Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
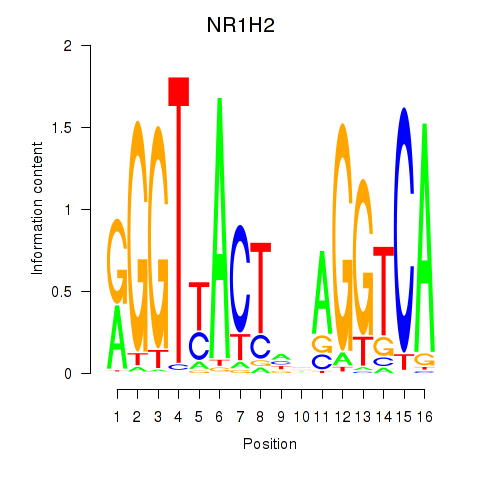
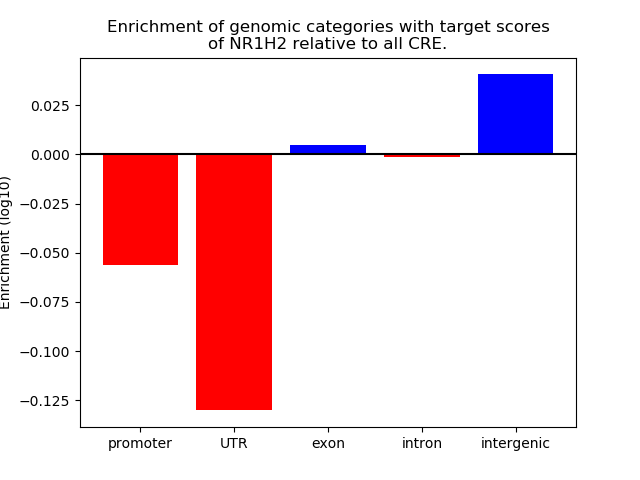
Results for NR1H2
Z-value: 0.73
Transcription factors associated with NR1H2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR1H2
|
ENSG00000131408.9 | nuclear receptor subfamily 1 group H member 2 |
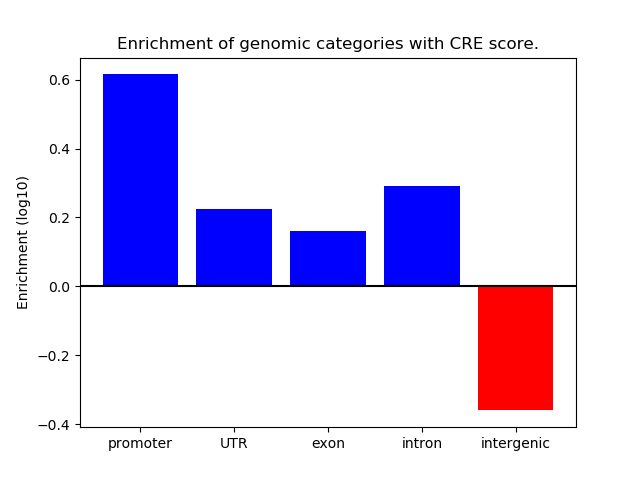
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr19_50881294_50881445 | NR1H2 | 678 | 0.476218 | 0.67 | 4.7e-02 | Click! |
| chr19_50834277_50834563 | NR1H2 | 1471 | 0.220966 | -0.59 | 9.5e-02 | Click! |
| chr19_50834588_50834739 | NR1H2 | 1714 | 0.193850 | -0.45 | 2.3e-01 | Click! |
| chr19_50880647_50880977 | NR1H2 | 121 | 0.914974 | -0.37 | 3.3e-01 | Click! |
| chr19_50879366_50879639 | NR1H2 | 213 | 0.855874 | -0.30 | 4.2e-01 | Click! |
Activity of the NR1H2 motif across conditions
Conditions sorted by the z-value of the NR1H2 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
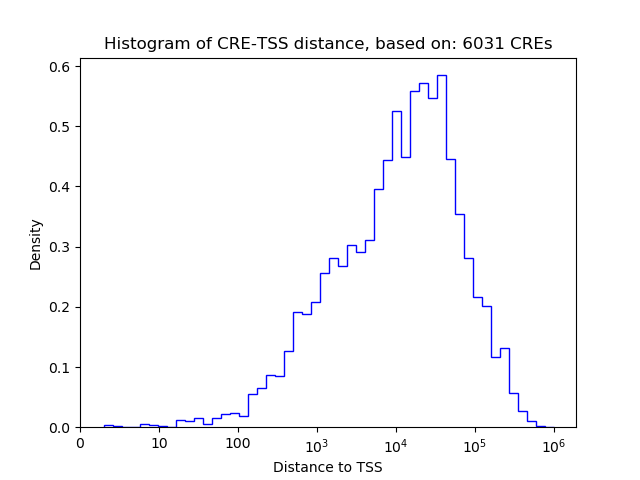
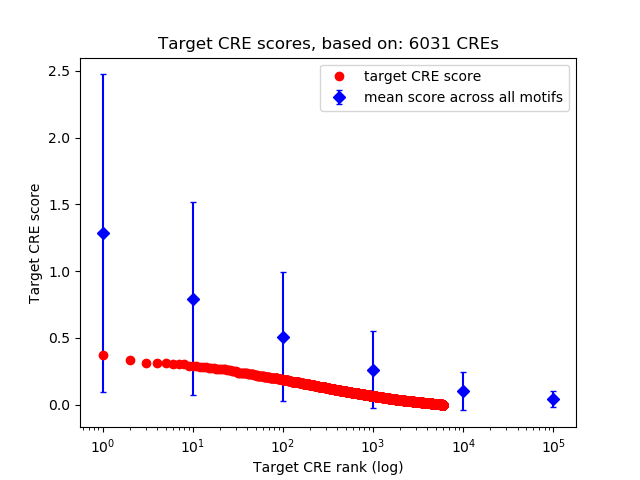
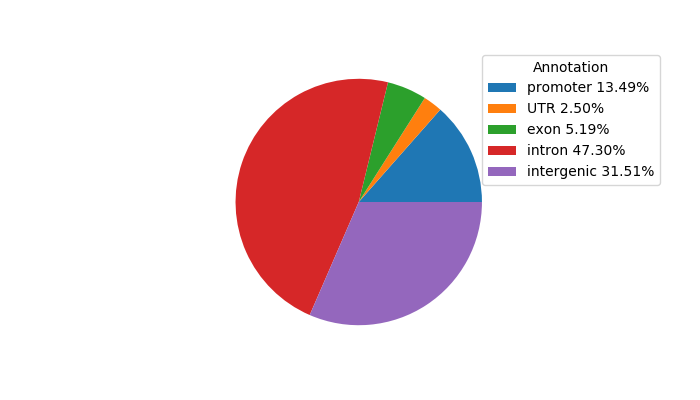
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr14_69411531_69411698 | 0.37 |
ACTN1 |
actinin, alpha 1 |
2651 |
0.33 |
| chr12_52585063_52585443 | 0.34 |
KRT80 |
keratin 80 |
531 |
0.69 |
| chr6_44018695_44019099 | 0.31 |
RP5-1120P11.1 |
|
23492 |
0.15 |
| chr20_16536753_16536975 | 0.31 |
KIF16B |
kinesin family member 16B |
17214 |
0.26 |
| chr14_69421781_69422763 | 0.31 |
ACTN1 |
actinin, alpha 1 |
8007 |
0.23 |
| chr20_30284083_30284439 | 0.31 |
RP11-243J16.7 |
|
10242 |
0.13 |
| chr14_92970056_92970406 | 0.30 |
RIN3 |
Ras and Rab interactor 3 |
9887 |
0.27 |
| chr9_107673636_107673787 | 0.30 |
RP11-217B7.2 |
|
16123 |
0.21 |
| chr3_69813399_69813761 | 0.29 |
MITF |
microphthalmia-associated transcription factor |
618 |
0.78 |
| chr10_77189776_77190205 | 0.29 |
RP11-399K21.10 |
|
1356 |
0.46 |
| chr3_183016185_183016744 | 0.29 |
B3GNT5 |
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5 |
33332 |
0.16 |
| chr1_183009603_183009956 | 0.28 |
LAMC1 |
laminin, gamma 1 (formerly LAMB2) |
17184 |
0.19 |
| chr22_46402410_46402690 | 0.28 |
WNT7B |
wingless-type MMTV integration site family, member 7B |
29541 |
0.1 |
| chr3_67698671_67698976 | 0.28 |
SUCLG2 |
succinate-CoA ligase, GDP-forming, beta subunit |
6208 |
0.34 |
| chr19_45428791_45429158 | 0.28 |
APOC1P1 |
apolipoprotein C-I pseudogene 1 |
1259 |
0.23 |
| chr17_75125652_75125803 | 0.28 |
SEC14L1 |
SEC14-like 1 (S. cerevisiae) |
1772 |
0.37 |
| chr17_42173180_42173449 | 0.27 |
HDAC5 |
histone deacetylase 5 |
2759 |
0.14 |
| chr9_4086696_4086970 | 0.27 |
RP11-70J12.1 |
|
14949 |
0.26 |
| chr15_57902216_57902367 | 0.27 |
MYZAP |
myocardial zonula adherens protein |
10682 |
0.2 |
| chr1_165827591_165827945 | 0.27 |
UCK2 |
uridine-cytidine kinase 2 |
30709 |
0.15 |
| chr19_6233179_6233473 | 0.27 |
CTC-503J8.4 |
|
25842 |
0.12 |
| chr16_81572532_81572683 | 0.26 |
CMIP |
c-Maf inducing protein |
43653 |
0.19 |
| chr1_68218506_68218657 | 0.26 |
ENSG00000238778 |
. |
19755 |
0.2 |
| chr13_80509662_80509813 | 0.26 |
SPRY2 |
sprouty homolog 2 (Drosophila) |
404057 |
0.01 |
| chr3_127415621_127415936 | 0.26 |
ABTB1 |
ankyrin repeat and BTB (POZ) domain containing 1 |
22825 |
0.15 |
| chr19_49004091_49004498 | 0.26 |
LMTK3 |
lemur tyrosine kinase 3 |
10767 |
0.1 |
| chr3_193590030_193590181 | 0.26 |
ENSG00000243991 |
. |
131595 |
0.05 |
| chr18_57223536_57223687 | 0.25 |
RP11-760L24.1 |
|
7330 |
0.26 |
| chr22_25979320_25979515 | 0.25 |
CTA-407F11.7 |
|
2068 |
0.28 |
| chr7_97982386_97982537 | 0.25 |
RP11-307C18.1 |
|
30296 |
0.16 |
| chr11_12307843_12308146 | 0.25 |
MICALCL |
MICAL C-terminal like |
453 |
0.82 |
| chr3_11178042_11178193 | 0.24 |
HRH1 |
histamine receptor H1 |
662 |
0.81 |
| chr12_81231723_81232151 | 0.24 |
ENSG00000207763 |
. |
5529 |
0.25 |
| chr1_162680021_162680172 | 0.24 |
DDR2 |
discoidin domain receptor tyrosine kinase 2 |
56931 |
0.11 |
| chr2_54556871_54557022 | 0.24 |
C2orf73 |
chromosome 2 open reading frame 73 |
225 |
0.96 |
| chr3_149356541_149356692 | 0.24 |
WWTR1-IT1 |
WWTR1 intronic transcript 1 (non-protein coding) |
11355 |
0.17 |
| chr12_13252440_13252619 | 0.24 |
GSG1 |
germ cell associated 1 |
3789 |
0.25 |
| chr8_144032439_144032590 | 0.23 |
CYP11B2 |
cytochrome P450, family 11, subfamily B, polypeptide 2 |
33255 |
0.13 |
| chr20_35110278_35110568 | 0.23 |
DLGAP4 |
discs, large (Drosophila) homolog-associated protein 4 |
20273 |
0.17 |
| chr5_138607548_138607819 | 0.23 |
MATR3 |
matrin 3 |
1758 |
0.23 |
| chr14_75080124_75080452 | 0.23 |
LTBP2 |
latent transforming growth factor beta binding protein 2 |
982 |
0.57 |
| chr17_47099426_47099577 | 0.23 |
RP11-501C14.6 |
|
8414 |
0.1 |
| chr6_123112103_123112308 | 0.23 |
SMPDL3A |
sphingomyelin phosphodiesterase, acid-like 3A |
1734 |
0.38 |
| chr15_41138763_41139692 | 0.23 |
SPINT1 |
serine peptidase inhibitor, Kunitz type 1 |
2354 |
0.15 |
| chr6_16367315_16367466 | 0.23 |
ENSG00000265642 |
. |
61364 |
0.14 |
| chr1_235242449_235242943 | 0.23 |
ENSG00000207181 |
. |
48556 |
0.13 |
| chr21_16594402_16594858 | 0.23 |
NRIP1 |
nuclear receptor interacting protein 1 |
157309 |
0.04 |
| chr12_58808774_58808925 | 0.22 |
RP11-362K2.2 |
Protein LOC100506869 |
129058 |
0.06 |
| chr9_125138658_125139005 | 0.22 |
PTGS1 |
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
1230 |
0.35 |
| chr3_149738658_149738809 | 0.22 |
PFN2 |
profilin 2 |
1865 |
0.34 |
| chr4_184560268_184560547 | 0.22 |
ENSG00000252157 |
. |
14422 |
0.14 |
| chr5_148352238_148352777 | 0.22 |
RP11-44B19.1 |
|
2604 |
0.3 |
| chr10_6763311_6763462 | 0.22 |
PRKCQ |
protein kinase C, theta |
141123 |
0.05 |
| chr4_7888432_7888664 | 0.22 |
AFAP1 |
actin filament associated protein 1 |
14508 |
0.22 |
| chr1_15610345_15610496 | 0.22 |
FHAD1 |
forkhead-associated (FHA) phosphopeptide binding domain 1 |
21838 |
0.17 |
| chr18_21296906_21297057 | 0.22 |
LAMA3 |
laminin, alpha 3 |
27333 |
0.2 |
| chr2_36565453_36565604 | 0.22 |
CRIM1 |
cysteine rich transmembrane BMP regulator 1 (chordin-like) |
17541 |
0.29 |
| chr12_50452001_50452282 | 0.21 |
ASIC1 |
acid-sensing (proton-gated) ion channel 1 |
654 |
0.59 |
| chr21_38711029_38711180 | 0.21 |
AP001437.1 |
|
27130 |
0.16 |
| chr8_38858707_38858858 | 0.21 |
ADAM9 |
ADAM metallopeptidase domain 9 |
4277 |
0.16 |
| chr3_149254018_149254169 | 0.21 |
ENSG00000252581 |
. |
10675 |
0.2 |
| chr8_146015607_146015758 | 0.21 |
RPL8 |
ribosomal protein L8 |
1856 |
0.19 |
| chr2_107618112_107618263 | 0.21 |
ST6GAL2 |
ST6 beta-galactosamide alpha-2,6-sialyltranferase 2 |
114623 |
0.07 |
| chr8_131687867_131688018 | 0.21 |
RP11-737F9.2 |
|
217089 |
0.02 |
| chr1_84719853_84720004 | 0.21 |
ENSG00000199959 |
. |
23076 |
0.21 |
| chr17_26793910_26794378 | 0.21 |
SLC13A2 |
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 |
6167 |
0.11 |
| chr8_129381988_129382255 | 0.21 |
ENSG00000201782 |
. |
149371 |
0.04 |
| chr11_67781609_67782080 | 0.21 |
ALDH3B1 |
aldehyde dehydrogenase 3 family, member B1 |
4054 |
0.13 |
| chr2_147743058_147743209 | 0.21 |
ENSG00000238860 |
. |
338406 |
0.01 |
| chr10_101946913_101947666 | 0.21 |
ERLIN1 |
ER lipid raft associated 1 |
802 |
0.51 |
| chr6_149491238_149491389 | 0.20 |
ENSG00000263481 |
. |
6617 |
0.24 |
| chr2_216529775_216530114 | 0.20 |
ENSG00000212055 |
. |
213698 |
0.02 |
| chr12_89328128_89328279 | 0.20 |
ENSG00000238302 |
. |
347859 |
0.01 |
| chr1_16466515_16466919 | 0.20 |
RP11-276H7.2 |
|
14989 |
0.11 |
| chr19_56618769_56618997 | 0.20 |
ENSG00000223060 |
. |
2098 |
0.22 |
| chr5_159107_159571 | 0.20 |
ENSG00000199540 |
. |
15027 |
0.14 |
| chr8_142244739_142244890 | 0.20 |
SLC45A4 |
solute carrier family 45, member 4 |
4367 |
0.18 |
| chr2_28078222_28078373 | 0.20 |
RBKS |
ribokinase |
12269 |
0.19 |
| chr13_52157550_52158046 | 0.20 |
WDFY2 |
WD repeat and FYVE domain containing 2 |
846 |
0.49 |
| chr1_39583247_39583398 | 0.20 |
ENSG00000206654 |
. |
3393 |
0.22 |
| chr11_10373135_10373286 | 0.20 |
AMPD3 |
adenosine monophosphate deaminase 3 |
33992 |
0.13 |
| chr5_154071675_154071826 | 0.20 |
ENSG00000221552 |
. |
6414 |
0.17 |
| chr11_75269832_75269983 | 0.20 |
SERPINH1 |
serpin peptidase inhibitor, clade H (heat shock protein 47), member 1, (collagen binding protein 1) |
3194 |
0.18 |
| chr22_50356250_50357150 | 0.20 |
PIM3 |
pim-3 oncogene |
2539 |
0.27 |
| chr2_234119413_234119564 | 0.20 |
ATG16L1 |
autophagy related 16-like 1 (S. cerevisiae) |
791 |
0.58 |
| chr13_45013936_45014201 | 0.20 |
TSC22D1 |
TSC22 domain family, member 1 |
3087 |
0.36 |
| chr10_102124571_102124984 | 0.20 |
SCD |
stearoyl-CoA desaturase (delta-9-desaturase) |
17896 |
0.13 |
| chr6_37023589_37023822 | 0.20 |
COX6A1P2 |
cytochrome c oxidase subunit VIa polypeptide 1 pseudogene 2 |
11098 |
0.2 |
| chr1_24434630_24434891 | 0.20 |
MYOM3 |
myomesin 3 |
3905 |
0.2 |
| chr1_9731771_9732062 | 0.19 |
RP11-558F24.4 |
|
15697 |
0.15 |
| chr2_62881644_62881884 | 0.19 |
AC092155.4 |
|
7999 |
0.23 |
| chr15_57615898_57616159 | 0.19 |
TCF12 |
transcription factor 12 |
41375 |
0.15 |
| chr17_43577703_43578072 | 0.19 |
RP11-798G7.5 |
|
2739 |
0.16 |
| chr10_14117012_14117163 | 0.19 |
RP11-397C18.2 |
|
804 |
0.73 |
| chr2_227199398_227199549 | 0.19 |
ENSG00000263363 |
. |
324036 |
0.01 |
| chr8_139856330_139856481 | 0.19 |
COL22A1 |
collagen, type XXII, alpha 1 |
64600 |
0.15 |
| chr7_40961790_40961941 | 0.19 |
AC005160.3 |
|
146708 |
0.05 |
| chr11_35058169_35058529 | 0.19 |
PDHX |
pyruvate dehydrogenase complex, component X |
59018 |
0.11 |
| chr10_128950431_128950582 | 0.19 |
FAM196A |
family with sequence similarity 196, member A |
24767 |
0.25 |
| chr6_108930285_108930625 | 0.19 |
FOXO3 |
forkhead box O3 |
47094 |
0.18 |
| chr10_96996414_96996565 | 0.19 |
RP11-310E22.4 |
|
5588 |
0.25 |
| chr1_167743848_167743999 | 0.19 |
MPZL1 |
myelin protein zero-like 1 |
9116 |
0.23 |
| chr10_80732581_80733347 | 0.19 |
ZMIZ1-AS1 |
ZMIZ1 antisense RNA 1 |
10813 |
0.3 |
| chr20_61469350_61469501 | 0.19 |
COL9A3 |
collagen, type IX, alpha 3 |
20262 |
0.11 |
| chr3_42077345_42077717 | 0.19 |
TRAK1 |
trafficking protein, kinesin binding 1 |
55031 |
0.14 |
| chr17_76677287_76677438 | 0.18 |
ENSG00000252391 |
. |
15517 |
0.19 |
| chr22_41000981_41001132 | 0.18 |
MKL1 |
megakaryoblastic leukemia (translocation) 1 |
31633 |
0.17 |
| chr14_76287931_76288578 | 0.18 |
RP11-270M14.4 |
|
15300 |
0.24 |
| chr19_16175455_16175640 | 0.18 |
TPM4 |
tropomyosin 4 |
2284 |
0.28 |
| chr19_10728070_10728416 | 0.18 |
SLC44A2 |
solute carrier family 44 (choline transporter), member 2 |
7691 |
0.11 |
| chr20_19983328_19983514 | 0.18 |
NAA20 |
N(alpha)-acetyltransferase 20, NatB catalytic subunit |
14339 |
0.2 |
| chr4_119260934_119261085 | 0.18 |
PRSS12 |
protease, serine, 12 (neurotrypsin, motopsin) |
13149 |
0.27 |
| chr6_13302624_13303087 | 0.18 |
RP1-257A7.4 |
|
7037 |
0.2 |
| chr6_43094189_43094340 | 0.18 |
PTK7 |
protein tyrosine kinase 7 |
4065 |
0.15 |
| chr2_69261565_69261716 | 0.18 |
ANTXR1 |
anthrax toxin receptor 1 |
21111 |
0.19 |
| chr6_52217103_52217564 | 0.18 |
PAQR8 |
progestin and adipoQ receptor family member VIII |
8886 |
0.23 |
| chr12_83113374_83113525 | 0.18 |
TMTC2 |
transmembrane and tetratricopeptide repeat containing 2 |
32669 |
0.25 |
| chr17_1391880_1392228 | 0.18 |
MYO1C |
myosin IC |
1700 |
0.27 |
| chr6_151990514_151990665 | 0.18 |
ESR1 |
estrogen receptor 1 |
21042 |
0.22 |
| chr6_7145383_7146135 | 0.18 |
RREB1 |
ras responsive element binding protein 1 |
6281 |
0.22 |
| chr10_114723539_114723817 | 0.18 |
RP11-57H14.2 |
|
12044 |
0.22 |
| chr7_92364918_92365200 | 0.18 |
ENSG00000206763 |
. |
33931 |
0.19 |
| chr1_92317205_92317356 | 0.18 |
TGFBR3 |
transforming growth factor, beta receptor III |
9870 |
0.22 |
| chr22_18373590_18373741 | 0.18 |
MICAL3 |
microtubule associated monooxygenase, calponin and LIM domain containing 3 |
4389 |
0.26 |
| chr11_122655696_122655847 | 0.17 |
CRTAM |
cytotoxic and regulatory T cell molecule |
53437 |
0.13 |
| chr4_185018477_185018810 | 0.17 |
STOX2 |
storkhead box 2 |
96165 |
0.07 |
| chr2_70823742_70823936 | 0.17 |
TGFA |
transforming growth factor, alpha |
42692 |
0.15 |
| chr20_13848549_13848700 | 0.17 |
NDUFAF5 |
NADH dehydrogenase (ubiquinone) complex I, assembly factor 5 |
82928 |
0.08 |
| chr8_142158040_142158331 | 0.17 |
DENND3 |
DENN/MADD domain containing 3 |
6883 |
0.21 |
| chr10_35595892_35596043 | 0.17 |
RP11-324I22.4 |
|
13959 |
0.18 |
| chr1_2321868_2322077 | 0.17 |
MORN1 |
MORN repeat containing 1 |
537 |
0.55 |
| chr1_178207539_178207690 | 0.17 |
RASAL2 |
RAS protein activator like 2 |
102992 |
0.08 |
| chr17_48502151_48502302 | 0.17 |
ACSF2 |
acyl-CoA synthetase family member 2 |
1293 |
0.29 |
| chr8_22604778_22604955 | 0.17 |
ENSG00000253125 |
. |
150 |
0.96 |
| chr1_26021848_26022016 | 0.17 |
MAN1C1 |
mannosidase, alpha, class 1C, member 1 |
14161 |
0.17 |
| chr9_114737008_114737159 | 0.17 |
ENSG00000222356 |
. |
21936 |
0.17 |
| chr19_3374038_3374830 | 0.17 |
NFIC |
nuclear factor I/C (CCAAT-binding transcription factor) |
7850 |
0.19 |
| chr3_66491690_66492240 | 0.17 |
LRIG1 |
leucine-rich repeats and immunoglobulin-like domains 1 |
59391 |
0.15 |
| chr20_47961516_47961667 | 0.17 |
ENSG00000212304 |
. |
64371 |
0.08 |
| chr22_32865711_32865862 | 0.17 |
FBXO7 |
F-box protein 7 |
4877 |
0.19 |
| chr7_20259929_20260378 | 0.17 |
MACC1 |
metastasis associated in colon cancer 1 |
3126 |
0.32 |
| chr14_24866543_24866694 | 0.17 |
NYNRIN |
NYN domain and retroviral integrase containing |
1374 |
0.25 |
| chr9_120482996_120483203 | 0.17 |
ENSG00000251847 |
. |
10650 |
0.2 |
| chr8_122369795_122369946 | 0.17 |
ENSG00000221644 |
. |
170738 |
0.03 |
| chr2_237796357_237796508 | 0.17 |
ENSG00000202341 |
. |
127516 |
0.05 |
| chr1_209381983_209382134 | 0.17 |
ENSG00000230937 |
. |
223420 |
0.02 |
| chrX_148577496_148577647 | 0.17 |
IDS |
iduronate 2-sulfatase |
5687 |
0.2 |
| chr19_15425157_15425308 | 0.17 |
BRD4 |
bromodomain containing 4 |
17485 |
0.16 |
| chr18_3649805_3649956 | 0.17 |
RP11-874J12.4 |
|
3530 |
0.24 |
| chr1_17871900_17872051 | 0.17 |
ARHGEF10L |
Rho guanine nucleotide exchange factor (GEF) 10-like |
5645 |
0.27 |
| chr13_94847942_94848231 | 0.17 |
GPC6-AS1 |
GPC6 antisense RNA 1 |
7841 |
0.32 |
| chr8_29449879_29450030 | 0.17 |
RP4-676L2.1 |
|
239267 |
0.02 |
| chr4_140871909_140872439 | 0.17 |
MAML3 |
mastermind-like 3 (Drosophila) |
60053 |
0.14 |
| chr2_43309506_43309703 | 0.16 |
ENSG00000207087 |
. |
9028 |
0.3 |
| chr3_184561796_184561947 | 0.16 |
VPS8 |
vacuolar protein sorting 8 homolog (S. cerevisiae) |
9408 |
0.29 |
| chr19_33686317_33686519 | 0.16 |
LRP3 |
low density lipoprotein receptor-related protein 3 |
928 |
0.46 |
| chr4_759415_759686 | 0.16 |
AC139887.4 |
|
688 |
0.55 |
| chr4_95481204_95481355 | 0.16 |
PDLIM5 |
PDZ and LIM domain 5 |
36403 |
0.23 |
| chr6_138084967_138085118 | 0.16 |
ENSG00000207300 |
. |
20469 |
0.21 |
| chr19_13957292_13957443 | 0.16 |
ENSG00000207980 |
. |
9894 |
0.08 |
| chr1_10447470_10447848 | 0.16 |
ENSG00000265945 |
. |
2400 |
0.2 |
| chr12_107256430_107256581 | 0.16 |
RP11-144F15.1 |
Uncharacterized protein |
87809 |
0.07 |
| chr2_56968245_56968396 | 0.16 |
ENSG00000238690 |
. |
274979 |
0.02 |
| chr16_67124269_67124420 | 0.16 |
C16orf70 |
chromosome 16 open reading frame 70 |
19517 |
0.1 |
| chr2_47371206_47371357 | 0.16 |
C2orf61 |
chromosome 2 open reading frame 61 |
7153 |
0.22 |
| chr21_46826449_46826600 | 0.16 |
COL18A1 |
collagen, type XVIII, alpha 1 |
1472 |
0.34 |
| chr16_72882320_72882471 | 0.16 |
ENSG00000251868 |
. |
26504 |
0.17 |
| chr1_233248528_233248728 | 0.16 |
PCNXL2 |
pecanex-like 2 (Drosophila) |
47425 |
0.19 |
| chr11_514795_514946 | 0.16 |
RNH1 |
ribonuclease/angiogenin inhibitor 1 |
7570 |
0.08 |
| chr1_94188465_94188866 | 0.16 |
BCAR3 |
breast cancer anti-estrogen resistance 3 |
41280 |
0.16 |
| chr10_79637661_79637812 | 0.16 |
AL391421.1 |
Uncharacterized protein; cDNA FLJ43696 fis, clone TBAES2007964 |
11103 |
0.18 |
| chr3_127538975_127539236 | 0.16 |
MGLL |
monoglyceride lipase |
1881 |
0.43 |
| chr3_159505457_159505608 | 0.16 |
IQCJ-SCHIP1-AS1 |
IQCJ-SCHIP1 readthrough antisense RNA 1 |
19131 |
0.15 |
| chr16_23540221_23540372 | 0.16 |
GGA2 |
golgi-associated, gamma adaptin ear containing, ARF binding protein 2 |
6980 |
0.14 |
| chr1_61916105_61916328 | 0.16 |
NFIA |
nuclear factor I/A |
43872 |
0.2 |
| chr13_97951105_97951256 | 0.16 |
MBNL2 |
muscleblind-like splicing regulator 2 |
22722 |
0.24 |
| chr19_41727336_41727487 | 0.16 |
CTD-2195B23.3 |
|
545 |
0.64 |
| chr20_32653471_32653622 | 0.16 |
RP1-64K7.4 |
|
15409 |
0.16 |
| chr12_71040955_71041106 | 0.16 |
PTPRB |
protein tyrosine phosphatase, receptor type, B |
9810 |
0.28 |
| chr6_11092979_11093838 | 0.16 |
SMIM13 |
small integral membrane protein 13 |
858 |
0.55 |
| chr5_53750809_53750960 | 0.16 |
HSPB3 |
heat shock 27kDa protein 3 |
561 |
0.84 |
| chr15_68682519_68683065 | 0.16 |
ITGA11 |
integrin, alpha 11 |
41700 |
0.18 |
| chr12_89549700_89550075 | 0.15 |
ENSG00000238302 |
. |
126175 |
0.06 |
| chr2_178119623_178120326 | 0.15 |
ENSG00000265396 |
. |
764 |
0.5 |
| chr4_74587786_74587937 | 0.15 |
IL8 |
interleukin 8 |
18362 |
0.22 |
| chr3_142572644_142572856 | 0.15 |
PCOLCE2 |
procollagen C-endopeptidase enhancer 2 |
34991 |
0.15 |
| chr1_85786366_85786652 | 0.15 |
ENSG00000264380 |
. |
36216 |
0.12 |
| chr3_12219225_12219376 | 0.15 |
TIMP4 |
TIMP metallopeptidase inhibitor 4 |
18449 |
0.25 |
| chr16_53133154_53133539 | 0.15 |
CHD9 |
chromodomain helicase DNA binding protein 9 |
274 |
0.94 |
| chr2_72162199_72162350 | 0.15 |
CYP26B1 |
cytochrome P450, family 26, subfamily B, polypeptide 1 |
207939 |
0.03 |
| chr17_38668560_38668716 | 0.15 |
TNS4 |
tensin 4 |
10789 |
0.16 |
| chr17_62174426_62174577 | 0.15 |
ERN1 |
endoplasmic reticulum to nucleus signaling 1 |
32984 |
0.13 |
| chr17_80343135_80343286 | 0.15 |
UTS2R |
urotensin 2 receptor |
11057 |
0.09 |
| chr1_94719949_94720119 | 0.15 |
ARHGAP29 |
Rho GTPase activating protein 29 |
16845 |
0.24 |
| chr10_30101231_30101626 | 0.15 |
SVIL |
supervillin |
76695 |
0.11 |
| chr17_1621771_1622017 | 0.15 |
WDR81 |
WD repeat domain 81 |
1999 |
0.18 |
| chr17_32999536_32999687 | 0.15 |
TMEM132E |
transmembrane protein 132E |
91843 |
0.08 |
| chr5_139641185_139641634 | 0.15 |
PFDN1 |
prefoldin subunit 1 |
41259 |
0.1 |
| chr15_38691031_38691182 | 0.15 |
FAM98B |
family with sequence similarity 98, member B |
55222 |
0.14 |
| chr4_102160089_102160359 | 0.15 |
ENSG00000221265 |
. |
91347 |
0.09 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.0 | 0.1 | GO:0071637 | monocyte chemotactic protein-1 production(GO:0071605) regulation of monocyte chemotactic protein-1 production(GO:0071637) |
| 0.0 | 0.1 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.1 | GO:0009169 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.0 | 0.3 | GO:0009629 | response to gravity(GO:0009629) |
| 0.0 | 0.1 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.0 | 0.1 | GO:1902745 | positive regulation of lamellipodium assembly(GO:0010592) positive regulation of lamellipodium organization(GO:1902745) |
| 0.0 | 0.1 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.0 | 0.0 | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.0 | 0.1 | GO:0061364 | apoptotic process involved in luteolysis(GO:0061364) |
| 0.0 | 0.2 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.1 | GO:0071731 | response to nitric oxide(GO:0071731) |
| 0.0 | 0.1 | GO:0051176 | positive regulation of sulfur metabolic process(GO:0051176) |
| 0.0 | 0.1 | GO:0044415 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.0 | 0.1 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.0 | 0.1 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.0 | 0.0 | GO:0032261 | purine nucleotide salvage(GO:0032261) |
| 0.0 | 0.0 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.0 | 0.1 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.0 | 0.1 | GO:0018101 | protein citrullination(GO:0018101) |
| 0.0 | 0.1 | GO:0001547 | antral ovarian follicle growth(GO:0001547) |
| 0.0 | 0.1 | GO:0010700 | negative regulation of norepinephrine secretion(GO:0010700) |
| 0.0 | 0.1 | GO:0002634 | regulation of germinal center formation(GO:0002634) |
| 0.0 | 0.1 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.0 | 0.0 | GO:0048319 | axial mesoderm morphogenesis(GO:0048319) |
| 0.0 | 0.0 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.0 | 0.0 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.1 | GO:0007028 | cytoplasm organization(GO:0007028) |
| 0.0 | 0.2 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.1 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.1 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.2 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.0 | 0.0 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 0.1 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.0 | GO:0055089 | fatty acid homeostasis(GO:0055089) |
| 0.0 | 0.1 | GO:0009649 | entrainment of circadian clock(GO:0009649) |
| 0.0 | 0.1 | GO:0050665 | hydrogen peroxide biosynthetic process(GO:0050665) |
| 0.0 | 0.1 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.0 | GO:0045989 | positive regulation of striated muscle contraction(GO:0045989) |
| 0.0 | 0.1 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.0 | GO:0045112 | integrin biosynthetic process(GO:0045112) |
| 0.0 | 0.0 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) |
| 0.0 | 0.1 | GO:0051549 | regulation of keratinocyte migration(GO:0051547) positive regulation of keratinocyte migration(GO:0051549) |
| 0.0 | 0.0 | GO:0060129 | thyroid-stimulating hormone-secreting cell differentiation(GO:0060129) |
| 0.0 | 0.1 | GO:0046504 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.0 | GO:0010591 | regulation of lamellipodium assembly(GO:0010591) |
| 0.0 | 0.0 | GO:0045077 | negative regulation of interferon-gamma biosynthetic process(GO:0045077) |
| 0.0 | 0.0 | GO:0071436 | sodium ion export(GO:0071436) |
| 0.0 | 0.1 | GO:0010510 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.0 | 0.0 | GO:0035581 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.0 | GO:0003077 | obsolete negative regulation of diuresis(GO:0003077) |
| 0.0 | 0.1 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.0 | GO:0032641 | lymphotoxin A production(GO:0032641) lymphotoxin A biosynthetic process(GO:0042109) |
| 0.0 | 0.0 | GO:1903332 | regulation of protein folding(GO:1903332) |
| 0.0 | 0.0 | GO:0030011 | maintenance of cell polarity(GO:0030011) |
| 0.0 | 0.0 | GO:0014819 | regulation of skeletal muscle contraction(GO:0014819) |
| 0.0 | 0.0 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.0 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.0 | 0.0 | GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0043619) |
| 0.0 | 0.1 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.0 | 0.1 | GO:0060710 | chorio-allantoic fusion(GO:0060710) |
| 0.0 | 0.0 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.0 | 0.0 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.0 | GO:0018243 | protein O-linked glycosylation via serine(GO:0018242) protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.1 | GO:0006000 | fructose metabolic process(GO:0006000) |
| 0.0 | 0.0 | GO:0035511 | oxidative DNA demethylation(GO:0035511) |
| 0.0 | 0.1 | GO:0008655 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine nucleoside salvage(GO:0043097) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.1 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.1 | GO:0043260 | laminin-11 complex(GO:0043260) |
| 0.0 | 0.1 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.1 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.1 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.1 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.0 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.0 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.1 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.0 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.1 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.1 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.0 | GO:0005588 | collagen type V trimer(GO:0005588) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.1 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.1 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 0.1 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.0 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.1 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) |
| 0.0 | 0.1 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.2 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.1 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.0 | 0.2 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.1 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.1 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.1 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.1 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.1 | GO:0042285 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.0 | GO:0016215 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.0 | 0.1 | GO:0004661 | protein geranylgeranyltransferase activity(GO:0004661) |
| 0.0 | 0.0 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.1 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.0 | GO:0042808 | obsolete neuronal Cdc2-like kinase binding(GO:0042808) |
| 0.0 | 0.1 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.1 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.0 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.1 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.0 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.1 | GO:0090482 | vitamin transmembrane transporter activity(GO:0090482) |
| 0.0 | 0.0 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.0 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.0 | GO:1901567 | icosanoid binding(GO:0050542) fatty acid derivative binding(GO:1901567) |
| 0.0 | 0.0 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.0 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.0 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.1 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.0 | 0.0 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.0 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.0 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.1 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.0 | 0.0 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.0 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.0 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.0 | 0.0 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.0 | GO:0035514 | DNA demethylase activity(GO:0035514) DNA-N1-methyladenine dioxygenase activity(GO:0043734) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.2 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.1 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.4 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.0 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 0.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.0 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.1 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.1 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |