Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
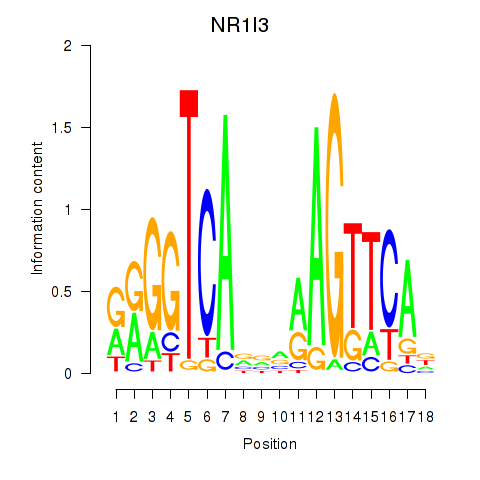
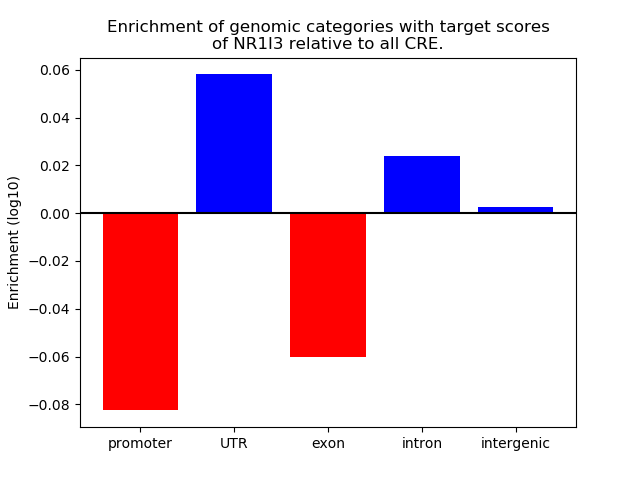
Results for NR1I3
Z-value: 0.70
Transcription factors associated with NR1I3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR1I3
|
ENSG00000143257.7 | nuclear receptor subfamily 1 group I member 3 |
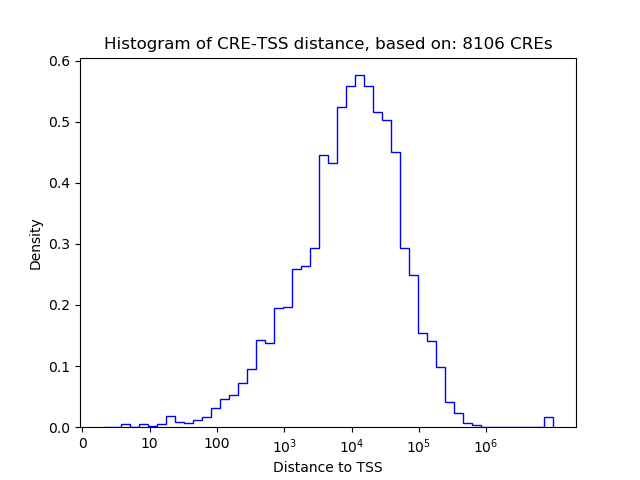
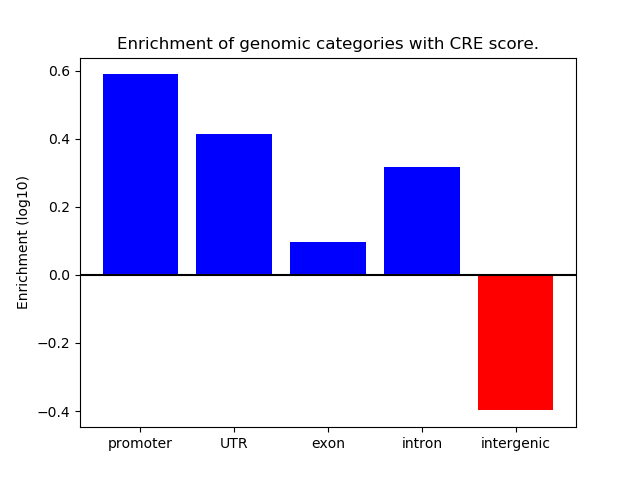
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr1_161211637_161212023 | NR1I3 | 3738 | 0.094204 | 0.14 | 7.2e-01 | Click! |
Activity of the NR1I3 motif across conditions
Conditions sorted by the z-value of the NR1I3 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
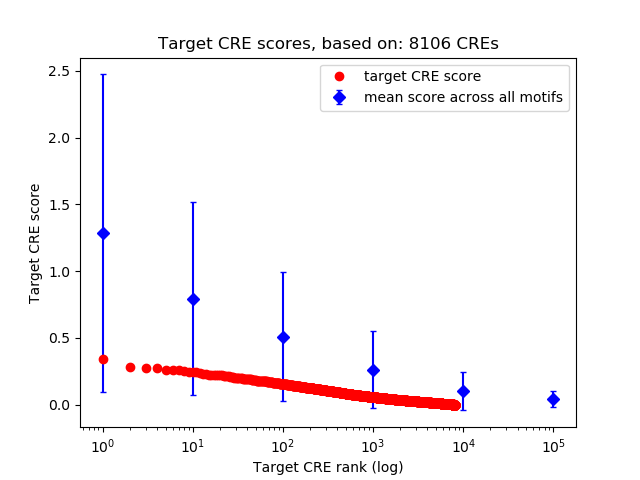
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr3_112178819_112179007 | 0.34 |
BTLA |
B and T lymphocyte associated |
39292 |
0.17 |
| chr5_67530096_67530377 | 0.28 |
PIK3R1 |
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
5490 |
0.28 |
| chr17_54679747_54680041 | 0.27 |
NOG |
noggin |
8834 |
0.3 |
| chr6_138137693_138137892 | 0.27 |
RP11-356I2.4 |
|
9239 |
0.22 |
| chr7_128777785_128778040 | 0.26 |
TSPAN33 |
tetraspanin 33 |
6800 |
0.16 |
| chr6_6593122_6593282 | 0.26 |
LY86 |
lymphocyte antigen 86 |
4300 |
0.27 |
| chr2_29233309_29233475 | 0.26 |
FAM179A |
family with sequence similarity 179, member A |
7735 |
0.16 |
| chr16_47515766_47515917 | 0.25 |
ITFG1 |
integrin alpha FG-GAP repeat containing 1 |
17781 |
0.15 |
| chr1_169662886_169663277 | 0.25 |
SELL |
selectin L |
17758 |
0.18 |
| chr7_24267550_24267736 | 0.24 |
AC004485.3 |
|
27695 |
0.18 |
| chr19_39819527_39819875 | 0.24 |
GMFG |
glia maturation factor, gamma |
6944 |
0.09 |
| chr1_65423839_65424016 | 0.24 |
JAK1 |
Janus kinase 1 |
8260 |
0.25 |
| chr2_45478028_45478179 | 0.23 |
SIX2 |
SIX homeobox 2 |
241534 |
0.02 |
| chr2_135049910_135050099 | 0.23 |
MGAT5 |
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase |
38174 |
0.2 |
| chr22_23482336_23482675 | 0.22 |
RTDR1 |
rhabdoid tumor deletion region gene 1 |
18 |
0.96 |
| chr10_134368978_134369207 | 0.22 |
INPP5A |
inositol polyphosphate-5-phosphatase, 40kDa |
17449 |
0.21 |
| chr8_8735555_8735713 | 0.22 |
MFHAS1 |
malignant fibrous histiocytoma amplified sequence 1 |
15521 |
0.19 |
| chr2_20094815_20094966 | 0.22 |
TTC32 |
tetratricopeptide repeat domain 32 |
6512 |
0.26 |
| chr1_46008468_46008619 | 0.22 |
AKR1A1 |
aldo-keto reductase family 1, member A1 (aldehyde reductase) |
7672 |
0.14 |
| chr16_27462069_27462270 | 0.22 |
IL21R-AS1 |
IL21R antisense RNA 1 |
2545 |
0.28 |
| chr19_5130688_5131043 | 0.22 |
CTC-482H14.5 |
|
47265 |
0.14 |
| chr10_4022036_4022300 | 0.22 |
KLF6 |
Kruppel-like factor 6 |
194695 |
0.03 |
| chr10_48460684_48460835 | 0.21 |
GDF10 |
growth differentiation factor 10 |
21783 |
0.18 |
| chr13_113806578_113806884 | 0.21 |
PROZ |
protein Z, vitamin K-dependent plasma glycoprotein |
6237 |
0.13 |
| chr2_16106193_16106413 | 0.21 |
ENSG00000243541 |
. |
15200 |
0.18 |
| chr20_2722250_2722401 | 0.21 |
EBF4 |
early B-cell factor 4 |
35438 |
0.09 |
| chr2_242881769_242881920 | 0.21 |
AC131097.3 |
|
5857 |
0.19 |
| chr15_60848077_60848282 | 0.20 |
CTD-2501E16.2 |
|
26007 |
0.16 |
| chr3_190414491_190414848 | 0.20 |
ENSG00000223117 |
. |
54717 |
0.15 |
| chr8_121762007_121762212 | 0.20 |
RP11-713M15.1 |
|
11384 |
0.25 |
| chr13_30041218_30041369 | 0.20 |
MTUS2-AS1 |
MTUS2 antisense RNA 1 |
20361 |
0.23 |
| chr3_15501836_15502208 | 0.20 |
EAF1-AS1 |
EAF1 antisense RNA 1 |
8913 |
0.12 |
| chr12_122027698_122027887 | 0.20 |
KDM2B |
lysine (K)-specific demethylase 2B |
8872 |
0.18 |
| chr9_130738343_130738494 | 0.20 |
FAM102A |
family with sequence similarity 102, member A |
4374 |
0.13 |
| chr10_7029346_7029497 | 0.20 |
PRKCQ |
protein kinase C, theta |
407158 |
0.01 |
| chr20_57723964_57724434 | 0.20 |
ZNF831 |
zinc finger protein 831 |
41876 |
0.16 |
| chr6_156955714_156956050 | 0.20 |
ARID1B |
AT rich interactive domain 1B (SWI1-like) |
143181 |
0.05 |
| chr2_175486588_175486745 | 0.19 |
WIPF1 |
WAS/WASL interacting protein family, member 1 |
12641 |
0.2 |
| chr2_58136289_58136440 | 0.19 |
VRK2 |
vaccinia related kinase 2 |
1578 |
0.56 |
| chr14_91502758_91502909 | 0.19 |
RPS6KA5 |
ribosomal protein S6 kinase, 90kDa, polypeptide 5 |
23645 |
0.17 |
| chrX_1600320_1600585 | 0.19 |
ASMTL |
acetylserotonin O-methyltransferase-like |
27797 |
0.16 |
| chr14_35303784_35304022 | 0.19 |
ENSG00000251726 |
. |
10663 |
0.17 |
| chrY_1550321_1550585 | 0.19 |
NA |
NA |
> 106 |
NA |
| chr2_197009997_197010148 | 0.19 |
RP11-347P5.1 |
|
5915 |
0.22 |
| chr1_27959886_27960101 | 0.19 |
FGR |
feline Gardner-Rasheed sarcoma viral oncogene homolog |
1734 |
0.27 |
| chr3_101832900_101833053 | 0.19 |
ZPLD1 |
zona pellucida-like domain containing 1 |
14888 |
0.29 |
| chr17_56456680_56457108 | 0.19 |
RNF43 |
ring finger protein 43 |
23188 |
0.11 |
| chr17_14096680_14097352 | 0.19 |
ENSG00000252305 |
. |
16568 |
0.19 |
| chr7_127054523_127055019 | 0.18 |
ZNF800 |
zinc finger protein 800 |
17207 |
0.24 |
| chr14_97586844_97587256 | 0.18 |
VRK1 |
vaccinia related kinase 1 |
244684 |
0.02 |
| chr2_69000085_69000318 | 0.18 |
ARHGAP25 |
Rho GTPase activating protein 25 |
1732 |
0.44 |
| chr2_38810640_38810791 | 0.18 |
HNRNPLL |
heterogeneous nuclear ribonucleoprotein L-like |
13737 |
0.19 |
| chr10_12631284_12631499 | 0.18 |
ENSG00000263584 |
. |
10639 |
0.27 |
| chr5_55884311_55884521 | 0.18 |
AC022431.2 |
Homo sapiens uncharacterized LOC101928448 (LOC101928448), mRNA. |
17643 |
0.22 |
| chr17_47809986_47810367 | 0.18 |
FAM117A |
family with sequence similarity 117, member A |
8287 |
0.15 |
| chr13_100071950_100072184 | 0.18 |
ENSG00000266207 |
. |
33656 |
0.15 |
| chr11_59535226_59535396 | 0.18 |
STX3 |
syntaxin 3 |
12386 |
0.14 |
| chr6_99938862_99939169 | 0.18 |
USP45 |
ubiquitin specific peptidase 45 |
8274 |
0.18 |
| chrX_1602588_1602855 | 0.18 |
ASMTL |
acetylserotonin O-methyltransferase-like |
30066 |
0.16 |
| chr19_50073795_50074168 | 0.18 |
ENSG00000207073 |
. |
4052 |
0.08 |
| chr15_29240397_29240548 | 0.18 |
APBA2 |
amyloid beta (A4) precursor protein-binding, family A, member 2 |
13035 |
0.23 |
| chr17_45297370_45297652 | 0.18 |
MYL4 |
myosin, light chain 4, alkali; atrial, embryonic |
115 |
0.84 |
| chrX_78416040_78416499 | 0.18 |
GPR174 |
G protein-coupled receptor 174 |
10200 |
0.32 |
| chr20_37505321_37505472 | 0.18 |
ENSG00000240474 |
. |
3983 |
0.24 |
| chr1_89595445_89595689 | 0.18 |
GBP2 |
guanylate binding protein 2, interferon-inducible |
3770 |
0.23 |
| chr4_148925524_148925723 | 0.17 |
ARHGAP10 |
Rho GTPase activating protein 10 |
122605 |
0.06 |
| chr3_44484489_44484640 | 0.17 |
ZNF445 |
zinc finger protein 445 |
34598 |
0.14 |
| chr6_111443474_111443841 | 0.17 |
ENSG00000200926 |
. |
31122 |
0.16 |
| chr3_196336038_196336189 | 0.17 |
LINC01063 |
long intergenic non-protein coding RNA 1063 |
23345 |
0.11 |
| chrX_78418820_78418971 | 0.17 |
GPR174 |
G protein-coupled receptor 174 |
7574 |
0.33 |
| chr11_13314702_13314853 | 0.17 |
ARNTL |
aryl hydrocarbon receptor nuclear translocator-like |
15345 |
0.25 |
| chr7_50364490_50364641 | 0.17 |
IKZF1 |
IKAROS family zinc finger 1 (Ikaros) |
2680 |
0.39 |
| chr14_22890396_22890547 | 0.17 |
ENSG00000251002 |
. |
11248 |
0.12 |
| chr1_160715023_160715174 | 0.17 |
SLAMF7 |
SLAM family member 7 |
6004 |
0.18 |
| chr4_113209820_113210146 | 0.17 |
TIFA |
TRAF-interacting protein with forkhead-associated domain |
2924 |
0.21 |
| chr1_168482288_168482558 | 0.17 |
XCL2 |
chemokine (C motif) ligand 2 |
30812 |
0.2 |
| chr3_23483889_23484071 | 0.17 |
ENSG00000265645 |
. |
38089 |
0.18 |
| chr1_110165621_110165808 | 0.17 |
AMPD2 |
adenosine monophosphate deaminase 2 |
722 |
0.46 |
| chr7_142941436_142941642 | 0.17 |
GSTK1 |
glutathione S-transferase kappa 1 |
353 |
0.79 |
| chr12_9144662_9144957 | 0.16 |
KLRG1 |
killer cell lectin-like receptor subfamily G, member 1 |
143 |
0.95 |
| chrY_1552539_1552862 | 0.16 |
NA |
NA |
> 106 |
NA |
| chr5_14983835_14984240 | 0.16 |
ENSG00000212036 |
. |
73288 |
0.11 |
| chr1_15483643_15483794 | 0.16 |
TMEM51 |
transmembrane protein 51 |
3433 |
0.27 |
| chr13_111188696_111188940 | 0.16 |
RAB20 |
RAB20, member RAS oncogene family |
25262 |
0.17 |
| chr7_106165541_106165692 | 0.16 |
CTB-111H14.1 |
|
20331 |
0.27 |
| chr18_72702111_72702262 | 0.16 |
ZADH2 |
zinc binding alcohol dehydrogenase domain containing 2 |
215282 |
0.02 |
| chr3_175075659_175075810 | 0.16 |
ENSG00000264974 |
. |
11595 |
0.25 |
| chr2_62430640_62430961 | 0.16 |
ENSG00000266097 |
. |
2161 |
0.29 |
| chr3_127348995_127349623 | 0.16 |
PODXL2 |
podocalyxin-like 2 |
1285 |
0.41 |
| chr2_85085633_85085887 | 0.16 |
TRABD2A |
TraB domain containing 2A |
22446 |
0.18 |
| chr13_30725272_30725600 | 0.16 |
ENSG00000266816 |
. |
55400 |
0.16 |
| chr17_56432340_56432491 | 0.16 |
SUPT4H1 |
suppressor of Ty 4 homolog 1 (S. cerevisiae) |
1961 |
0.22 |
| chr8_119292042_119292193 | 0.16 |
AC023590.1 |
Uncharacterized protein |
2364 |
0.46 |
| chr15_76141909_76142060 | 0.16 |
UBE2Q2 |
ubiquitin-conjugating enzyme E2Q family member 2 |
5599 |
0.16 |
| chr17_76779813_76780089 | 0.16 |
CYTH1 |
cytohesin 1 |
1572 |
0.38 |
| chr7_142001773_142001924 | 0.16 |
PRSS3P3 |
protease, serine, 3 pseudogene 3 |
12239 |
0.22 |
| chr12_9889248_9889774 | 0.16 |
CLECL1 |
C-type lectin-like 1 |
3616 |
0.19 |
| chr12_55355288_55355457 | 0.16 |
TESPA1 |
thymocyte expressed, positive selection associated 1 |
1109 |
0.58 |
| chr17_20156757_20156908 | 0.16 |
ENSG00000252971 |
. |
36738 |
0.14 |
| chr8_8713745_8714044 | 0.16 |
MFHAS1 |
malignant fibrous histiocytoma amplified sequence 1 |
37261 |
0.16 |
| chr19_14279308_14279579 | 0.16 |
LPHN1 |
latrophilin 1 |
7095 |
0.12 |
| chr6_37017673_37017845 | 0.16 |
COX6A1P2 |
cytochrome c oxidase subunit VIa polypeptide 1 pseudogene 2 |
5152 |
0.22 |
| chr22_40621973_40622124 | 0.16 |
TNRC6B |
trinucleotide repeat containing 6B |
38959 |
0.18 |
| chr9_95786581_95786807 | 0.15 |
FGD3 |
FYVE, RhoGEF and PH domain containing 3 |
9352 |
0.19 |
| chr2_16105641_16106067 | 0.15 |
ENSG00000243541 |
. |
14751 |
0.18 |
| chr8_59924469_59924620 | 0.15 |
RP11-328K2.1 |
|
19047 |
0.25 |
| chr10_123598584_123598735 | 0.15 |
ENSG00000201884 |
. |
7456 |
0.26 |
| chr4_143492069_143492229 | 0.15 |
INPP4B |
inositol polyphosphate-4-phosphatase, type II, 105kDa |
10327 |
0.33 |
| chr9_33147549_33147791 | 0.15 |
RP11-326F20.5 |
|
19303 |
0.14 |
| chr14_35699557_35699724 | 0.15 |
PSMA6 |
proteasome (prosome, macropain) subunit, alpha type, 6 |
48199 |
0.14 |
| chr2_207396183_207396430 | 0.15 |
ADAM23 |
ADAM metallopeptidase domain 23 |
63253 |
0.11 |
| chr9_130787624_130787958 | 0.15 |
RP11-379C10.1 |
|
31850 |
0.08 |
| chr5_35826936_35827087 | 0.15 |
CTD-2113L7.1 |
|
109 |
0.97 |
| chr11_128541166_128541342 | 0.15 |
RP11-744N12.3 |
|
15069 |
0.18 |
| chr11_95434433_95434612 | 0.15 |
FAM76B |
family with sequence similarity 76, member B |
85447 |
0.09 |
| chr14_90009937_90010173 | 0.15 |
FOXN3-AS2 |
FOXN3 antisense RNA 2 |
32505 |
0.14 |
| chr2_112190871_112191169 | 0.15 |
ENSG00000266139 |
. |
112352 |
0.07 |
| chr16_68116183_68116430 | 0.15 |
NFATC3 |
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 3 |
2941 |
0.15 |
| chr2_25556807_25556990 | 0.15 |
ENSG00000221445 |
. |
5308 |
0.23 |
| chr9_129159177_129159360 | 0.15 |
ENSG00000253079 |
. |
11421 |
0.2 |
| chr3_45735930_45736127 | 0.15 |
SACM1L |
SAC1 suppressor of actin mutations 1-like (yeast) |
5077 |
0.19 |
| chrX_39195070_39195348 | 0.15 |
ENSG00000207122 |
. |
254671 |
0.02 |
| chr1_50882030_50882181 | 0.15 |
DMRTA2 |
DMRT-like family A2 |
5199 |
0.26 |
| chr3_46411260_46411477 | 0.15 |
CCR5 |
chemokine (C-C motif) receptor 5 (gene/pseudogene) |
265 |
0.9 |
| chr1_25308637_25308788 | 0.15 |
RUNX3 |
runt-related transcription factor 3 |
17211 |
0.18 |
| chr11_47417148_47417745 | 0.14 |
RP11-750H9.5 |
|
255 |
0.83 |
| chr3_13456823_13457198 | 0.14 |
NUP210 |
nucleoporin 210kDa |
4799 |
0.27 |
| chr4_170745007_170745217 | 0.14 |
ENSG00000264713 |
. |
38681 |
0.18 |
| chr2_235055184_235055371 | 0.14 |
SPP2 |
secreted phosphoprotein 2, 24kDa |
95846 |
0.08 |
| chrX_65240625_65241043 | 0.14 |
ENSG00000207939 |
. |
2122 |
0.33 |
| chr10_11214521_11214672 | 0.14 |
RP3-323N1.2 |
|
1257 |
0.5 |
| chr17_37930387_37930595 | 0.14 |
IKZF3 |
IKAROS family zinc finger 3 (Aiolos) |
3987 |
0.16 |
| chr6_110711697_110711922 | 0.14 |
DDO |
D-aspartate oxidase |
24956 |
0.16 |
| chr2_135425836_135426057 | 0.14 |
TMEM163 |
transmembrane protein 163 |
50624 |
0.17 |
| chr3_66444911_66445177 | 0.14 |
ENSG00000241587 |
. |
70765 |
0.12 |
| chr11_128478831_128478982 | 0.14 |
ETS1 |
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
21453 |
0.21 |
| chr8_129665336_129665520 | 0.14 |
ENSG00000221351 |
. |
166612 |
0.04 |
| chr6_131605644_131605795 | 0.14 |
AKAP7 |
A kinase (PRKA) anchor protein 7 |
34149 |
0.23 |
| chr4_153007039_153007190 | 0.14 |
ENSG00000253077 |
. |
114349 |
0.06 |
| chr7_142197913_142198064 | 0.14 |
MTRNR2L6 |
MT-RNR2-like 6 |
176116 |
0.03 |
| chr2_242712064_242712447 | 0.14 |
GAL3ST2 |
galactose-3-O-sulfotransferase 2 |
3985 |
0.12 |
| chrX_16612049_16612225 | 0.14 |
ENSG00000265144 |
. |
33071 |
0.16 |
| chr1_186268373_186268524 | 0.14 |
PRG4 |
proteoglycan 4 |
3037 |
0.25 |
| chrX_131613764_131613952 | 0.14 |
MBNL3 |
muscleblind-like splicing regulator 3 |
9185 |
0.29 |
| chr12_122812012_122812295 | 0.14 |
CLIP1 |
CAP-GLY domain containing linker protein 1 |
36411 |
0.15 |
| chr17_33774331_33774488 | 0.14 |
SLFN13 |
schlafen family member 13 |
4 |
0.97 |
| chr2_203530973_203531252 | 0.14 |
FAM117B |
family with sequence similarity 117, member B |
31201 |
0.23 |
| chr11_117097868_117098019 | 0.14 |
PCSK7 |
proprotein convertase subtilisin/kexin type 7 |
3013 |
0.18 |
| chr20_62712580_62712905 | 0.14 |
OPRL1 |
opiate receptor-like 1 |
1193 |
0.24 |
| chr15_52022956_52023209 | 0.14 |
LYSMD2 |
LysM, putative peptidoglycan-binding, domain containing 2 |
7236 |
0.14 |
| chr12_89727726_89727877 | 0.14 |
DUSP6 |
dual specificity phosphatase 6 |
17147 |
0.24 |
| chr2_85050169_85050320 | 0.14 |
TRABD2A |
TraB domain containing 2A |
57962 |
0.11 |
| chr6_109357060_109357211 | 0.14 |
ENSG00000199944 |
. |
23682 |
0.16 |
| chr10_8466384_8466535 | 0.14 |
ENSG00000212505 |
. |
232335 |
0.02 |
| chr13_33514702_33514853 | 0.14 |
KL |
klotho |
75430 |
0.11 |
| chr3_186757183_186757348 | 0.14 |
ST6GAL1 |
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
4266 |
0.28 |
| chr7_101388868_101389019 | 0.14 |
CUX1 |
cut-like homeobox 1 |
70016 |
0.11 |
| chr17_39496485_39496762 | 0.14 |
KRT33A |
keratin 33A |
10441 |
0.08 |
| chr12_22691459_22691610 | 0.13 |
C2CD5 |
C2 calcium-dependent domain containing 5 |
5653 |
0.3 |
| chr1_121258953_121259201 | 0.13 |
FCGR1B |
Fc fragment of IgG, high affinity Ib, receptor (CD64) |
323140 |
0.01 |
| chr12_46629041_46629192 | 0.13 |
SLC38A1 |
solute carrier family 38, member 1 |
32368 |
0.23 |
| chr1_12149159_12149332 | 0.13 |
ENSG00000201135 |
. |
11204 |
0.15 |
| chr7_104850801_104851099 | 0.13 |
SRPK2 |
SRSF protein kinase 2 |
58512 |
0.12 |
| chr20_37460991_37461142 | 0.13 |
PPP1R16B |
protein phosphatase 1, regulatory subunit 16B |
26699 |
0.16 |
| chr2_12417779_12417930 | 0.13 |
ENSG00000264089 |
. |
78598 |
0.11 |
| chr14_91698344_91698868 | 0.13 |
CTD-2547L24.3 |
HCG1816139; Uncharacterized protein |
10497 |
0.16 |
| chr1_45162405_45162569 | 0.13 |
C1orf228 |
chromosome 1 open reading frame 228 |
6981 |
0.11 |
| chr11_65319407_65320254 | 0.13 |
LTBP3 |
latent transforming growth factor beta binding protein 3 |
1368 |
0.23 |
| chr15_50389614_50389935 | 0.13 |
ATP8B4 |
ATPase, class I, type 8B, member 4 |
16927 |
0.23 |
| chr1_144360487_144360771 | 0.13 |
PPIAL4B |
peptidylprolyl isomerase A (cyclophilin A)-like 4B |
3617 |
0.25 |
| chr3_131104918_131105242 | 0.13 |
NUDT16 |
nudix (nucleoside diphosphate linked moiety X)-type motif 16 |
4451 |
0.19 |
| chr5_49929906_49930306 | 0.13 |
PARP8 |
poly (ADP-ribose) polymerase family, member 8 |
31627 |
0.25 |
| chr6_112060261_112060435 | 0.13 |
FYN |
FYN oncogene related to SRC, FGR, YES |
19083 |
0.24 |
| chr9_130768286_130768549 | 0.13 |
FAM102A |
family with sequence similarity 102, member A |
25625 |
0.1 |
| chr19_49594900_49595051 | 0.13 |
SNRNP70 |
small nuclear ribonucleoprotein 70kDa (U1) |
5238 |
0.07 |
| chr20_57733456_57733718 | 0.13 |
ZNF831 |
zinc finger protein 831 |
32488 |
0.19 |
| chr11_102319316_102319512 | 0.13 |
TMEM123 |
transmembrane protein 123 |
769 |
0.6 |
| chr2_240139272_240139549 | 0.13 |
HDAC4 |
histone deacetylase 4 |
26658 |
0.16 |
| chr15_32653399_32653683 | 0.13 |
ENSG00000221444 |
. |
32132 |
0.1 |
| chr2_39604885_39605036 | 0.13 |
MAP4K3 |
mitogen-activated protein kinase kinase kinase kinase 3 |
1990 |
0.32 |
| chr9_138372034_138372554 | 0.13 |
PPP1R26-AS1 |
PPP1R26 antisense RNA 1 |
270 |
0.72 |
| chr2_2689850_2690051 | 0.13 |
ENSG00000263570 |
. |
331152 |
0.01 |
| chr1_192854593_192854744 | 0.13 |
ENSG00000223075 |
. |
9553 |
0.26 |
| chr16_89362946_89363099 | 0.13 |
AC137932.5 |
|
894 |
0.47 |
| chr5_139035453_139035988 | 0.13 |
CXXC5 |
CXXC finger protein 5 |
3420 |
0.27 |
| chr1_100929192_100929425 | 0.13 |
RP5-837M10.4 |
|
22245 |
0.2 |
| chrX_48913511_48913662 | 0.13 |
CCDC120 |
coiled-coil domain containing 120 |
2485 |
0.14 |
| chr4_40195311_40195826 | 0.13 |
RHOH |
ras homolog family member H |
285 |
0.92 |
| chr9_35639292_35639607 | 0.13 |
RP11-331F9.3 |
|
3062 |
0.1 |
| chr14_35595819_35596045 | 0.13 |
KIAA0391 |
