | 0.04 |
RP11-452C13.1 |
|
13322 |
0.23 |
| chr5_169066038_169066189 | 0.04 |
DOCK2 |
dedicator of cytokinesis 2 |
1862 |
0.43 |
| chr5_61874985_61875174 | 0.04 |
LRRC70 |
leucine rich repeat containing 70 |
377 |
0.53 |
| chr1_235439552_235439703 | 0.04 |
GGPS1 |
geranylgeranyl diphosphate synthase 1 |
51038 |
0.11 |
| chr4_78777778_78777929 | 0.04 |
MRPL1 |
mitochondrial ribosomal protein L1 |
5821 |
0.28 |
| chr11_87248503_87248654 | 0.04 |
ENSG00000223015 |
. |
69097 |
0.13 |
| chr1_100862907_100863178 | 0.04 |
ENSG00000216067 |
. |
18711 |
0.19 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.0 | 0.1 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 0.1 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.0 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.0 | 0.0 | GO:0042996 | regulation of Golgi to plasma membrane protein transport(GO:0042996) |
| 0.0 | 0.0 | GO:0072182 | nephron tubule epithelial cell differentiation(GO:0072160) regulation of nephron tubule epithelial cell differentiation(GO:0072182) regulation of epithelial cell differentiation involved in kidney development(GO:2000696) |
| 0.0 | 0.1 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.0 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 0.0 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.0 | 0.1 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.0 | GO:0072216 | positive regulation of metanephros development(GO:0072216) |
| 0.0 | 0.1 | GO:0045066 | regulatory T cell differentiation(GO:0045066) |
| 0.0 | 0.0 | GO:2000758 | positive regulation of histone acetylation(GO:0035066) positive regulation of protein acetylation(GO:1901985) positive regulation of peptidyl-lysine acetylation(GO:2000758) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.1 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.0 | GO:0042608 | T cell receptor binding(GO:0042608) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.1 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |