Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
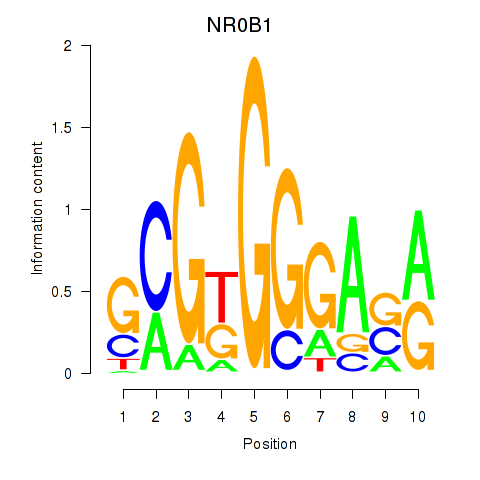
Results for NR0B1
Z-value: 1.34
Transcription factors associated with NR0B1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR0B1
|
ENSG00000169297.6 | nuclear receptor subfamily 0 group B member 1 |
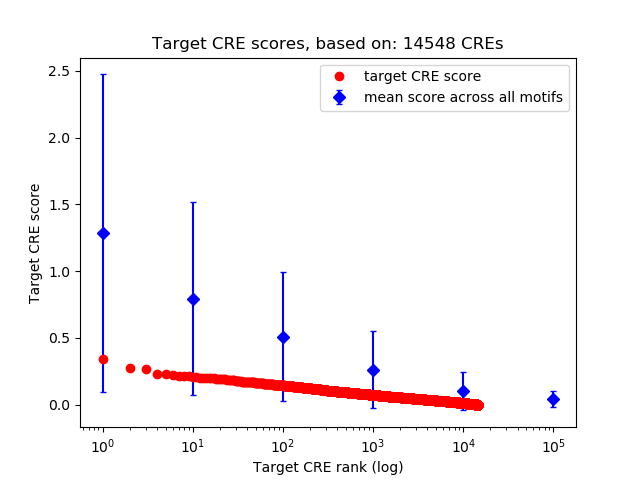
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chrX_30327610_30327761 | NR0B1 | 30 | 0.977016 | 0.30 | 4.4e-01 | Click! |
Activity of the NR0B1 motif across conditions
Conditions sorted by the z-value of the NR0B1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
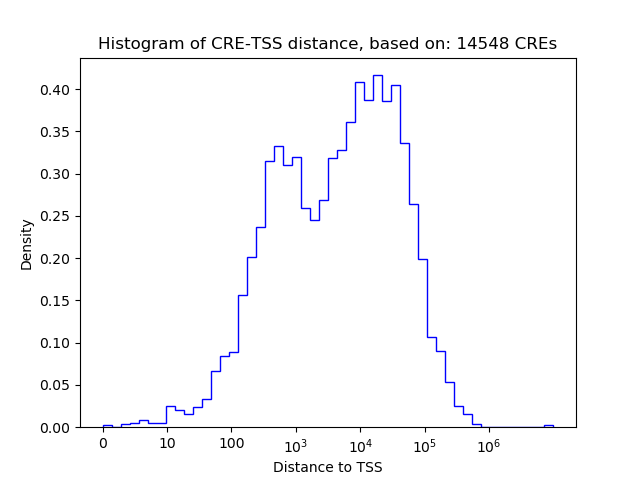
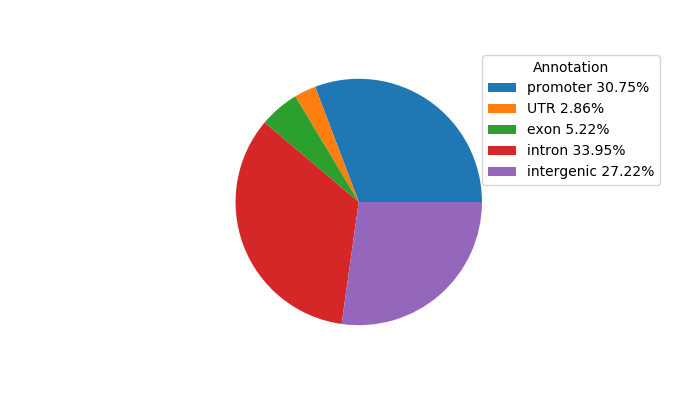
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr10_134387680_134387844 | 0.35 |
INPP5A |
inositol polyphosphate-5-phosphatase, 40kDa |
33668 |
0.18 |
| chr12_67753116_67753506 | 0.27 |
CAND1 |
cullin-associated and neddylation-dissociated 1 |
60559 |
0.16 |
| chrX_102719262_102719533 | 0.27 |
LL0XNC01-250H12.3 |
|
33054 |
0.14 |
| chr9_109623180_109623558 | 0.23 |
ZNF462 |
zinc finger protein 462 |
2009 |
0.38 |
| chr21_47603311_47603605 | 0.23 |
SPATC1L |
spermatogenesis and centriole associated 1-like |
915 |
0.34 |
| chr3_35680980_35681355 | 0.22 |
ARPP21 |
cAMP-regulated phosphoprotein, 21kDa |
86 |
0.98 |
| chr3_123601595_123601750 | 0.22 |
MYLK |
myosin light chain kinase |
1477 |
0.48 |
| chr1_54147270_54147460 | 0.22 |
ENSG00000239007 |
. |
7120 |
0.23 |
| chr17_3795838_3795989 | 0.21 |
CAMKK1 |
calcium/calmodulin-dependent protein kinase kinase 1, alpha |
424 |
0.81 |
| chr3_193857613_193857764 | 0.21 |
HES1 |
hes family bHLH transcription factor 1 |
3754 |
0.23 |
| chr17_62061939_62062090 | 0.21 |
SCN4A |
sodium channel, voltage-gated, type IV, alpha subunit |
11736 |
0.13 |
| chr7_30265884_30266155 | 0.20 |
ZNRF2 |
zinc and ring finger 2 |
57904 |
0.1 |
| chr8_133685594_133685859 | 0.20 |
LRRC6 |
leucine rich repeat containing 6 |
2079 |
0.33 |
| chr3_53529546_53529812 | 0.20 |
CACNA1D |
calcium channel, voltage-dependent, L type, alpha 1D subunit |
485 |
0.83 |
| chr6_15281229_15281559 | 0.20 |
JARID2 |
jumonji, AT rich interactive domain 2 |
32251 |
0.14 |
| chr9_125765061_125765504 | 0.20 |
RABGAP1 |
RAB GTPase activating protein 1 |
30506 |
0.12 |
| chr5_369667_369964 | 0.20 |
AHRR |
aryl-hydrocarbon receptor repressor |
26172 |
0.15 |
| chr7_138666516_138666785 | 0.20 |
KIAA1549 |
KIAA1549 |
586 |
0.8 |
| chr18_43915409_43915560 | 0.20 |
RNF165 |
ring finger protein 165 |
1297 |
0.59 |
| chr3_3841565_3841901 | 0.20 |
LRRN1 |
leucine rich repeat neuronal 1 |
612 |
0.85 |
| chr1_85463137_85463288 | 0.20 |
MCOLN2 |
mucolipin 2 |
417 |
0.8 |
| chr19_41828944_41829156 | 0.19 |
CCDC97 |
coiled-coil domain containing 97 |
3412 |
0.13 |
| chr19_1214948_1215099 | 0.19 |
STK11 |
serine/threonine kinase 11 |
6189 |
0.1 |
| chr13_41239473_41239624 | 0.19 |
FOXO1 |
forkhead box O1 |
1186 |
0.56 |
| chr12_68043974_68044273 | 0.19 |
DYRK2 |
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 2 |
661 |
0.78 |
| chr7_100487586_100487737 | 0.19 |
UFSP1 |
UFM1-specific peptidase 1 (non-functional) |
322 |
0.77 |
| chr12_54780838_54781095 | 0.19 |
ZNF385A |
zinc finger protein 385A |
1414 |
0.25 |
| chr12_27702595_27702869 | 0.18 |
PPFIBP1 |
PTPRF interacting protein, binding protein 1 (liprin beta 1) |
11676 |
0.23 |
| chr6_133562644_133562795 | 0.18 |
EYA4 |
eyes absent homolog 4 (Drosophila) |
5 |
0.99 |
| chr1_12227573_12227724 | 0.18 |
TNFRSF1B |
tumor necrosis factor receptor superfamily, member 1B |
588 |
0.71 |
| chr5_56741137_56741395 | 0.18 |
ENSG00000264748 |
. |
36464 |
0.16 |
| chr2_206595324_206595734 | 0.18 |
AC007362.3 |
|
33148 |
0.2 |
| chr17_45401427_45401685 | 0.18 |
EFCAB13 |
EF-hand calcium binding domain 13 |
165 |
0.84 |
| chr3_120278487_120278806 | 0.17 |
NDUFB4 |
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 4, 15kDa |
36510 |
0.19 |
| chr19_41083149_41083300 | 0.17 |
SHKBP1 |
SH3KBP1 binding protein 1 |
160 |
0.94 |
| chr3_37964439_37964907 | 0.17 |
CTDSPL |
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like |
41397 |
0.12 |
| chr9_127005358_127005801 | 0.17 |
NEK6 |
NIMA-related kinase 6 |
14306 |
0.17 |
| chr11_64322674_64322825 | 0.17 |
SLC22A11 |
solute carrier family 22 (organic anion/urate transporter), member 11 |
349 |
0.87 |
| chr3_42306280_42306453 | 0.17 |
CCK |
cholecystokinin |
29 |
0.98 |
| chr2_42721697_42722094 | 0.17 |
MTA3 |
metastasis associated 1 family, member 3 |
186 |
0.85 |
| chr1_156867514_156867913 | 0.17 |
PEAR1 |
platelet endothelial aggregation receptor 1 |
4190 |
0.15 |
| chr11_117185411_117185562 | 0.17 |
CEP164 |
centrosomal protein 164kDa |
213 |
0.87 |
| chr12_96588798_96589434 | 0.17 |
ELK3 |
ELK3, ETS-domain protein (SRF accessory protein 2) |
724 |
0.7 |
| chr9_131218778_131219020 | 0.17 |
ODF2 |
outer dense fiber of sperm tails 2 |
154 |
0.92 |
| chr3_194117055_194117206 | 0.17 |
GP5 |
glycoprotein V (platelet) |
1953 |
0.3 |
| chr8_1992676_1993045 | 0.17 |
MYOM2 |
myomesin 2 |
295 |
0.94 |
| chr16_4723224_4723375 | 0.17 |
NUDT16L1 |
nudix (nucleoside diphosphate linked moiety X)-type motif 16-like 1 |
20396 |
0.1 |
| chr17_40673227_40673704 | 0.17 |
ENSG00000265611 |
. |
7259 |
0.08 |
| chr12_11653096_11653254 | 0.17 |
RP11-711K1.7 |
|
13563 |
0.19 |
| chr19_8794292_8794443 | 0.17 |
ACTL9 |
actin-like 9 |
14805 |
0.19 |
| chr14_68864676_68864966 | 0.16 |
RAD51B |
RAD51 paralog B |
13406 |
0.29 |
| chr12_77718872_77719219 | 0.16 |
ENSG00000238769 |
. |
162240 |
0.04 |
| chr19_56109651_56109802 | 0.16 |
FIZ1 |
FLT3-interacting zinc finger 1 |
428 |
0.61 |
| chr3_189826682_189826960 | 0.16 |
ENSG00000265045 |
. |
4902 |
0.18 |
| chr3_120364035_120364285 | 0.16 |
HGD |
homogentisate 1,2-dioxygenase |
1022 |
0.62 |
| chr2_28615239_28615407 | 0.16 |
FOSL2 |
FOS-like antigen 2 |
346 |
0.84 |
| chr12_111074662_111075000 | 0.16 |
TCTN1 |
tectonic family member 1 |
4023 |
0.17 |
| chr19_41857694_41857871 | 0.16 |
TMEM91 |
transmembrane protein 91 |
966 |
0.33 |
| chr19_55720445_55720596 | 0.16 |
PTPRH |
protein tyrosine phosphatase, receptor type, H |
304 |
0.77 |
| chr22_45622079_45622539 | 0.16 |
KIAA0930 |
KIAA0930 |
475 |
0.8 |
| chr22_18452853_18453098 | 0.16 |
ENSG00000207780 |
. |
10752 |
0.16 |
| chr17_48233037_48233205 | 0.16 |
PPP1R9B |
protein phosphatase 1, regulatory subunit 9B |
5244 |
0.1 |
| chr5_169064615_169064766 | 0.16 |
DOCK2 |
dedicator of cytokinesis 2 |
439 |
0.88 |
| chr10_89577780_89577931 | 0.16 |
ATAD1 |
ATPase family, AAA domain containing 1 |
62 |
0.97 |
| chr1_55008801_55008989 | 0.16 |
ACOT11 |
acyl-CoA thioesterase 11 |
5006 |
0.19 |
| chr2_66803103_66803386 | 0.16 |
MEIS1 |
Meis homeobox 1 |
67185 |
0.13 |
| chr5_68389500_68389651 | 0.16 |
SLC30A5 |
solute carrier family 30 (zinc transporter), member 5 |
102 |
0.97 |
| chr17_1080828_1080990 | 0.15 |
ABR |
active BCR-related |
2169 |
0.3 |
| chr19_15333671_15333847 | 0.15 |
EPHX3 |
epoxide hydrolase 3 |
9484 |
0.16 |
| chr12_124508725_124508964 | 0.15 |
ZNF664 |
zinc finger protein 664 |
51002 |
0.07 |
| chr10_45455297_45455490 | 0.15 |
RASSF4 |
Ras association (RalGDS/AF-6) domain family member 4 |
114 |
0.63 |
| chr15_39802419_39802570 | 0.15 |
THBS1 |
thrombospondin 1 |
70786 |
0.1 |
| chr3_85008969_85009125 | 0.15 |
CADM2 |
cell adhesion molecule 2 |
350 |
0.93 |
| chr5_149883084_149883321 | 0.15 |
NDST1 |
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
4472 |
0.21 |
| chr17_48501944_48502107 | 0.15 |
ACSF2 |
acyl-CoA synthetase family member 2 |
1494 |
0.25 |
| chr2_182715012_182715242 | 0.15 |
RP11-366L5.1 |
|
41263 |
0.15 |
| chr8_129197888_129198179 | 0.15 |
ENSG00000201782 |
. |
34717 |
0.18 |
| chr20_43987047_43987198 | 0.15 |
SYS1 |
SYS1 Golgi-localized integral membrane protein homolog (S. cerevisiae) |
3455 |
0.15 |
| chr10_101283342_101283569 | 0.15 |
RP11-129J12.2 |
|
3745 |
0.22 |
| chr16_12552047_12552229 | 0.15 |
RP11-165M1.2 |
|
85458 |
0.07 |
| chr11_126152650_126152801 | 0.15 |
TIRAP |
toll-interleukin 1 receptor (TIR) domain containing adaptor protein |
257 |
0.79 |
| chr12_109089717_109090214 | 0.15 |
CORO1C |
coronin, actin binding protein, 1C |
213 |
0.92 |
| chr6_42110106_42110370 | 0.15 |
C6orf132 |
chromosome 6 open reading frame 132 |
56 |
0.97 |
| chr10_74034261_74034412 | 0.15 |
DDIT4 |
DNA-damage-inducible transcript 4 |
658 |
0.58 |
| chr10_75758429_75758627 | 0.15 |
VCL |
vinculin |
654 |
0.69 |
| chr7_23401357_23401759 | 0.15 |
ENSG00000212264 |
. |
34507 |
0.16 |
| chr19_1667467_1667618 | 0.15 |
TCF3 |
transcription factor 3 |
14938 |
0.12 |
| chr12_51781847_51782170 | 0.15 |
GALNT6 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 6 (GalNAc-T6) |
3075 |
0.2 |
| chr18_46247007_46247395 | 0.15 |
RP11-426J5.2 |
|
32411 |
0.19 |
| chr15_43478099_43478250 | 0.15 |
CCNDBP1 |
cyclin D-type binding-protein 1 |
537 |
0.55 |
| chr1_229364834_229364985 | 0.15 |
RP5-1061H20.5 |
|
1600 |
0.36 |
| chr10_44806204_44806393 | 0.15 |
RP11-20J15.3 |
|
17141 |
0.24 |
| chr13_42615279_42615552 | 0.15 |
DGKH |
diacylglycerol kinase, eta |
1239 |
0.48 |
| chr3_12044959_12045206 | 0.15 |
ENSG00000263988 |
. |
50523 |
0.15 |
| chr14_23289177_23289429 | 0.15 |
SLC7A7 |
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
283 |
0.79 |
| chr17_48049156_48049307 | 0.15 |
DLX4 |
distal-less homeobox 4 |
899 |
0.52 |
| chr9_114659712_114659863 | 0.15 |
UGCG |
UDP-glucose ceramide glucosyltransferase |
741 |
0.68 |
| chr4_1754002_1754153 | 0.15 |
TACC3 |
transforming, acidic coiled-coil containing protein 3 |
23946 |
0.13 |
| chr15_70434952_70435153 | 0.15 |
TLE3 |
transducin-like enhancer of split 3 (E(sp1) homolog, Drosophila) |
44537 |
0.16 |
| chr20_43590008_43590159 | 0.15 |
TOMM34 |
translocase of outer mitochondrial membrane 34 |
956 |
0.48 |
| chr10_104389202_104389369 | 0.14 |
TRIM8 |
tripartite motif containing 8 |
14968 |
0.16 |
| chrX_39964958_39965304 | 0.14 |
BCOR |
BCL6 corepressor |
8475 |
0.32 |
| chr6_3624272_3624465 | 0.14 |
PXDC1 |
PX domain containing 1 |
114760 |
0.06 |
| chr20_3758571_3758722 | 0.14 |
SPEF1 |
sperm flagellar 1 |
3449 |
0.15 |
| chr17_27894487_27894949 | 0.14 |
TP53I13 |
tumor protein p53 inducible protein 13 |
327 |
0.58 |
| chr21_30617757_30617942 | 0.14 |
BACH1 |
BTB and CNC homology 1, basic leucine zipper transcription factor 1 |
51381 |
0.1 |
| chr6_7431781_7431972 | 0.14 |
CAGE1 |
cancer antigen 1 |
41900 |
0.15 |
| chr1_11796539_11796690 | 0.14 |
AGTRAP |
angiotensin II receptor-associated protein |
382 |
0.79 |
| chr1_208137980_208138131 | 0.14 |
CD34 |
CD34 molecule |
53308 |
0.17 |
| chr1_3050486_3050709 | 0.14 |
RP1-163G9.2 |
|
324 |
0.92 |
| chr21_36042164_36042315 | 0.14 |
CLIC6 |
chloride intracellular channel 6 |
551 |
0.82 |
| chr11_67781609_67782080 | 0.14 |
ALDH3B1 |
aldehyde dehydrogenase 3 family, member B1 |
4054 |
0.13 |
| chr19_56620271_56620526 | 0.14 |
ENSG00000223060 |
. |
3613 |
0.16 |
| chr9_5846170_5846325 | 0.14 |
ERMP1 |
endoplasmic reticulum metallopeptidase 1 |
13130 |
0.19 |
| chr18_77138968_77139119 | 0.14 |
NFATC1 |
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1 |
16813 |
0.2 |
| chr8_103255015_103255398 | 0.14 |
KB-431C1.4 |
|
631 |
0.67 |
| chr3_85007560_85007788 | 0.14 |
CADM2 |
cell adhesion molecule 2 |
458 |
0.89 |
| chr9_124985233_124985384 | 0.14 |
LHX6 |
LIM homeobox 6 |
1289 |
0.37 |
| chr4_75240215_75240456 | 0.14 |
EREG |
epiregulin |
9475 |
0.2 |
| chr3_177651904_177652275 | 0.14 |
ENSG00000199858 |
. |
73478 |
0.13 |
| chr9_694654_694862 | 0.14 |
RP11-130C19.3 |
|
9203 |
0.22 |
| chr4_10126213_10126364 | 0.14 |
WDR1 |
WD repeat domain 1 |
7715 |
0.18 |
| chr1_26145213_26145423 | 0.14 |
MTFR1L |
mitochondrial fission regulator 1-like |
187 |
0.83 |
| chr5_36241274_36241468 | 0.14 |
NADK2 |
NAD kinase 2, mitochondrial |
122 |
0.96 |
| chr22_45095245_45095396 | 0.14 |
PRR5 |
proline rich 5 (renal) |
2758 |
0.23 |
| chr11_27723424_27723575 | 0.14 |
BDNF |
brain-derived neurotrophic factor |
319 |
0.92 |
| chr1_203155609_203155963 | 0.14 |
CHI3L1 |
chitinase 3-like 1 (cartilage glycoprotein-39) |
91 |
0.96 |
| chr11_12790666_12790817 | 0.14 |
TEAD1 |
TEA domain family member 1 (SV40 transcriptional enhancer factor) |
24153 |
0.21 |
| chr16_68334547_68335007 | 0.14 |
SLC7A6OS |
solute carrier family 7, member 6 opposite strand |
3355 |
0.11 |
| chr1_48937375_48937526 | 0.14 |
SPATA6 |
spermatogenesis associated 6 |
232 |
0.95 |
| chr16_57671558_57671817 | 0.14 |
GPR56 |
G protein-coupled receptor 56 |
1520 |
0.31 |
| chr4_86935791_86936081 | 0.14 |
RP13-514E23.1 |
|
909 |
0.62 |
| chr10_105211829_105211980 | 0.14 |
CALHM2 |
calcium homeostasis modulator 2 |
171 |
0.82 |
| chr6_163681290_163681441 | 0.14 |
ENSG00000239136 |
. |
33423 |
0.21 |
| chr11_44971760_44972074 | 0.14 |
TP53I11 |
tumor protein p53 inducible protein 11 |
158 |
0.97 |
| chr3_159903422_159903573 | 0.13 |
IL12A-AS1 |
IL12A antisense RNA 1 |
21382 |
0.18 |
| chr10_49815677_49815878 | 0.13 |
ARHGAP22 |
Rho GTPase activating protein 22 |
2639 |
0.3 |
| chr6_163776181_163776490 | 0.13 |
QKI |
QKI, KH domain containing, RNA binding |
59340 |
0.15 |
| chr12_26380208_26380359 | 0.13 |
SSPN |
sarcospan |
31677 |
0.16 |
| chr7_150659770_150659921 | 0.13 |
KCNH2 |
potassium voltage-gated channel, subfamily H (eag-related), member 2 |
1881 |
0.27 |
| chr12_124877793_124878051 | 0.13 |
NCOR2 |
nuclear receptor corepressor 2 |
4552 |
0.32 |
| chr11_1913396_1913650 | 0.13 |
C11orf89 |
chromosome 11 open reading frame 89 |
1439 |
0.25 |
| chr1_28517417_28517723 | 0.13 |
PTAFR |
platelet-activating factor receptor |
2814 |
0.15 |
| chr20_3652796_3652947 | 0.13 |
GFRA4 |
GDNF family receptor alpha 4 |
8825 |
0.15 |
| chr2_172945273_172945521 | 0.13 |
DLX1 |
distal-less homeobox 1 |
4071 |
0.25 |
| chr1_50888417_50888568 | 0.13 |
DMRTA2 |
DMRT-like family A2 |
680 |
0.75 |
| chr6_15276422_15276621 | 0.13 |
JARID2 |
jumonji, AT rich interactive domain 2 |
27378 |
0.16 |
| chr17_39942503_39942654 | 0.13 |
JUP |
junction plakoglobin |
117 |
0.93 |
| chr21_47603648_47603799 | 0.13 |
SPATC1L |
spermatogenesis and centriole associated 1-like |
650 |
0.46 |
| chr5_139034464_139034615 | 0.13 |
CXXC5 |
CXXC finger protein 5 |
3965 |
0.26 |
| chr17_8383287_8383438 | 0.13 |
NDEL1 |
nudE neurodevelopment protein 1-like 1 |
29263 |
0.12 |
| chr8_28347877_28348093 | 0.13 |
FBXO16 |
F-box protein 16 |
150 |
0.94 |
| chr10_80938884_80939035 | 0.13 |
ZMIZ1 |
zinc finger, MIZ-type containing 1 |
110167 |
0.06 |
| chr6_21847212_21847442 | 0.13 |
ENSG00000222515 |
. |
238446 |
0.02 |
| chr8_8408574_8408871 | 0.13 |
ENSG00000263616 |
. |
31443 |
0.21 |
| chr1_19639347_19639591 | 0.13 |
PQLC2 |
PQ loop repeat containing 2 |
634 |
0.43 |
| chr12_132337369_132337604 | 0.13 |
MMP17 |
matrix metallopeptidase 17 (membrane-inserted) |
7646 |
0.18 |
| chr15_26084539_26084690 | 0.13 |
ENSG00000266517 |
. |
9358 |
0.19 |
| chr6_41754836_41755024 | 0.13 |
TOMM6 |
translocase of outer mitochondrial membrane 6 homolog (yeast) |
470 |
0.49 |
| chr16_29616402_29616553 | 0.13 |
ENSG00000266758 |
. |
5891 |
0.16 |
| chr15_77707876_77708432 | 0.13 |
PEAK1 |
pseudopodium-enriched atypical kinase 1 |
4288 |
0.26 |
| chr12_7126708_7126912 | 0.13 |
LPCAT3 |
lysophosphatidylcholine acyltransferase 3 |
996 |
0.35 |
| chr3_59761256_59761407 | 0.13 |
NPCDR1 |
nasopharyngeal carcinoma, down-regulated 1 |
196252 |
0.03 |
| chr2_242499276_242499525 | 0.13 |
BOK-AS1 |
BOK antisense RNA 1 |
1008 |
0.35 |
| chrX_54948494_54948718 | 0.13 |
TRO |
trophinin |
196 |
0.94 |
| chr5_172266910_172267216 | 0.13 |
ERGIC1 |
endoplasmic reticulum-golgi intermediate compartment (ERGIC) 1 |
5649 |
0.2 |
| chr10_45474416_45474772 | 0.13 |
C10orf10 |
chromosome 10 open reading frame 10 |
336 |
0.82 |
| chr3_16006227_16006629 | 0.13 |
ENSG00000207815 |
. |
91150 |
0.08 |
| chr19_13905830_13905981 | 0.13 |
ZSWIM4 |
zinc finger, SWIM-type containing 4 |
369 |
0.68 |
| chr2_172380569_172381265 | 0.13 |
CYBRD1 |
cytochrome b reductase 1 |
1328 |
0.53 |
| chr1_206845534_206845685 | 0.13 |
ENSG00000252853 |
. |
9699 |
0.14 |
| chr19_4878563_4878757 | 0.13 |
ENSG00000264899 |
. |
1936 |
0.21 |
| chr5_692826_693134 | 0.13 |
TPPP |
tubulin polymerization promoting protein |
530 |
0.74 |
| chr3_194600486_194600791 | 0.13 |
FAM43A |
family with sequence similarity 43, member A |
194016 |
0.02 |
| chr1_36786899_36787105 | 0.13 |
RP11-268J15.5 |
|
2333 |
0.2 |
| chr5_158179607_158179937 | 0.13 |
CTD-2363C16.1 |
|
230242 |
0.02 |
| chr22_37416048_37416272 | 0.13 |
MPST |
mercaptopyruvate sulfurtransferase |
347 |
0.54 |
| chr4_57909352_57909761 | 0.13 |
ENSG00000251703 |
. |
40698 |
0.11 |
| chr20_50180187_50180357 | 0.13 |
NFATC2 |
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2 |
902 |
0.72 |
| chr17_78237788_78237939 | 0.13 |
RNF213 |
ring finger protein 213 |
3194 |
0.2 |
| chr12_106695790_106695941 | 0.13 |
TCP11L2 |
t-complex 11, testis-specific-like 2 |
158 |
0.94 |
| chr11_76411277_76411428 | 0.13 |
GUCY2EP |
guanylate cyclase 2E, pseudogene |
7333 |
0.18 |
| chr6_56517195_56517397 | 0.12 |
DST |
dystonin |
9502 |
0.31 |
| chr14_29254769_29255066 | 0.12 |
RP11-966I7.2 |
|
76 |
0.97 |
| chr2_217354341_217354722 | 0.12 |
AC098820.4 |
|
2612 |
0.22 |
| chr2_29678241_29678392 | 0.12 |
ENSG00000266310 |
. |
48543 |
0.18 |
| chr22_36822490_36822658 | 0.12 |
ENSG00000252575 |
. |
12397 |
0.15 |
| chr7_5632818_5633083 | 0.12 |
FSCN1 |
fascin homolog 1, actin-bundling protein (Strongylocentrotus purpuratus) |
496 |
0.76 |
| chr7_5595160_5595387 | 0.12 |
CTB-161C1.1 |
|
1089 |
0.41 |
| chr12_113298325_113298476 | 0.12 |
RPH3A |
rabphilin 3A homolog (mouse) |
32276 |
0.17 |
| chr1_64626493_64626644 | 0.12 |
RP11-24J23.2 |
|
10412 |
0.21 |
| chr5_123964374_123964729 | 0.12 |
RP11-436H11.2 |
|
99973 |
0.06 |
| chr17_46012490_46012796 | 0.12 |
RP11-6N17.3 |
|
2171 |
0.16 |
| chr12_50020089_50020240 | 0.12 |
PRPF40B |
PRP40 pre-mRNA processing factor 40 homolog B (S. cerevisiae) |
2761 |
0.2 |
| chr18_46422994_46423234 | 0.12 |
SMAD7 |
SMAD family member 7 |
51761 |
0.14 |
| chr1_41290137_41290288 | 0.12 |
KCNQ4 |
potassium voltage-gated channel, KQT-like subfamily, member 4 |
7275 |
0.15 |
| chr3_149095823_149096214 | 0.12 |
TM4SF1-AS1 |
TM4SF1 antisense RNA 1 |
12 |
0.86 |
| chr4_87889842_87889993 | 0.12 |
AFF1 |
AF4/FMR2 family, member 1 |
32355 |
0.19 |
| chr10_104380767_104380980 | 0.12 |
TRIM8 |
tripartite motif containing 8 |
23380 |
0.14 |
| chr10_126707561_126707832 | 0.12 |
CTBP2 |
C-terminal binding protein 2 |
8763 |
0.2 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0031061 | negative regulation of histone methylation(GO:0031061) |
| 0.1 | 0.2 | GO:0007028 | cytoplasm organization(GO:0007028) |
| 0.1 | 0.2 | GO:0060831 | smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:0060831) |
| 0.0 | 0.1 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
| 0.0 | 0.1 | GO:0034135 | regulation of toll-like receptor 2 signaling pathway(GO:0034135) |
| 0.0 | 0.3 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.2 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.0 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.0 | 0.2 | GO:1904181 | positive regulation of mitochondrial depolarization(GO:0051901) positive regulation of membrane depolarization(GO:1904181) |
| 0.0 | 0.0 | GO:0046083 | adenine salvage(GO:0006168) purine nucleobase salvage(GO:0043096) adenine metabolic process(GO:0046083) adenine biosynthetic process(GO:0046084) |
| 0.0 | 0.2 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.1 | GO:0032071 | regulation of endodeoxyribonuclease activity(GO:0032071) |
| 0.0 | 0.3 | GO:0021799 | cerebral cortex radially oriented cell migration(GO:0021799) |
| 0.0 | 0.1 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.0 | 0.1 | GO:0010193 | response to ozone(GO:0010193) |
| 0.0 | 0.1 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.2 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.2 | GO:0006824 | cobalt ion transport(GO:0006824) |
| 0.0 | 0.1 | GO:0003010 | voluntary skeletal muscle contraction(GO:0003010) twitch skeletal muscle contraction(GO:0014721) |
| 0.0 | 0.2 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.2 | GO:0046855 | phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) |
| 0.0 | 0.1 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 0.1 | GO:0021569 | rhombomere 3 development(GO:0021569) |
| 0.0 | 0.1 | GO:0002085 | inhibition of neuroepithelial cell differentiation(GO:0002085) |
| 0.0 | 0.1 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.1 | GO:0072393 | microtubule anchoring at microtubule organizing center(GO:0072393) |
| 0.0 | 0.1 | GO:0046599 | regulation of centriole replication(GO:0046599) |
| 0.0 | 0.1 | GO:0071436 | sodium ion export(GO:0071436) |
| 0.0 | 0.1 | GO:0001766 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 0.1 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.0 | 0.2 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.0 | 0.1 | GO:0070561 | vitamin D receptor signaling pathway(GO:0070561) |
| 0.0 | 0.1 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.1 | GO:0070874 | negative regulation of glycogen metabolic process(GO:0070874) |
| 0.0 | 0.1 | GO:0051503 | adenine nucleotide transport(GO:0051503) |
| 0.0 | 0.1 | GO:0006565 | L-serine catabolic process(GO:0006565) |
| 0.0 | 0.1 | GO:0090206 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.0 | 0.1 | GO:0006925 | inflammatory cell apoptotic process(GO:0006925) |
| 0.0 | 0.1 | GO:0002407 | dendritic cell chemotaxis(GO:0002407) dendritic cell migration(GO:0036336) |
| 0.0 | 0.2 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.1 | GO:0060586 | multicellular organismal iron ion homeostasis(GO:0060586) |
| 0.0 | 0.1 | GO:0060789 | hair follicle placode formation(GO:0060789) |
| 0.0 | 0.1 | GO:0042637 | catagen(GO:0042637) |
| 0.0 | 0.1 | GO:0046697 | decidualization(GO:0046697) |
| 0.0 | 0.4 | GO:0043392 | negative regulation of DNA binding(GO:0043392) |
| 0.0 | 0.1 | GO:0090292 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.1 | GO:0042816 | pyridoxine metabolic process(GO:0008614) pyridoxine biosynthetic process(GO:0008615) vitamin B6 metabolic process(GO:0042816) vitamin B6 biosynthetic process(GO:0042819) |
| 0.0 | 0.1 | GO:0060083 | smooth muscle contraction involved in micturition(GO:0060083) |
| 0.0 | 0.0 | GO:0033505 | floor plate formation(GO:0021508) floor plate morphogenesis(GO:0033505) |
| 0.0 | 0.0 | GO:0010980 | regulation of vitamin D 24-hydroxylase activity(GO:0010979) positive regulation of vitamin D 24-hydroxylase activity(GO:0010980) |
| 0.0 | 0.1 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.0 | 0.3 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.1 | GO:0046831 | regulation of RNA export from nucleus(GO:0046831) |
| 0.0 | 0.1 | GO:0002328 | pro-B cell differentiation(GO:0002328) |
| 0.0 | 0.1 | GO:0010766 | negative regulation of sodium ion transport(GO:0010766) |
| 0.0 | 0.1 | GO:0010586 | miRNA metabolic process(GO:0010586) miRNA catabolic process(GO:0010587) |
| 0.0 | 0.1 | GO:0042832 | defense response to protozoan(GO:0042832) |
| 0.0 | 0.1 | GO:0032277 | negative regulation of gonadotropin secretion(GO:0032277) |
| 0.0 | 0.1 | GO:0045759 | negative regulation of action potential(GO:0045759) |
| 0.0 | 0.1 | GO:0009186 | deoxyribonucleoside diphosphate metabolic process(GO:0009186) |
| 0.0 | 0.1 | GO:0046476 | glycosylceramide biosynthetic process(GO:0046476) |
| 0.0 | 0.1 | GO:0072148 | epithelial cell fate commitment(GO:0072148) |
| 0.0 | 0.1 | GO:0061030 | epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.0 | 0.0 | GO:0033688 | regulation of osteoblast proliferation(GO:0033688) |
| 0.0 | 0.1 | GO:0014049 | positive regulation of glutamate secretion(GO:0014049) |
| 0.0 | 0.0 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 0.1 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.0 | 0.1 | GO:0071072 | negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 0.0 | 0.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.0 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.1 | GO:0043497 | regulation of protein heterodimerization activity(GO:0043497) |
| 0.0 | 0.1 | GO:0043116 | negative regulation of vascular permeability(GO:0043116) |
| 0.0 | 0.1 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.1 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.1 | GO:0019720 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.0 | 0.1 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.0 | GO:0072201 | negative regulation of mesenchymal cell proliferation(GO:0072201) |
| 0.0 | 0.0 | GO:0060298 | positive regulation of sarcomere organization(GO:0060298) |
| 0.0 | 0.1 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 | 0.1 | GO:0001547 | antral ovarian follicle growth(GO:0001547) |
| 0.0 | 0.1 | GO:0070120 | ciliary neurotrophic factor-mediated signaling pathway(GO:0070120) |
| 0.0 | 0.1 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.1 | GO:0070059 | intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress(GO:0070059) |
| 0.0 | 0.1 | GO:2000178 | negative regulation of neuroblast proliferation(GO:0007406) negative regulation of neural precursor cell proliferation(GO:2000178) |
| 0.0 | 0.0 | GO:0031558 | obsolete induction of apoptosis in response to chemical stimulus(GO:0031558) |
| 0.0 | 0.0 | GO:0001821 | histamine secretion(GO:0001821) |
| 0.0 | 0.1 | GO:0021548 | pons development(GO:0021548) |
| 0.0 | 0.1 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000794) |
| 0.0 | 0.1 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.0 | GO:0048368 | lateral mesoderm development(GO:0048368) |
| 0.0 | 0.0 | GO:0046940 | nucleoside monophosphate phosphorylation(GO:0046940) |
| 0.0 | 0.0 | GO:0019049 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.0 | 0.1 | GO:0002093 | auditory receptor cell morphogenesis(GO:0002093) auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.1 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 0.0 | 0.1 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.0 | 0.0 | GO:0003099 | positive regulation of the force of heart contraction by chemical signal(GO:0003099) |
| 0.0 | 0.0 | GO:0030540 | female genitalia development(GO:0030540) |
| 0.0 | 0.1 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.0 | GO:0033024 | mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) |
| 0.0 | 0.1 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.0 | GO:0014041 | regulation of neuron maturation(GO:0014041) regulation of cell maturation(GO:1903429) |
| 0.0 | 0.0 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.0 | GO:0061047 | foregut regionalization(GO:0060423) lung field specification(GO:0060424) lung induction(GO:0060492) positive regulation of branching involved in lung morphogenesis(GO:0061047) |
| 0.0 | 0.1 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.0 | 0.1 | GO:0042448 | progesterone metabolic process(GO:0042448) |
| 0.0 | 0.0 | GO:0045112 | integrin biosynthetic process(GO:0045112) |
| 0.0 | 0.1 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.0 | 0.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.0 | GO:0048840 | otolith development(GO:0048840) |
| 0.0 | 0.0 | GO:0010957 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.0 | 0.1 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.0 | GO:0003352 | regulation of cilium movement(GO:0003352) regulation of cilium beat frequency(GO:0003356) cilium movement involved in cell motility(GO:0060294) regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.0 | 0.0 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.0 | 0.0 | GO:0071545 | inositol phosphate catabolic process(GO:0071545) |
| 0.0 | 0.1 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.0 | 0.0 | GO:0045007 | depurination(GO:0045007) |
| 0.0 | 0.0 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.1 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.0 | 0.0 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.4 | GO:0048146 | positive regulation of fibroblast proliferation(GO:0048146) |
| 0.0 | 0.0 | GO:0018119 | protein nitrosylation(GO:0017014) peptidyl-cysteine S-nitrosylation(GO:0018119) |
| 0.0 | 0.1 | GO:0046337 | phosphatidylethanolamine biosynthetic process(GO:0006646) phosphatidylethanolamine metabolic process(GO:0046337) |
| 0.0 | 0.0 | GO:0021615 | glossopharyngeal nerve development(GO:0021563) glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.1 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 | 0.2 | GO:0048844 | artery morphogenesis(GO:0048844) |
| 0.0 | 0.0 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.0 | 0.0 | GO:0042339 | keratan sulfate metabolic process(GO:0042339) |
| 0.0 | 0.0 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.0 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.0 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) |
| 0.0 | 0.0 | GO:0072143 | mesangial cell differentiation(GO:0072007) kidney interstitial fibroblast differentiation(GO:0072071) renal interstitial fibroblast development(GO:0072141) mesangial cell development(GO:0072143) pericyte cell differentiation(GO:1904238) |
| 0.0 | 0.0 | GO:0060157 | urinary bladder development(GO:0060157) |
| 0.0 | 0.0 | GO:0070366 | regulation of hepatocyte differentiation(GO:0070366) |
| 0.0 | 0.0 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.0 | 0.1 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.0 | 0.0 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) chloride ion homeostasis(GO:0055064) |
| 0.0 | 0.0 | GO:0002689 | negative regulation of leukocyte chemotaxis(GO:0002689) |
| 0.0 | 0.0 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.0 | 0.0 | GO:0032291 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.0 | 0.2 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.0 | 0.1 | GO:0009886 | post-embryonic morphogenesis(GO:0009886) |
| 0.0 | 0.0 | GO:0042536 | negative regulation of tumor necrosis factor biosynthetic process(GO:0042536) |
| 0.0 | 0.0 | GO:1903299 | regulation of glucokinase activity(GO:0033131) positive regulation of glucokinase activity(GO:0033133) regulation of hexokinase activity(GO:1903299) positive regulation of hexokinase activity(GO:1903301) |
| 0.0 | 0.1 | GO:0043276 | anoikis(GO:0043276) |
| 0.0 | 0.0 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.1 | GO:0046457 | prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.0 | 0.2 | GO:0042490 | mechanoreceptor differentiation(GO:0042490) |
| 0.0 | 0.0 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.0 | 0.0 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.0 | 0.0 | GO:0072338 | allantoin metabolic process(GO:0000255) creatinine metabolic process(GO:0046449) cellular lactam metabolic process(GO:0072338) |
| 0.0 | 0.2 | GO:0001706 | endoderm formation(GO:0001706) |
| 0.0 | 0.0 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.0 | 0.0 | GO:0021794 | thalamus development(GO:0021794) |
| 0.0 | 0.1 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.0 | 0.0 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.0 | GO:1903726 | negative regulation of phospholipid metabolic process(GO:1903726) |
| 0.0 | 0.0 | GO:2000328 | T-helper cell lineage commitment(GO:0002295) alpha-beta T cell lineage commitment(GO:0002363) CD4-positive, alpha-beta T cell lineage commitment(GO:0043373) memory T cell differentiation(GO:0043379) regulation of memory T cell differentiation(GO:0043380) positive regulation of memory T cell differentiation(GO:0043382) T-helper 17 type immune response(GO:0072538) T-helper 17 cell differentiation(GO:0072539) T-helper 17 cell lineage commitment(GO:0072540) regulation of T-helper 17 type immune response(GO:2000316) positive regulation of T-helper 17 type immune response(GO:2000318) regulation of T-helper 17 cell differentiation(GO:2000319) positive regulation of T-helper 17 cell differentiation(GO:2000321) regulation of T-helper 17 cell lineage commitment(GO:2000328) positive regulation of T-helper 17 cell lineage commitment(GO:2000330) |
| 0.0 | 0.1 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.0 | 0.1 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.0 | 0.1 | GO:1901976 | regulation of cell cycle checkpoint(GO:1901976) |
| 0.0 | 0.1 | GO:0009125 | nucleoside monophosphate catabolic process(GO:0009125) |
| 0.0 | 0.1 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 0.0 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.0 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.0 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.0 | 0.1 | GO:0030033 | microvillus assembly(GO:0030033) microvillus organization(GO:0032528) |
| 0.0 | 0.1 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) |
| 0.0 | 0.0 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 0.0 | GO:0072133 | kidney mesenchyme morphogenesis(GO:0072131) metanephric mesenchyme morphogenesis(GO:0072133) |
| 0.0 | 0.1 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.0 | 0.0 | GO:0032632 | interleukin-3 production(GO:0032632) |
| 0.0 | 0.0 | GO:0030011 | maintenance of cell polarity(GO:0030011) |
| 0.0 | 0.1 | GO:0002829 | negative regulation of type 2 immune response(GO:0002829) |
| 0.0 | 0.0 | GO:0072526 | pyridine-containing compound catabolic process(GO:0072526) |
| 0.0 | 0.1 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.0 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.0 | GO:0085020 | protein K6-linked ubiquitination(GO:0085020) |
| 0.0 | 0.0 | GO:0090009 | primitive streak formation(GO:0090009) |
| 0.0 | 0.1 | GO:0090083 | regulation of inclusion body assembly(GO:0090083) |
| 0.0 | 0.0 | GO:0010823 | negative regulation of mitochondrion organization(GO:0010823) negative regulation of release of cytochrome c from mitochondria(GO:0090201) negative regulation of apoptotic signaling pathway(GO:2001234) |
| 0.0 | 0.1 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 0.1 | GO:0046487 | glyoxylate metabolic process(GO:0046487) |
| 0.0 | 0.1 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.1 | GO:0032410 | negative regulation of transporter activity(GO:0032410) |
| 0.0 | 0.1 | GO:0050872 | white fat cell differentiation(GO:0050872) |
| 0.0 | 0.0 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.0 | GO:0045657 | positive regulation of monocyte differentiation(GO:0045657) |
| 0.0 | 0.1 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.0 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.2 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.0 | 0.1 | GO:0042976 | activation of Janus kinase activity(GO:0042976) |
| 0.0 | 0.2 | GO:0006636 | unsaturated fatty acid biosynthetic process(GO:0006636) |
| 0.0 | 0.0 | GO:0033137 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) |
| 0.0 | 0.0 | GO:0060014 | granulosa cell differentiation(GO:0060014) |
| 0.0 | 0.0 | GO:0051124 | synaptic growth at neuromuscular junction(GO:0051124) |
| 0.0 | 0.0 | GO:0034379 | very-low-density lipoprotein particle assembly(GO:0034379) |
| 0.0 | 0.0 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.1 | GO:0060263 | regulation of respiratory burst(GO:0060263) |
| 0.0 | 0.1 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.0 | 0.0 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.2 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 0.1 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.1 | GO:0007141 | male meiosis I(GO:0007141) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.1 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.1 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.2 | GO:0031512 | motile primary cilium(GO:0031512) |
| 0.0 | 0.2 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.1 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.1 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.1 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.1 | GO:0044453 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 0.1 | GO:0031332 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 0.1 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.1 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.1 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.1 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.1 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.0 | 0.0 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.1 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.1 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.0 | GO:0060200 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.0 | 0.0 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.1 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.2 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.1 | GO:0002178 | palmitoyltransferase complex(GO:0002178) serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.1 | GO:0008274 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.0 | 0.1 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.1 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.1 | GO:0043205 | fibril(GO:0043205) |
| 0.0 | 0.1 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 0.2 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.1 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.1 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.0 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 0.1 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.1 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.0 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.0 | 0.2 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.0 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.1 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.1 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.1 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.0 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 0.1 | GO:0061200 | clathrin-sculpted gamma-aminobutyric acid transport vesicle(GO:0061200) clathrin-sculpted gamma-aminobutyric acid transport vesicle membrane(GO:0061202) |
| 0.0 | 0.0 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.0 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.0 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.1 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.1 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.0 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.1 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 0.2 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.1 | 0.2 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.1 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.0 | 0.1 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.0 | 0.1 | GO:0052659 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) inositol-1,3,4,5-tetrakisphosphate 5-phosphatase activity(GO:0052659) |
| 0.0 | 0.2 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.1 | GO:0052743 | inositol tetrakisphosphate phosphatase activity(GO:0052743) |
| 0.0 | 0.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.1 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.1 | GO:0004748 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.2 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.3 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.2 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.1 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.0 | 0.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.1 | GO:0000987 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.0 | 0.1 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.1 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.2 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) |
| 0.0 | 0.1 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.2 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.2 | GO:0004437 | obsolete inositol or phosphatidylinositol phosphatase activity(GO:0004437) |
| 0.0 | 0.1 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.1 | GO:0042153 | obsolete RPTP-like protein binding(GO:0042153) |
| 0.0 | 0.1 | GO:0004992 | platelet activating factor receptor activity(GO:0004992) |
| 0.0 | 0.1 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.0 | 0.3 | GO:0005035 | death receptor activity(GO:0005035) |
| 0.0 | 0.1 | GO:0008443 | phosphofructokinase activity(GO:0008443) |
| 0.0 | 0.1 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.1 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.1 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.1 | GO:0015216 | adenine nucleotide transmembrane transporter activity(GO:0000295) purine nucleotide transmembrane transporter activity(GO:0015216) |
| 0.0 | 0.0 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) phosphatidylinositol phosphate 5-phosphatase activity(GO:0034595) |
| 0.0 | 0.0 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.0 | 0.1 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.0 | 0.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.1 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.1 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.0 | 0.1 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.0 | 0.1 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.1 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.1 | GO:0042285 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.1 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.1 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.1 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.0 | 0.1 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.1 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.0 | 0.2 | GO:0030553 | cGMP binding(GO:0030553) |
| 0.0 | 0.2 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.1 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.1 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.1 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.2 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 0.1 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.0 | 0.1 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.1 | GO:0050543 | icosatetraenoic acid binding(GO:0050543) |
| 0.0 | 0.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.1 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.0 | 0.0 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.1 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.1 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.0 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.0 | 0.2 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.0 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.0 | 0.0 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.1 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.1 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.0 | 0.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.2 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 0.1 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.1 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.0 | GO:0042808 | obsolete neuronal Cdc2-like kinase binding(GO:0042808) |
| 0.0 | 0.0 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.0 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 0.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.0 | GO:0097493 | extracellular matrix constituent conferring elasticity(GO:0030023) structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.1 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.0 | GO:0030343 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.0 | 0.0 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.1 | GO:0016901 | oxidoreductase activity, acting on the CH-OH group of donors, quinone or similar compound as acceptor(GO:0016901) |
| 0.0 | 0.1 | GO:0050780 | dopamine receptor binding(GO:0050780) |
| 0.0 | 0.2 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.0 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.0 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.0 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.1 | GO:0016717 | oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water(GO:0016717) |
| 0.0 | 0.2 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 0.1 | GO:0004945 | angiotensin receptor activity(GO:0001595) angiotensin type II receptor activity(GO:0004945) |
| 0.0 | 0.1 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.0 | 0.1 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 0.1 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.1 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 0.0 | 0.1 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.1 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.1 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.1 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.0 | 0.0 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.0 | 0.1 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 0.1 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.1 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.0 | GO:0051380 | norepinephrine binding(GO:0051380) |
| 0.0 | 0.0 | GO:0004607 | phosphatidylcholine-sterol O-acyltransferase activity(GO:0004607) |
| 0.0 | 0.0 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 0.1 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 0.0 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.0 | 0.0 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.0 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.0 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.0 | 0.0 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.1 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.0 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 0.0 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.0 | 0.0 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.0 | 0.0 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.1 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.0 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.0 | GO:0004470 | malic enzyme activity(GO:0004470) |
| 0.0 | 0.3 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.1 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.0 | 0.1 | GO:0001640 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) G-protein coupled glutamate receptor activity(GO:0098988) |
| 0.0 | 0.1 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.0 | 0.0 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.1 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.0 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.0 | 0.0 | GO:0004114 | 3',5'-cyclic-nucleotide phosphodiesterase activity(GO:0004114) |
| 0.0 | 0.0 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.0 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.0 | 0.0 | GO:0005047 | signal recognition particle binding(GO:0005047) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 0.0 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 0.1 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.1 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.0 | 0.2 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.0 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.0 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.0 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.4 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.2 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.2 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.3 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME DOWNSTREAM SIGNALING OF ACTIVATED FGFR | Genes involved in Downstream signaling of activated FGFR |
| 0.0 | 0.0 | REACTOME NCAM SIGNALING FOR NEURITE OUT GROWTH | Genes involved in NCAM signaling for neurite out-growth |
| 0.0 | 0.1 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.0 | 0.5 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.0 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.0 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.0 | 0.4 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.2 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.5 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.4 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.0 | REACTOME PLATELET AGGREGATION PLUG FORMATION | Genes involved in Platelet Aggregation (Plug Formation) |
| 0.0 | 0.0 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.2 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.2 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.2 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.2 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.1 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.2 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.3 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.2 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 0.2 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.1 | REACTOME NRIF SIGNALS CELL DEATH FROM THE NUCLEUS | Genes involved in NRIF signals cell death from the nucleus |
| 0.0 | 0.1 | REACTOME SIGNALING BY NOTCH3 | Genes involved in Signaling by NOTCH3 |
| 0.0 | 0.1 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 0.0 | REACTOME APC C CDC20 MEDIATED DEGRADATION OF MITOTIC PROTEINS | Genes involved in APC/C:Cdc20 mediated degradation of mitotic proteins |
| 0.0 | 0.1 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.1 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.1 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.1 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.1 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.3 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.0 | REACTOME PI3K CASCADE | Genes involved in PI3K Cascade |