KIAA0391 |
3967 |
0.23 |
| chr19_51221635_51221873 | 0.13 |
SHANK1 |
SH3 and multiple ankyrin repeat domains 1 |
953 |
0.4 |
| chr2_42523029_42523463 | 0.13 |
EML4 |
echinoderm microtubule associated protein like 4 |
5147 |
0.28 |
| chr14_99691611_99691762 | 0.13 |
AL109767.1 |
|
37599 |
0.16 |
| chr4_39030218_39030369 | 0.13 |
TMEM156 |
transmembrane protein 156 |
3748 |
0.25 |
| chr2_32133230_32133428 | 0.13 |
ENSG00000221326 |
. |
11689 |
0.21 |
| chr10_104338876_104339209 | 0.13 |
ENSG00000207029 |
. |
12239 |
0.15 |
| chr6_152043719_152044019 | 0.13 |
ESR1 |
estrogen receptor 1 |
32238 |
0.21 |
| chrY_7598458_7598609 | 0.13 |
RFTN1P1 |
raftlin, lipid raft linker 1 pseudogene 1 |
46215 |
0.21 |
| chr8_100549940_100550091 | 0.13 |
ENSG00000216069 |
. |
926 |
0.39 |
| chr11_30348563_30348857 | 0.13 |
ARL14EP |
ADP-ribosylation factor-like 14 effector protein |
873 |
0.73 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.0 | 0.1 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.0 | 0.1 | GO:1900121 | regulation of receptor binding(GO:1900120) negative regulation of receptor binding(GO:1900121) |
| 0.0 | 0.1 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) regulation of store-operated calcium channel activity(GO:1901339) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.1 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.1 | GO:0030186 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.0 | 0.1 | GO:0010867 | positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.0 | 0.1 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.0 | 0.1 | GO:0007343 | egg activation(GO:0007343) |
| 0.0 | 0.0 | GO:0002513 | tolerance induction to self antigen(GO:0002513) |
| 0.0 | 0.1 | GO:0032506 | cytokinetic process(GO:0032506) |
| 0.0 | 0.1 | GO:0090116 | DNA methylation on cytosine(GO:0032776) C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 0.1 | GO:0003077 | obsolete negative regulation of diuresis(GO:0003077) |
| 0.0 | 0.1 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.1 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 0.1 | GO:0045591 | positive regulation of regulatory T cell differentiation(GO:0045591) |
| 0.0 | 0.0 | GO:0045589 | regulation of regulatory T cell differentiation(GO:0045589) |
| 0.0 | 0.1 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.0 | 0.1 | GO:0061756 | leukocyte tethering or rolling(GO:0050901) leukocyte adhesion to vascular endothelial cell(GO:0061756) |
| 0.0 | 0.1 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.0 | 0.1 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.0 | 0.0 | GO:0015801 | aromatic amino acid transport(GO:0015801) |
| 0.0 | 0.0 | GO:0045341 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) |
| 0.0 | 0.0 | GO:0048289 | isotype switching to IgE isotypes(GO:0048289) regulation of isotype switching to IgE isotypes(GO:0048293) |
| 0.0 | 0.0 | GO:0051409 | response to nitrosative stress(GO:0051409) |
| 0.0 | 0.2 | GO:0019059 | obsolete initiation of viral infection(GO:0019059) |
| 0.0 | 0.1 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.0 | 0.1 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.0 | 0.1 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.0 | 0.1 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.1 | GO:0021898 | commitment of multipotent stem cells to neuronal lineage in forebrain(GO:0021898) |
| 0.0 | 0.0 | GO:0045425 | positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) |
| 0.0 | 0.1 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 | 0.1 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.0 | 0.0 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.1 | GO:0003056 | regulation of vascular smooth muscle contraction(GO:0003056) |
| 0.0 | 0.1 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.1 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.0 | GO:0009048 | dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 0.0 | GO:0002313 | mature B cell differentiation involved in immune response(GO:0002313) |
| 0.0 | 0.1 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) meiotic chromosome separation(GO:0051307) |
| 0.0 | 0.0 | GO:0035268 | protein mannosylation(GO:0035268) protein O-linked mannosylation(GO:0035269) mannosylation(GO:0097502) |
| 0.0 | 0.0 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.0 | 0.0 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 0.0 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.0 | 0.0 | GO:0052251 | induction by organism of defense response of other organism involved in symbiotic interaction(GO:0052251) |
| 0.0 | 0.1 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.0 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.0 | 0.0 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.0 | GO:0031442 | positive regulation of mRNA 3'-end processing(GO:0031442) positive regulation of mRNA processing(GO:0050685) positive regulation of mRNA metabolic process(GO:1903313) |
| 0.0 | 0.0 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.0 | 0.1 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.0 | 0.0 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.0 | 0.1 | GO:2000193 | regulation of icosanoid secretion(GO:0032303) positive regulation of icosanoid secretion(GO:0032305) regulation of prostaglandin secretion(GO:0032306) positive regulation of prostaglandin secretion(GO:0032308) prostaglandin secretion(GO:0032310) positive regulation of fatty acid transport(GO:2000193) |
| 0.0 | 0.1 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.0 | 0.0 | GO:0072600 | protein targeting to Golgi(GO:0000042) establishment of protein localization to Golgi(GO:0072600) |
| 0.0 | 0.1 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.0 | 0.0 | GO:0045046 | protein import into peroxisome membrane(GO:0045046) |
| 0.0 | 0.0 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.0 | 0.0 | GO:0032621 | interleukin-18 production(GO:0032621) regulation of interleukin-18 production(GO:0032661) |
| 0.0 | 0.0 | GO:0061299 | retina vasculature development in camera-type eye(GO:0061298) retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 | 0.0 | GO:0002829 | negative regulation of type 2 immune response(GO:0002829) |
| 0.0 | 0.3 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 0.1 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.2 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.1 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.1 | GO:0005954 | calcium- and calmodulin-dependent protein kinase complex(GO:0005954) |
| 0.0 | 0.1 | GO:0030288 | outer membrane-bounded periplasmic space(GO:0030288) periplasmic space(GO:0042597) |
| 0.0 | 0.2 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 0.1 | GO:0071778 | obsolete WINAC complex(GO:0071778) |
| 0.0 | 0.0 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.0 | GO:0044462 | external encapsulating structure part(GO:0044462) |
| 0.0 | 0.1 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.2 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.1 | GO:0000791 | euchromatin(GO:0000791) |
| 0.0 | 0.0 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.1 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.1 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.0 | GO:0030312 | external encapsulating structure(GO:0030312) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.3 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.1 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.1 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.1 | GO:0052813 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.1 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.1 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.0 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) inositol-1,3,4,5-tetrakisphosphate 5-phosphatase activity(GO:0052659) |
| 0.0 | 0.1 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.1 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.0 | 0.1 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.1 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.0 | 0.0 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.0 | 0.1 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 0.3 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.1 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.0 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.0 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.0 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.0 | 0.0 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.1 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.1 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.2 | GO:0008373 | sialyltransferase activity(GO:0008373) |
| 0.0 | 0.0 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.0 | 0.0 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.0 | 0.1 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.1 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.1 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.0 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.1 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.2 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.1 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 0.1 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.1 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.4 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.1 | PID ALK2 PATHWAY | ALK2 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.1 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.3 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.0 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.2 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.0 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 0.1 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.2 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.0 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.0 | REACTOME POST NMDA RECEPTOR ACTIVATION EVENTS | Genes involved in Post NMDA receptor activation events |
| 0.0 | 0.0 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.2 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.3 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.0 | 0.2 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.1 | REACTOME ABORTIVE ELONGATION OF HIV1 TRANSCRIPT IN THE ABSENCE OF TAT | Genes involved in Abortive elongation of HIV-1 transcript in the absence of Tat |
| 0.0 | 0.2 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |