Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
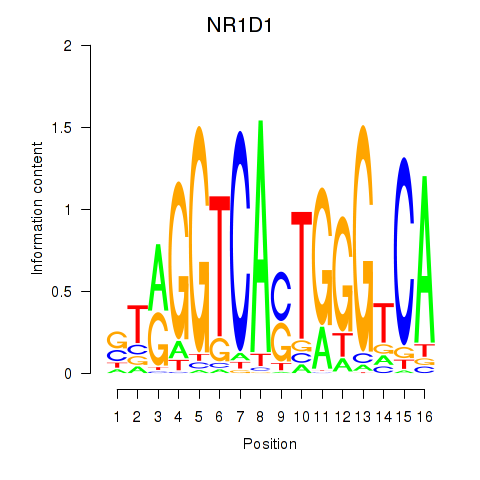
Results for NR1D1
Z-value: 0.99
Transcription factors associated with NR1D1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR1D1
|
ENSG00000126368.5 | nuclear receptor subfamily 1 group D member 1 |
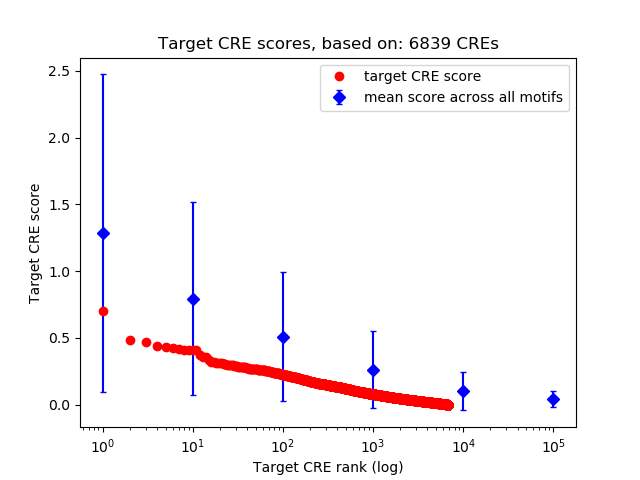
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr17_38267210_38267680 | NR1D1 | 10467 | 0.111988 | -0.87 | 2.5e-03 | Click! |
| chr17_38256127_38256278 | NR1D1 | 776 | 0.511252 | -0.84 | 4.2e-03 | Click! |
| chr17_38267680_38267831 | NR1D1 | 10777 | 0.111446 | -0.83 | 5.9e-03 | Click! |
| chr17_38257796_38257963 | NR1D1 | 901 | 0.448949 | -0.82 | 7.1e-03 | Click! |
| chr17_38263559_38264452 | NR1D1 | 7027 | 0.119812 | -0.72 | 2.7e-02 | Click! |
Activity of the NR1D1 motif across conditions
Conditions sorted by the z-value of the NR1D1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
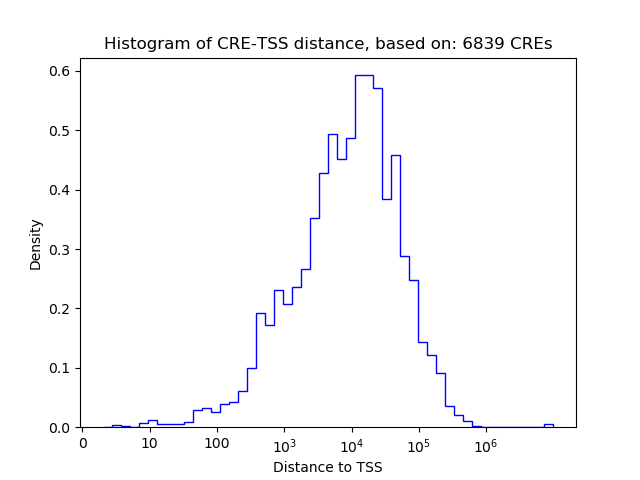
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr20_62240080_62240231 | 0.70 |
GMEB2 |
glucocorticoid modulatory element binding protein 2 |
3900 |
0.13 |
| chr16_16168105_16168644 | 0.49 |
ABCC1 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 1 |
64661 |
0.11 |
| chr11_48057502_48058059 | 0.47 |
AC103828.1 |
|
20373 |
0.19 |
| chr7_508753_509105 | 0.44 |
PDGFA |
platelet-derived growth factor alpha polypeptide |
49216 |
0.14 |
| chr3_98275754_98275968 | 0.43 |
GPR15 |
G protein-coupled receptor 15 |
25118 |
0.13 |
| chr20_62240442_62240593 | 0.43 |
GMEB2 |
glucocorticoid modulatory element binding protein 2 |
4262 |
0.12 |
| chr1_206751930_206752246 | 0.41 |
RASSF5 |
Ras association (RalGDS/AF-6) domain family member 5 |
21595 |
0.14 |
| chr22_30679087_30679350 | 0.41 |
GATSL3 |
GATS protein-like 3 |
6378 |
0.1 |
| chrX_54442126_54442496 | 0.41 |
TSR2 |
TSR2, 20S rRNA accumulation, homolog (S. cerevisiae) |
24523 |
0.19 |
| chr9_140215104_140215809 | 0.41 |
NRARP |
NOTCH-regulated ankyrin repeat protein |
18753 |
0.09 |
| chr16_56999551_56999979 | 0.41 |
CETP |
cholesteryl ester transfer protein, plasma |
3746 |
0.14 |
| chr11_47433734_47434023 | 0.37 |
SLC39A13 |
solute carrier family 39 (zinc transporter), member 13 |
2242 |
0.14 |
| chr11_326945_327265 | 0.36 |
IFITM3 |
interferon induced transmembrane protein 3 |
432 |
0.62 |
| chr20_62241205_62241356 | 0.36 |
GMEB2 |
glucocorticoid modulatory element binding protein 2 |
5025 |
0.12 |
| chr19_13204892_13205479 | 0.34 |
LYL1 |
lymphoblastic leukemia derived sequence 1 |
8496 |
0.1 |
| chr2_113930541_113930896 | 0.32 |
PSD4 |
pleckstrin and Sec7 domain containing 4 |
830 |
0.5 |
| chr11_73086432_73086886 | 0.32 |
RELT |
RELT tumor necrosis factor receptor |
650 |
0.64 |
| chr9_92055035_92055186 | 0.32 |
SEMA4D |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D |
3712 |
0.3 |
| chr15_76023964_76024375 | 0.32 |
AC019294.1 |
|
6142 |
0.1 |
| chr2_114662697_114662848 | 0.31 |
ACTR3 |
ARP3 actin-related protein 3 homolog (yeast) |
14597 |
0.22 |
| chr2_19105837_19105988 | 0.31 |
NT5C1B |
5'-nucleotidase, cytosolic IB |
335074 |
0.01 |
| chr5_169058393_169058544 | 0.31 |
DOCK2 |
dedicator of cytokinesis 2 |
5783 |
0.26 |
| chr4_6930967_6931437 | 0.30 |
TBC1D14 |
TBC1 domain family, member 14 |
19227 |
0.15 |
| chr21_47972385_47972571 | 0.30 |
ENSG00000272283 |
. |
16769 |
0.17 |
| chr12_12628244_12628725 | 0.30 |
DUSP16 |
dual specificity phosphatase 16 |
45575 |
0.15 |
| chr3_71573965_71574178 | 0.30 |
ENSG00000221264 |
. |
17169 |
0.2 |
| chr1_153514792_153515209 | 0.30 |
S100A5 |
S100 calcium binding protein A5 |
759 |
0.41 |
| chr1_59407152_59407509 | 0.29 |
JUN |
jun proto-oncogene |
157545 |
0.04 |
| chr2_219676341_219676492 | 0.29 |
CYP27A1 |
cytochrome P450, family 27, subfamily A, polypeptide 1 |
5779 |
0.15 |
| chr12_6557124_6557320 | 0.29 |
CD27 |
CD27 molecule |
3189 |
0.12 |
| chr5_67515227_67515407 | 0.29 |
PIK3R1 |
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
3713 |
0.29 |
| chr14_51574865_51575105 | 0.28 |
TRIM9 |
tripartite motif containing 9 |
12206 |
0.21 |
| chr18_13438233_13438460 | 0.28 |
LDLRAD4-AS1 |
LDLRAD4 antisense RNA 1 |
10867 |
0.12 |
| chr4_177756596_177756747 | 0.28 |
VEGFC |
vascular endothelial growth factor C |
42790 |
0.21 |
| chr5_32838212_32838391 | 0.28 |
AC026703.1 |
|
49356 |
0.16 |
| chr16_29212717_29212868 | 0.28 |
CTB-134H23.3 |
|
94026 |
0.05 |
| chr19_54874397_54874958 | 0.28 |
LAIR1 |
leukocyte-associated immunoglobulin-like receptor 1 |
1737 |
0.22 |
| chr10_89953557_89953785 | 0.28 |
ENSG00000200891 |
. |
199219 |
0.03 |
| chr6_112362238_112362440 | 0.28 |
WISP3 |
WNT1 inducible signaling pathway protein 3 |
12936 |
0.2 |
| chr11_60859183_60859572 | 0.28 |
CD5 |
CD5 molecule |
10490 |
0.18 |
| chr4_2299878_2300029 | 0.27 |
RP11-478C1.7 |
|
2642 |
0.2 |
| chr22_38850403_38851076 | 0.27 |
KCNJ4 |
potassium inwardly-rectifying channel, subfamily J, member 4 |
466 |
0.74 |
| chr10_105671782_105672044 | 0.27 |
OBFC1 |
oligonucleotide/oligosaccharide-binding fold containing 1 |
5514 |
0.19 |
| chr1_160635144_160635480 | 0.27 |
RP11-404F10.2 |
|
5256 |
0.17 |
| chr1_198632560_198632794 | 0.27 |
RP11-553K8.5 |
|
3513 |
0.3 |
| chr17_57442951_57443122 | 0.27 |
ENSG00000263857 |
. |
408 |
0.84 |
| chr4_3204868_3205180 | 0.27 |
MSANTD1 |
Myb/SANT-like DNA-binding domain containing 1 |
41072 |
0.15 |
| chr8_144489216_144489574 | 0.27 |
RP11-909N17.2 |
|
5524 |
0.11 |
| chr9_92054735_92055023 | 0.27 |
SEMA4D |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D |
3481 |
0.31 |
| chr14_98689187_98689819 | 0.27 |
ENSG00000222066 |
. |
108584 |
0.07 |
| chr12_49371240_49371781 | 0.26 |
WNT1 |
wingless-type MMTV integration site family, member 1 |
888 |
0.29 |
| chr19_55016899_55017050 | 0.26 |
LAIR2 |
leukocyte-associated immunoglobulin-like receptor 2 |
2851 |
0.15 |
| chr17_75876491_75876757 | 0.26 |
FLJ45079 |
|
2035 |
0.39 |
| chr6_157388395_157388707 | 0.26 |
RP1-137K2.2 |
|
52289 |
0.17 |
| chr11_117844189_117844444 | 0.26 |
IL10RA |
interleukin 10 receptor, alpha |
12747 |
0.18 |
| chr21_46390444_46390672 | 0.26 |
FAM207A |
family with sequence similarity 207, member A |
30618 |
0.1 |
| chr19_18722693_18723176 | 0.26 |
TMEM59L |
transmembrane protein 59-like |
748 |
0.45 |
| chr16_66524550_66524737 | 0.26 |
AC132186.1 |
CDNA FLJ27243 fis, clone SYN08134; Uncharacterized protein |
4896 |
0.13 |
| chr5_149464844_149464995 | 0.26 |
CSF1R |
colony stimulating factor 1 receptor |
1071 |
0.46 |
| chr5_139049687_139050600 | 0.26 |
CXXC5 |
CXXC finger protein 5 |
4878 |
0.24 |
| chr5_148746761_148747114 | 0.26 |
RP11-394O4.3 |
|
4800 |
0.13 |
| chr14_55222326_55222477 | 0.26 |
SAMD4A |
sterile alpha motif domain containing 4A |
850 |
0.72 |
| chr22_24829765_24830186 | 0.26 |
ADORA2A |
adenosine A2a receptor |
1167 |
0.48 |
| chr4_129405273_129405424 | 0.26 |
PGRMC2 |
progesterone receptor membrane component 2 |
195364 |
0.03 |
| chr8_122001527_122001678 | 0.25 |
ENSG00000252093 |
. |
80026 |
0.11 |
| chr17_38495361_38495755 | 0.25 |
RARA |
retinoic acid receptor, alpha |
2082 |
0.19 |
| chr1_167442877_167443113 | 0.25 |
RP11-104L21.2 |
|
15097 |
0.2 |
| chr5_148884105_148884256 | 0.25 |
CSNK1A1 |
casein kinase 1, alpha 1 |
8540 |
0.18 |
| chr19_1396678_1396882 | 0.25 |
AC005329.7 |
|
314 |
0.74 |
| chr9_4082585_4082736 | 0.25 |
RP11-70J12.1 |
|
10776 |
0.27 |
| chr11_75218502_75218831 | 0.25 |
RP11-939C17.4 |
|
770 |
0.59 |
| chr19_934349_934769 | 0.25 |
ARID3A |
AT rich interactive domain 3A (BRIGHT-like) |
2050 |
0.15 |
| chr13_49435917_49436145 | 0.25 |
ENSG00000265585 |
. |
68317 |
0.11 |
| chr22_38029281_38029617 | 0.25 |
SH3BP1 |
SH3-domain binding protein 1 |
6033 |
0.11 |
| chr12_56414258_56414409 | 0.24 |
IKZF4 |
IKAROS family zinc finger 4 (Eos) |
499 |
0.6 |
| chr21_36408185_36408474 | 0.24 |
RUNX1 |
runt-related transcription factor 1 |
13133 |
0.31 |
| chr20_30457298_30457514 | 0.24 |
DUSP15 |
dual specificity phosphatase 15 |
395 |
0.71 |
| chr7_151048271_151048422 | 0.24 |
NUB1 |
negative regulator of ubiquitin-like proteins 1 |
5915 |
0.16 |
| chr2_113404081_113404232 | 0.24 |
SLC20A1 |
solute carrier family 20 (phosphate transporter), member 1 |
722 |
0.69 |
| chr1_42272500_42272651 | 0.24 |
ENSG00000264896 |
. |
47763 |
0.18 |
| chr12_94600310_94600885 | 0.24 |
RP11-74K11.2 |
|
20777 |
0.19 |
| chr5_133879974_133880666 | 0.24 |
JADE2 |
jade family PHD finger 2 |
17993 |
0.15 |
| chr7_27938817_27939020 | 0.24 |
ENSG00000265382 |
. |
18812 |
0.24 |
| chr2_231977221_231977372 | 0.24 |
HTR2B |
5-hydroxytryptamine (serotonin) receptor 2B, G protein-coupled |
12536 |
0.15 |
| chr7_5649047_5649443 | 0.24 |
FSCN1 |
fascin homolog 1, actin-bundling protein (Strongylocentrotus purpuratus) |
14928 |
0.16 |
| chr12_124978539_124978958 | 0.24 |
NCOR2 |
nuclear receptor corepressor 2 |
1050 |
0.68 |
| chr11_1885891_1886314 | 0.24 |
LSP1 |
lymphocyte-specific protein 1 |
293 |
0.69 |
| chr19_41329835_41330195 | 0.23 |
EGLN2 |
egl-9 family hypoxia-inducible factor 2 |
16867 |
0.11 |
| chr5_172200499_172201004 | 0.23 |
DUSP1 |
dual specificity phosphatase 1 |
2553 |
0.2 |
| chr12_53578247_53578590 | 0.23 |
CSAD |
cysteine sulfinic acid decarboxylase |
3283 |
0.14 |
| chrX_128892305_128892642 | 0.23 |
XPNPEP2 |
X-prolyl aminopeptidase (aminopeptidase P) 2, membrane-bound |
19475 |
0.18 |
| chrX_106175914_106176065 | 0.23 |
CLDN2 |
claudin 2 |
12355 |
0.2 |
| chr2_7148460_7148835 | 0.23 |
RNF144A |
ring finger protein 144A |
11576 |
0.24 |
| chr18_21966009_21966160 | 0.23 |
OSBPL1A |
oxysterol binding protein-like 1A |
11668 |
0.15 |
| chr2_28586534_28586721 | 0.23 |
FOSL2 |
FOS-like antigen 2 |
29042 |
0.15 |
| chr9_129887874_129888025 | 0.23 |
ANGPTL2 |
angiopoietin-like 2 |
2787 |
0.33 |
| chr13_32434206_32434357 | 0.23 |
EEF1DP3 |
eukaryotic translation elongation factor 1 delta pseudogene 3 |
92436 |
0.09 |
| chr12_106476343_106476494 | 0.23 |
NUAK1 |
NUAK family, SNF1-like kinase, 1 |
1393 |
0.5 |
| chr16_19138994_19139145 | 0.23 |
RP11-626G11.3 |
|
6118 |
0.17 |
| chr2_197066450_197066719 | 0.23 |
ENSG00000239161 |
. |
13462 |
0.18 |
| chr20_57733456_57733718 | 0.22 |
ZNF831 |
zinc finger protein 831 |
32488 |
0.19 |
| chr6_76459106_76459699 | 0.22 |
MYO6 |
myosin VI |
353 |
0.88 |
| chr16_27248607_27249099 | 0.22 |
NSMCE1 |
non-SMC element 1 homolog (S. cerevisiae) |
4454 |
0.19 |
| chr5_115844005_115844156 | 0.22 |
SEMA6A |
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
28044 |
0.22 |
| chr16_57161336_57161487 | 0.22 |
CPNE2 |
copine II |
8300 |
0.15 |
| chr1_19394717_19394868 | 0.22 |
UBR4 |
ubiquitin protein ligase E3 component n-recognin 4 |
13942 |
0.19 |
| chr3_126217149_126217300 | 0.22 |
UROC1 |
urocanate hydratase 1 |
19372 |
0.13 |
| chr15_67219603_67219754 | 0.22 |
SMAD3 |
SMAD family member 3 |
136423 |
0.05 |
| chr6_100055589_100055740 | 0.22 |
PRDM13 |
PR domain containing 13 |
1058 |
0.51 |
| chr11_35327376_35327706 | 0.22 |
SLC1A2 |
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
4466 |
0.23 |
| chr16_57167278_57167429 | 0.22 |
CPNE2 |
copine II |
14242 |
0.14 |
| chr2_131132025_131132424 | 0.22 |
PTPN18 |
protein tyrosine phosphatase, non-receptor type 18 (brain-derived) |
4562 |
0.17 |
| chr2_27296602_27297161 | 0.22 |
OST4 |
oligosaccharyltransferase 4 homolog (S. cerevisiae) |
2240 |
0.12 |
| chr8_1901926_1902200 | 0.22 |
KBTBD11 |
kelch repeat and BTB (POZ) domain containing 11 |
19981 |
0.23 |
| chr1_25831625_25831776 | 0.22 |
LDLRAP1 |
low density lipoprotein receptor adaptor protein 1 |
38371 |
0.14 |
| chr3_107819281_107819678 | 0.22 |
CD47 |
CD47 molecule |
9607 |
0.3 |
| chr9_4760903_4761054 | 0.21 |
AK3 |
adenylate kinase 3 |
18935 |
0.17 |
| chr16_86803007_86803158 | 0.21 |
FOXL1 |
forkhead box L1 |
190967 |
0.03 |
| chr14_81630336_81630487 | 0.21 |
RP11-114N19.3 |
|
6347 |
0.25 |
| chr17_42844116_42844267 | 0.21 |
ADAM11 |
ADAM metallopeptidase domain 11 |
7498 |
0.14 |
| chr10_33284606_33284757 | 0.21 |
ITGB1 |
integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12) |
3290 |
0.32 |
| chr3_52267957_52268256 | 0.21 |
TLR9 |
TLR9 |
2900 |
0.13 |
| chr11_122599812_122599963 | 0.21 |
ENSG00000239079 |
. |
2868 |
0.34 |
| chr8_37822610_37823607 | 0.21 |
ADRB3 |
adrenoceptor beta 3 |
1375 |
0.34 |
| chr1_151117119_151117270 | 0.21 |
SEMA6C |
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6C |
1739 |
0.18 |
| chr19_4473935_4474715 | 0.21 |
HDGFRP2 |
Hepatoma-derived growth factor-related protein 2 |
564 |
0.54 |
| chr22_45404880_45405307 | 0.21 |
PHF21B |
PHD finger protein 21B |
219 |
0.95 |
| chr3_190414491_190414848 | 0.21 |
ENSG00000223117 |
. |
54717 |
0.15 |
| chr18_2975917_2976088 | 0.21 |
LPIN2 |
lipin 2 |
6869 |
0.16 |
| chr20_37079512_37079663 | 0.21 |
ENSG00000199266 |
. |
1574 |
0.2 |
| chr15_86128935_86129221 | 0.21 |
RP11-815J21.2 |
|
5669 |
0.19 |
| chr16_85963082_85963233 | 0.21 |
IRF8 |
interferon regulatory factor 8 |
15238 |
0.24 |
| chr10_105665248_105665814 | 0.21 |
OBFC1 |
oligonucleotide/oligosaccharide-binding fold containing 1 |
11896 |
0.17 |
| chr22_17538917_17539068 | 0.21 |
IL17RA |
interleukin 17 receptor A |
26857 |
0.14 |
| chr2_30937309_30937483 | 0.21 |
CAPN13 |
calpain 13 |
92915 |
0.09 |
| chr9_92076120_92076631 | 0.21 |
SEMA4D |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D |
5507 |
0.26 |
| chr15_70049707_70049903 | 0.20 |
ENSG00000238870 |
. |
26644 |
0.2 |
| chr6_166900667_166901285 | 0.20 |
ENSG00000222958 |
. |
21945 |
0.16 |
| chr3_16384271_16384422 | 0.20 |
RP11-415F23.4 |
|
2287 |
0.27 |
| chr8_1320967_1321354 | 0.20 |
DLGAP2 |
discs, large (Drosophila) homolog-associated protein 2 |
128372 |
0.06 |
| chr10_103226335_103226486 | 0.20 |
ENSG00000239091 |
. |
94701 |
0.06 |
| chr9_278055_278426 | 0.20 |
DOCK8 |
dedicator of cytokinesis 8 |
5170 |
0.21 |
| chr16_75661991_75662169 | 0.20 |
ADAT1 |
adenosine deaminase, tRNA-specific 1 |
4882 |
0.15 |
| chr13_100071950_100072184 | 0.20 |
ENSG00000266207 |
. |
33656 |
0.15 |
| chr19_15400617_15401337 | 0.20 |
BRD4 |
bromodomain containing 4 |
9715 |
0.18 |
| chr9_139014504_139014903 | 0.20 |
C9orf69 |
chromosome 9 open reading frame 69 |
3972 |
0.23 |
| chr15_91230173_91230369 | 0.20 |
RP11-387D10.3 |
|
26301 |
0.12 |
| chr6_159625174_159625325 | 0.20 |
FNDC1 |
fibronectin type III domain containing 1 |
6770 |
0.22 |
| chr7_37305599_37305750 | 0.20 |
ELMO1 |
engulfment and cell motility 1 |
43381 |
0.17 |
| chr6_158087850_158088001 | 0.20 |
ZDHHC14 |
zinc finger, DHHC-type containing 14 |
73884 |
0.11 |
| chr7_92267627_92267778 | 0.20 |
FAM133B |
family with sequence similarity 133, member B |
47994 |
0.14 |
| chr10_90656340_90656541 | 0.20 |
STAMBPL1 |
STAM binding protein-like 1 |
4392 |
0.2 |
| chr12_124044061_124044270 | 0.20 |
ENSG00000266655 |
. |
23209 |
0.11 |
| chrX_153606522_153606774 | 0.19 |
EMD |
emerin |
909 |
0.35 |
| chr16_85169897_85170486 | 0.19 |
FAM92B |
family with sequence similarity 92, member B |
24077 |
0.18 |
| chr5_112107941_112108092 | 0.19 |
ENSG00000212370 |
. |
6044 |
0.23 |
| chrX_20280109_20280362 | 0.19 |
RPS6KA3 |
ribosomal protein S6 kinase, 90kDa, polypeptide 3 |
4525 |
0.3 |
| chr3_194929559_194929710 | 0.19 |
ENSG00000206600 |
. |
5882 |
0.18 |
| chr11_128421567_128421718 | 0.19 |
RP11-1007G5.2 |
|
25605 |
0.2 |
| chr11_35294355_35294642 | 0.19 |
SLC1A2 |
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
7206 |
0.18 |
| chr1_213097977_213098128 | 0.19 |
VASH2 |
vasohibin 2 |
25810 |
0.18 |
| chr17_17078547_17078698 | 0.19 |
MPRIP |
myosin phosphatase Rho interacting protein |
56 |
0.95 |
| chr2_112132645_112132888 | 0.19 |
ENSG00000266139 |
. |
54098 |
0.18 |
| chr19_47731951_47732818 | 0.19 |
BBC3 |
BCL2 binding component 3 |
2067 |
0.22 |
| chr19_42706331_42706855 | 0.19 |
ENSG00000265122 |
. |
9363 |
0.09 |
| chr9_139298088_139298507 | 0.19 |
SDCCAG3 |
serologically defined colon cancer antigen 3 |
3340 |
0.13 |
| chr10_54413332_54413483 | 0.19 |
RP11-556E13.1 |
|
92542 |
0.09 |
| chr10_52304911_52305098 | 0.19 |
ENSG00000251950 |
. |
40459 |
0.14 |
| chr10_90552652_90552880 | 0.19 |
LIPM |
lipase, family member M |
9721 |
0.18 |
| chr1_202132763_202133787 | 0.19 |
PTPN7 |
protein tyrosine phosphatase, non-receptor type 7 |
2559 |
0.21 |
| chr14_91814952_91815599 | 0.19 |
ENSG00000265856 |
. |
15218 |
0.22 |
| chr2_100478654_100479136 | 0.19 |
AFF3 |
AF4/FMR2 family, member 3 |
242097 |
0.02 |
| chr8_81992943_81993251 | 0.19 |
PAG1 |
phosphoprotein associated with glycosphingolipid microdomains 1 |
31206 |
0.23 |
| chr11_64121545_64121911 | 0.19 |
RPS6KA4 |
ribosomal protein S6 kinase, 90kDa, polypeptide 4 |
4892 |
0.09 |
| chr16_1351228_1351529 | 0.19 |
UBE2I |
ubiquitin-conjugating enzyme E2I |
7503 |
0.09 |
| chr22_46691822_46692448 | 0.19 |
GTSE1 |
G-2 and S-phase expressed 1 |
503 |
0.76 |
| chr17_76412475_76412647 | 0.18 |
AC061992.1 |
Uncharacterized protein |
9848 |
0.14 |
| chr10_30668096_30668247 | 0.18 |
ENSG00000239625 |
. |
12924 |
0.16 |
| chr3_113933161_113933548 | 0.18 |
RP11-553L6.2 |
|
116 |
0.96 |
| chr17_72588858_72589016 | 0.18 |
CD300LD |
CD300 molecule-like family member d |
515 |
0.68 |
| chr11_59822676_59822875 | 0.18 |
MS4A3 |
membrane-spanning 4-domains, subfamily A, member 3 (hematopoietic cell-specific) |
1285 |
0.44 |
| chr1_227731215_227731366 | 0.18 |
ENSG00000202264 |
. |
17592 |
0.2 |
| chrX_128774957_128775108 | 0.18 |
APLN |
apelin |
7710 |
0.26 |
| chr11_73721865_73722409 | 0.18 |
UCP3 |
uncoupling protein 3 (mitochondrial, proton carrier) |
1657 |
0.32 |
| chr2_197944268_197944419 | 0.18 |
ANKRD44 |
ankyrin repeat domain 44 |
31225 |
0.22 |
| chr8_9022198_9022349 | 0.18 |
RP11-10A14.4 |
Uncharacterized protein |
12960 |
0.17 |
| chr9_140162312_140162640 | 0.18 |
TOR4A |
torsin family 4, member A |
9725 |
0.07 |
| chr7_138747161_138747312 | 0.18 |
ZC3HAV1 |
zinc finger CCCH-type, antiviral 1 |
16777 |
0.19 |
| chr1_16751681_16751832 | 0.18 |
SPATA21 |
spermatogenesis associated 21 |
11929 |
0.14 |
| chr18_10060647_10060798 | 0.18 |
ENSG00000263630 |
. |
55528 |
0.15 |
| chr14_75428743_75429108 | 0.18 |
PGF |
placental growth factor |
6438 |
0.15 |
| chr10_71721356_71721785 | 0.18 |
COL13A1 |
collagen, type XIII, alpha 1 |
31361 |
0.19 |
| chr9_4621034_4621322 | 0.18 |
PPAPDC2 |
phosphatidic acid phosphatase type 2 domain containing 2 |
41120 |
0.13 |
| chr1_33723200_33724119 | 0.18 |
ZNF362 |
zinc finger protein 362 |
1485 |
0.38 |
| chr3_63917329_63917607 | 0.18 |
ATXN7 |
ataxin 7 |
19193 |
0.16 |
| chr16_79313034_79313388 | 0.18 |
ENSG00000222244 |
. |
14860 |
0.29 |
| chr5_53902252_53902403 | 0.18 |
SNX18 |
sorting nexin 18 |
88734 |
0.09 |
| chr8_22581370_22581593 | 0.18 |
ENSG00000253125 |
. |
23535 |
0.13 |
| chr1_40557010_40557161 | 0.18 |
PPT1 |
palmitoyl-protein thioesterase 1 |
678 |
0.7 |
| chr7_37354921_37355641 | 0.18 |
ELMO1 |
engulfment and cell motility 1 |
27086 |
0.19 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0007042 | lysosomal lumen acidification(GO:0007042) regulation of lysosomal lumen pH(GO:0035751) |
| 0.0 | 0.2 | GO:0001820 | serotonin secretion(GO:0001820) |
| 0.0 | 0.2 | GO:0043116 | negative regulation of vascular permeability(GO:0043116) |
| 0.0 | 0.1 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.1 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.1 | GO:0034392 | negative regulation of smooth muscle cell apoptotic process(GO:0034392) |
| 0.0 | 0.1 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.1 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.0 | 0.1 | GO:1902745 | positive regulation of lamellipodium assembly(GO:0010592) positive regulation of lamellipodium organization(GO:1902745) |
| 0.0 | 0.1 | GO:0005997 | xylulose metabolic process(GO:0005997) |
| 0.0 | 0.1 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.0 | 0.1 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 0.2 | GO:0030853 | negative regulation of granulocyte differentiation(GO:0030853) |
| 0.0 | 0.2 | GO:0016264 | gap junction assembly(GO:0016264) |
| 0.0 | 0.2 | GO:0002710 | negative regulation of T cell mediated immunity(GO:0002710) |
| 0.0 | 0.2 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.1 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.0 | 0.1 | GO:0051938 | L-glutamate import(GO:0051938) |
| 0.0 | 0.0 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.0 | 0.0 | GO:0071803 | regulation of podosome assembly(GO:0071801) positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.1 | GO:0009138 | pyrimidine nucleoside diphosphate metabolic process(GO:0009138) |
| 0.0 | 0.2 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.2 | GO:0030851 | granulocyte differentiation(GO:0030851) |
| 0.0 | 0.1 | GO:0045906 | negative regulation of vasoconstriction(GO:0045906) |
| 0.0 | 0.0 | GO:0002313 | mature B cell differentiation involved in immune response(GO:0002313) |
| 0.0 | 0.1 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.0 | 0.1 | GO:0060754 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 0.2 | GO:0000085 | mitotic G2 phase(GO:0000085) G2 phase(GO:0051319) |
| 0.0 | 0.0 | GO:0006154 | adenosine catabolic process(GO:0006154) |
| 0.0 | 0.1 | GO:0006924 | activation-induced cell death of T cells(GO:0006924) |
| 0.0 | 0.1 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 | 0.1 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.1 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 | 0.1 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.0 | 0.1 | GO:0006498 | N-terminal protein lipidation(GO:0006498) |
| 0.0 | 0.1 | GO:0009301 | snRNA transcription(GO:0009301) |
| 0.0 | 0.0 | GO:2000794 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000794) |
| 0.0 | 0.0 | GO:0072007 | mesangial cell differentiation(GO:0072007) kidney interstitial fibroblast differentiation(GO:0072071) renal interstitial fibroblast development(GO:0072141) mesangial cell development(GO:0072143) pericyte cell differentiation(GO:1904238) |
| 0.0 | 0.2 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.0 | 0.1 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.0 | GO:0046940 | nucleoside monophosphate phosphorylation(GO:0046940) |
| 0.0 | 0.1 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.0 | 0.0 | GO:2000188 | regulation of cholesterol homeostasis(GO:2000188) positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.1 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.1 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.0 | 0.1 | GO:0055091 | phospholipid homeostasis(GO:0055091) |
| 0.0 | 0.1 | GO:0051569 | regulation of histone H3-K4 methylation(GO:0051569) |
| 0.0 | 0.0 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 0.1 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.0 | 0.0 | GO:0010511 | regulation of phosphatidylinositol biosynthetic process(GO:0010511) |
| 0.0 | 0.1 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.0 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.1 | GO:0019987 | obsolete negative regulation of anti-apoptosis(GO:0019987) |
| 0.0 | 0.1 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.1 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) regulation of store-operated calcium channel activity(GO:1901339) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.1 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) |
| 0.0 | 0.2 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 0.0 | GO:1901534 | positive regulation of megakaryocyte differentiation(GO:0045654) positive regulation of hematopoietic progenitor cell differentiation(GO:1901534) |
| 0.0 | 0.0 | GO:0010831 | positive regulation of myotube differentiation(GO:0010831) |
| 0.0 | 0.0 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
| 0.0 | 0.0 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.0 | 0.0 | GO:0002238 | response to molecule of fungal origin(GO:0002238) |
| 0.0 | 0.0 | GO:0051971 | positive regulation of transmission of nerve impulse(GO:0051971) |
| 0.0 | 0.0 | GO:0043301 | negative regulation of myeloid leukocyte mediated immunity(GO:0002887) negative regulation of leukocyte degranulation(GO:0043301) |
| 0.0 | 0.1 | GO:0008089 | anterograde axonal transport(GO:0008089) |
| 0.0 | 0.1 | GO:0042832 | response to protozoan(GO:0001562) defense response to protozoan(GO:0042832) |
| 0.0 | 0.0 | GO:0072010 | renal filtration cell differentiation(GO:0061318) glomerular epithelium development(GO:0072010) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.0 | 0.0 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.0 | 0.0 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.0 | 0.0 | GO:0032898 | neurotrophin production(GO:0032898) |
| 0.0 | 0.2 | GO:0032011 | ARF protein signal transduction(GO:0032011) |
| 0.0 | 0.1 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.0 | 0.2 | GO:0034199 | activation of protein kinase A activity(GO:0034199) |
| 0.0 | 0.0 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.0 | 0.1 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.0 | 0.1 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 0.0 | GO:0002606 | positive regulation of dendritic cell antigen processing and presentation(GO:0002606) |
| 0.0 | 0.0 | GO:0060437 | lung growth(GO:0060437) |
| 0.0 | 0.0 | GO:0032661 | interleukin-18 production(GO:0032621) regulation of interleukin-18 production(GO:0032661) |
| 0.0 | 0.0 | GO:0052805 | histidine catabolic process(GO:0006548) imidazole-containing compound catabolic process(GO:0052805) |
| 0.0 | 0.0 | GO:0002295 | T-helper cell lineage commitment(GO:0002295) alpha-beta T cell lineage commitment(GO:0002363) CD4-positive, alpha-beta T cell lineage commitment(GO:0043373) memory T cell differentiation(GO:0043379) regulation of memory T cell differentiation(GO:0043380) positive regulation of memory T cell differentiation(GO:0043382) T-helper 17 type immune response(GO:0072538) T-helper 17 cell differentiation(GO:0072539) T-helper 17 cell lineage commitment(GO:0072540) regulation of T-helper 17 type immune response(GO:2000316) positive regulation of T-helper 17 type immune response(GO:2000318) regulation of T-helper 17 cell differentiation(GO:2000319) positive regulation of T-helper 17 cell differentiation(GO:2000321) regulation of T-helper 17 cell lineage commitment(GO:2000328) positive regulation of T-helper 17 cell lineage commitment(GO:2000330) |
| 0.0 | 0.0 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.1 | GO:0033632 | regulation of cell-cell adhesion mediated by integrin(GO:0033632) |
| 0.0 | 0.1 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.1 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.0 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.0 | 0.0 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 | 0.1 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0044462 | external encapsulating structure part(GO:0044462) |
| 0.0 | 0.0 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.2 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.2 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.1 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.1 | GO:0000306 | extrinsic component of vacuolar membrane(GO:0000306) extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.1 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.1 | GO:0036454 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.0 | 0.1 | GO:0070188 | obsolete Stn1-Ten1 complex(GO:0070188) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.1 | GO:0060200 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.0 | 0.1 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.0 | 0.0 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.1 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.1 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.0 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.0 | 0.0 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.1 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 0.3 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.0 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.0 | GO:0000800 | lateral element(GO:0000800) |
| 0.0 | 0.1 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.2 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.0 | GO:0032009 | early phagosome(GO:0032009) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0015065 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.0 | 0.1 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.1 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.1 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.1 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.1 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.0 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) |
| 0.0 | 0.1 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.1 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.1 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.0 | 0.1 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.1 | GO:0004476 | mannose-6-phosphate isomerase activity(GO:0004476) |
| 0.0 | 0.1 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.0 | 0.1 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.1 | GO:0098847 | single-stranded telomeric DNA binding(GO:0043047) sequence-specific single stranded DNA binding(GO:0098847) |
| 0.0 | 0.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.1 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.1 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.0 | 0.1 | GO:0016670 | oxidoreductase activity, acting on a sulfur group of donors, oxygen as acceptor(GO:0016670) |
| 0.0 | 0.1 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.1 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.1 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.1 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.3 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.0 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.1 | GO:0004308 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) |
| 0.0 | 0.1 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.0 | 0.1 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.2 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.0 | GO:0070643 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.0 | 0.0 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.0 | 0.1 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.0 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.1 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.0 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.0 | 0.1 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 0.0 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.0 | 0.1 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.0 | 0.2 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 0.0 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.1 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.0 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.0 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.1 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.1 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.1 | GO:0015651 | quaternary ammonium group transmembrane transporter activity(GO:0015651) |
| 0.0 | 0.1 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) inositol trisphosphate phosphatase activity(GO:0046030) |
| 0.0 | 0.1 | GO:0016530 | metallochaperone activity(GO:0016530) |
| 0.0 | 0.0 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.0 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.1 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.2 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.0 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.1 | GO:0001637 | G-protein coupled chemoattractant receptor activity(GO:0001637) chemokine receptor activity(GO:0004950) |
| 0.0 | 0.1 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.0 | 0.1 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.4 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.2 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.1 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.0 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 0.3 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.2 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 0.3 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.3 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.0 | 0.1 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.1 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.0 | REACTOME ACYL CHAIN REMODELLING OF PC | Genes involved in Acyl chain remodelling of PC |
| 0.0 | 0.2 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.2 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.1 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.3 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.1 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.2 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.0 | REACTOME SIGNALING BY WNT | Genes involved in Signaling by Wnt |
| 0.0 | 0.1 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.1 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 0.2 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.1 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |