Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
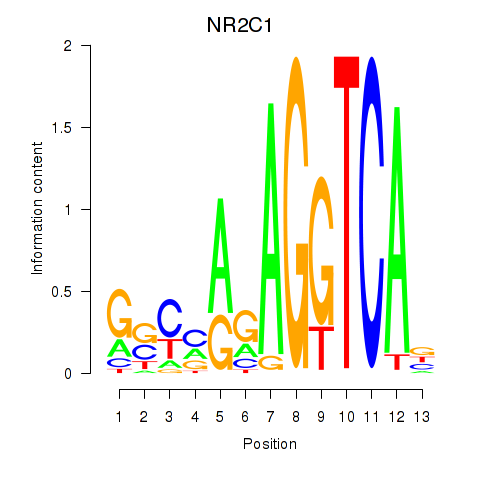
Results for NR2C1
Z-value: 1.18
Transcription factors associated with NR2C1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR2C1
|
ENSG00000120798.12 | nuclear receptor subfamily 2 group C member 1 |
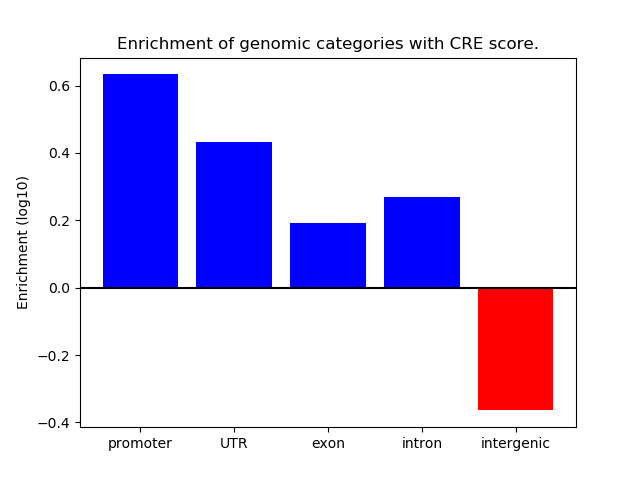
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr12_95434630_95434781 | NR2C1 | 331 | 0.906752 | -0.70 | 3.7e-02 | Click! |
| chr12_95465235_95465386 | NR2C1 | 1483 | 0.446330 | -0.69 | 3.9e-02 | Click! |
| chr12_95429746_95429897 | NR2C1 | 4553 | 0.251226 | -0.50 | 1.7e-01 | Click! |
| chr12_95462168_95462319 | NR2C1 | 4550 | 0.245510 | 0.42 | 2.6e-01 | Click! |
| chr12_95467454_95467605 | NR2C1 | 50 | 0.979550 | 0.37 | 3.3e-01 | Click! |
Activity of the NR2C1 motif across conditions
Conditions sorted by the z-value of the NR2C1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
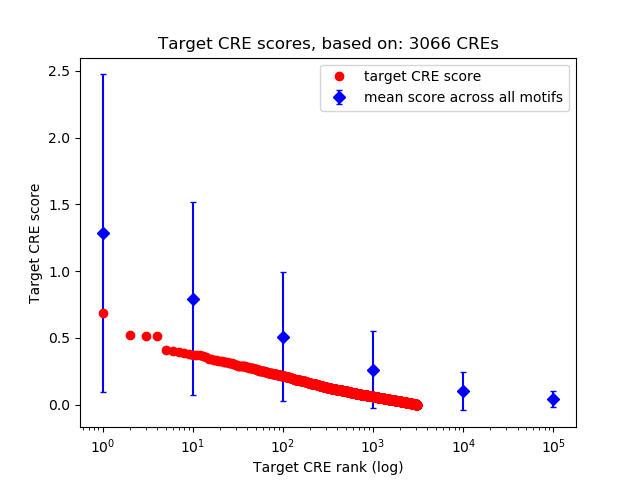
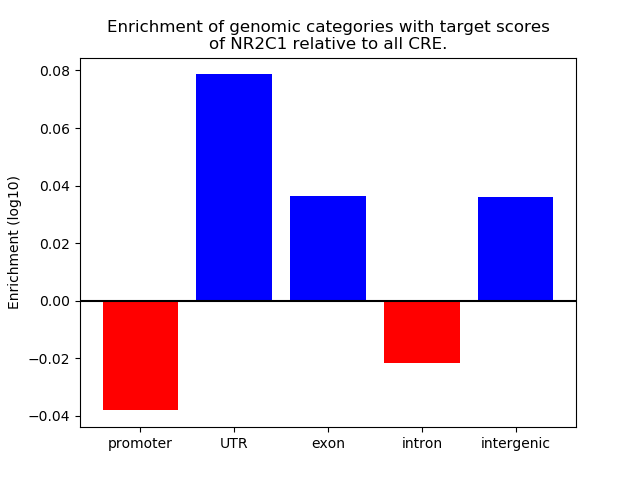
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr11_65257627_65258285 | 0.69 |
SCYL1 |
SCY1-like 1 (S. cerevisiae) |
34592 |
0.08 |
| chr9_34253979_34254272 | 0.52 |
ENSG00000222426 |
. |
28543 |
0.11 |
| chr1_94730682_94730833 | 0.52 |
ARHGAP29 |
Rho GTPase activating protein 29 |
27568 |
0.21 |
| chr13_114543410_114543586 | 0.51 |
GAS6 |
growth arrest-specific 6 |
4481 |
0.26 |
| chr18_56247923_56248074 | 0.41 |
ENSG00000252284 |
. |
19865 |
0.14 |
| chr15_74232933_74233272 | 0.40 |
LOXL1-AS1 |
LOXL1 antisense RNA 1 |
12513 |
0.14 |
| chr20_23812897_23813048 | 0.39 |
CST2 |
cystatin SA |
5604 |
0.22 |
| chr3_14514350_14514501 | 0.39 |
SLC6A6 |
solute carrier family 6 (neurotransmitter transporter), member 6 |
675 |
0.76 |
| chr1_94128945_94129096 | 0.38 |
BCAR3 |
breast cancer anti-estrogen resistance 3 |
17906 |
0.22 |
| chr7_101258153_101258304 | 0.38 |
MYL10 |
myosin, light chain 10, regulatory |
14348 |
0.27 |
| chr20_48755585_48755736 | 0.37 |
TMEM189 |
transmembrane protein 189 |
7991 |
0.17 |
| chr1_5998398_5998549 | 0.37 |
ENSG00000266687 |
. |
44489 |
0.12 |
| chr17_42621489_42621640 | 0.36 |
FZD2 |
frizzled family receptor 2 |
13361 |
0.17 |
| chr2_37969328_37969479 | 0.36 |
CDC42EP3 |
CDC42 effector protein (Rho GTPase binding) 3 |
3792 |
0.35 |
| chr18_10058872_10059075 | 0.35 |
ENSG00000263630 |
. |
53779 |
0.15 |
| chr1_115734054_115734205 | 0.34 |
RP4-663N10.1 |
|
91526 |
0.08 |
| chr2_43152776_43153425 | 0.34 |
HAAO |
3-hydroxyanthranilate 3,4-dioxygenase |
133368 |
0.05 |
| chr15_35998583_35998734 | 0.34 |
DPH6 |
diphthamine biosynthesis 6 |
160265 |
0.04 |
| chr5_134581986_134582263 | 0.33 |
C5orf66 |
chromosome 5 open reading frame 66 |
91505 |
0.08 |
| chr17_17642775_17643230 | 0.33 |
RAI1-AS1 |
RAI1 antisense RNA 1 |
31133 |
0.12 |
| chr15_80466917_80467209 | 0.32 |
FAH |
fumarylacetoacetate hydrolase (fumarylacetoacetase) |
18770 |
0.21 |
| chr11_113545454_113545605 | 0.32 |
TMPRSS5 |
transmembrane protease, serine 5 |
31495 |
0.18 |
| chr17_18138665_18138878 | 0.32 |
LLGL1 |
lethal giant larvae homolog 1 (Drosophila) |
9870 |
0.11 |
| chr3_194930783_194931056 | 0.32 |
ENSG00000206600 |
. |
4597 |
0.2 |
| chr1_246023169_246023412 | 0.31 |
RP11-83A16.1 |
|
173563 |
0.03 |
| chr11_10948038_10948189 | 0.31 |
ZBED5-AS1 |
ZBED5 antisense RNA 1 |
61305 |
0.11 |
| chr12_78412506_78412657 | 0.31 |
NAV3 |
neuron navigator 3 |
52525 |
0.17 |
| chr13_24001441_24001592 | 0.30 |
SACS |
spastic ataxia of Charlevoix-Saguenay (sacsin) |
6325 |
0.32 |
| chr9_129949574_129949725 | 0.30 |
RALGPS1 |
Ral GEF with PH domain and SH3 binding motif 1 |
23375 |
0.16 |
| chr8_42330645_42330848 | 0.30 |
SLC20A2 |
solute carrier family 20 (phosphate transporter), member 2 |
28015 |
0.16 |
| chrX_68357230_68357381 | 0.30 |
PJA1 |
praja ring finger 1, E3 ubiquitin protein ligase |
25232 |
0.27 |
| chr5_95193163_95193314 | 0.29 |
ENSG00000250362 |
. |
4252 |
0.16 |
| chr10_69915938_69916089 | 0.29 |
ENSG00000222371 |
. |
1823 |
0.38 |
| chr17_79107219_79107370 | 0.29 |
ENSG00000221025 |
. |
186 |
0.79 |
| chr7_48134998_48135206 | 0.29 |
UPP1 |
uridine phosphorylase 1 |
6129 |
0.24 |
| chr1_1476104_1476623 | 0.29 |
TMEM240 |
transmembrane protein 240 |
530 |
0.63 |
| chr20_36901417_36901587 | 0.29 |
KIAA1755 |
KIAA1755 |
12328 |
0.16 |
| chr20_36824784_36825086 | 0.29 |
TGM2 |
transglutaminase 2 |
29955 |
0.14 |
| chr2_20383859_20384010 | 0.29 |
ENSG00000200763 |
. |
8368 |
0.17 |
| chr16_84863200_84863351 | 0.28 |
RP11-254F19.2 |
|
1406 |
0.4 |
| chr1_223745292_223745443 | 0.28 |
CAPN8 |
calpain 8 |
20987 |
0.25 |
| chr2_235935219_235935383 | 0.28 |
SH3BP4 |
SH3-domain binding protein 4 |
31817 |
0.26 |
| chr11_35572250_35572401 | 0.28 |
PAMR1 |
peptidase domain containing associated with muscle regeneration 1 |
20477 |
0.22 |
| chr14_68989421_68989674 | 0.27 |
CTD-2325P2.4 |
|
105615 |
0.07 |
| chrY_18008517_18008676 | 0.27 |
ENSG00000252664 |
. |
166050 |
0.04 |
| chr5_133774585_133774736 | 0.27 |
CDKN2AIPNL |
CDKN2A interacting protein N-terminal like |
27071 |
0.14 |
| chr1_178620335_178620486 | 0.27 |
ENSG00000266417 |
. |
26474 |
0.17 |
| chr16_85319654_85319805 | 0.27 |
ENSG00000266307 |
. |
20202 |
0.23 |
| chr21_33828676_33828827 | 0.27 |
EVA1C |
eva-1 homolog C (C. elegans) |
28109 |
0.14 |
| chr1_183247214_183247365 | 0.27 |
NMNAT2 |
nicotinamide nucleotide adenylyltransferase 2 |
26720 |
0.22 |
| chr7_25110058_25110209 | 0.27 |
CYCS |
cytochrome c, somatic |
54735 |
0.14 |
| chr7_46525977_46526128 | 0.26 |
AC011294.3 |
Uncharacterized protein |
210668 |
0.03 |
| chr15_58836323_58836474 | 0.26 |
RP11-50C13.2 |
|
22888 |
0.16 |
| chr7_130576045_130576324 | 0.26 |
ENSG00000226380 |
. |
13886 |
0.26 |
| chr15_52386582_52387272 | 0.26 |
CTD-2184D3.5 |
|
5793 |
0.16 |
| chr16_66653317_66653503 | 0.25 |
CMTM3 |
CKLF-like MARVEL transmembrane domain containing 3 |
13986 |
0.11 |
| chr3_142226508_142226659 | 0.25 |
RP11-383G6.3 |
|
35404 |
0.13 |
| chr19_41829897_41830246 | 0.25 |
CCDC97 |
coiled-coil domain containing 97 |
4433 |
0.12 |
| chr17_48287279_48287648 | 0.25 |
COL1A1 |
collagen, type I, alpha 1 |
8470 |
0.12 |
| chr15_39283697_39283848 | 0.25 |
RP11-624L4.1 |
|
127142 |
0.06 |
| chr1_19217476_19217627 | 0.25 |
ALDH4A1 |
aldehyde dehydrogenase 4 family, member A1 |
166 |
0.94 |
| chr7_77827088_77827239 | 0.25 |
RP5-1185I7.1 |
|
207426 |
0.03 |
| chr19_583116_583267 | 0.25 |
HCN2 |
hyperpolarization activated cyclic nucleotide-gated potassium channel 2 |
6702 |
0.09 |
| chr19_1749097_1749397 | 0.25 |
ONECUT3 |
one cut homeobox 3 |
3125 |
0.18 |
| chr22_47331632_47331783 | 0.25 |
ENSG00000221672 |
. |
87904 |
0.09 |
| chr4_6345648_6345799 | 0.24 |
PPP2R2C |
protein phosphatase 2, regulatory subunit B, gamma |
37874 |
0.18 |
| chr22_50356250_50357150 | 0.24 |
PIM3 |
pim-3 oncogene |
2539 |
0.27 |
| chr7_105137285_105137436 | 0.24 |
PUS7 |
pseudouridylate synthase 7 homolog (S. cerevisiae) |
11222 |
0.21 |
| chr5_135389966_135390117 | 0.24 |
TGFBI |
transforming growth factor, beta-induced, 68kDa |
4497 |
0.23 |
| chr7_69680701_69680852 | 0.24 |
AUTS2 |
autism susceptibility candidate 2 |
513349 |
0.0 |
| chr3_161134127_161134278 | 0.24 |
SPTSSB |
serine palmitoyltransferase, small subunit B |
43534 |
0.19 |
| chr16_27252390_27252716 | 0.24 |
NSMCE1 |
non-SMC element 1 homolog (S. cerevisiae) |
8154 |
0.17 |
| chr14_56954824_56954975 | 0.24 |
TMEM260 |
transmembrane protein 260 |
173 |
0.97 |
| chr20_45754256_45754424 | 0.24 |
ENSG00000264901 |
. |
41269 |
0.18 |
| chr1_10958636_10958867 | 0.23 |
C1orf127 |
chromosome 1 open reading frame 127 |
49176 |
0.12 |
| chr1_116610144_116610295 | 0.23 |
MAB21L3 |
mab-21-like 3 (C. elegans) |
44157 |
0.15 |
| chr2_96347293_96347444 | 0.23 |
TRIM43 |
tripartite motif containing 43 |
89602 |
0.08 |
| chr3_58045428_58045645 | 0.23 |
FLNB |
filamin B, beta |
18520 |
0.24 |
| chr3_52229790_52230208 | 0.23 |
ALAS1 |
aminolevulinate, delta-, synthase 1 |
2103 |
0.21 |
| chr17_45955898_45956049 | 0.23 |
SP2 |
Sp2 transcription factor |
17543 |
0.1 |
| chr19_19746511_19746662 | 0.23 |
GMIP |
GEM interacting protein |
2410 |
0.18 |
| chr6_82649464_82649641 | 0.23 |
ENSG00000206886 |
. |
175811 |
0.03 |
| chr1_233750521_233750914 | 0.23 |
KCNK1 |
potassium channel, subfamily K, member 1 |
967 |
0.63 |
| chr8_37672483_37672634 | 0.23 |
GPR124 |
G protein-coupled receptor 124 |
17784 |
0.14 |
| chr2_40661252_40661403 | 0.23 |
SLC8A1 |
solute carrier family 8 (sodium/calcium exchanger), member 1 |
3883 |
0.37 |
| chr1_8087716_8087867 | 0.23 |
ERRFI1 |
ERBB receptor feedback inhibitor 1 |
1423 |
0.46 |
| chr2_217555244_217555395 | 0.23 |
AC007563.5 |
|
3868 |
0.2 |
| chr16_58154872_58155238 | 0.22 |
CTB-134F13.1 |
|
4266 |
0.14 |
| chr6_25359520_25359732 | 0.22 |
ENSG00000207490 |
. |
24201 |
0.16 |
| chr12_106476343_106476494 | 0.22 |
NUAK1 |
NUAK family, SNF1-like kinase, 1 |
1393 |
0.5 |
| chr4_10109523_10110061 | 0.22 |
ENSG00000223086 |
. |
7716 |
0.16 |
| chr6_33383896_33384047 | 0.22 |
PHF1 |
PHD finger protein 1 |
1129 |
0.29 |
| chr22_37960029_37960180 | 0.22 |
CDC42EP1 |
CDC42 effector protein (Rho GTPase binding) 1 |
309 |
0.83 |
| chr19_11313952_11314103 | 0.22 |
CTC-510F12.2 |
|
277 |
0.81 |
| chr17_45302121_45302272 | 0.22 |
ENSG00000252088 |
. |
4188 |
0.14 |
| chr1_31381768_31382084 | 0.22 |
SDC3 |
syndecan 3 |
318 |
0.87 |
| chr18_19863132_19863283 | 0.22 |
ENSG00000238907 |
. |
22668 |
0.2 |
| chr12_116820731_116820960 | 0.22 |
ENSG00000264037 |
. |
45278 |
0.17 |
| chr1_243664702_243664853 | 0.22 |
RP11-269F20.1 |
|
44057 |
0.19 |
| chr19_43356078_43356229 | 0.22 |
PSG10P |
pregnancy specific beta-1-glycoprotein 10, pseudogene |
3618 |
0.2 |
| chr19_19740386_19740663 | 0.22 |
LPAR2 |
lysophosphatidic acid receptor 2 |
785 |
0.49 |
| chr18_43901802_43901953 | 0.22 |
RNF165 |
ring finger protein 165 |
4895 |
0.31 |
| chr4_120655944_120656095 | 0.21 |
PDE5A |
phosphodiesterase 5A, cGMP-specific |
105873 |
0.07 |
| chr6_109561464_109561868 | 0.21 |
ENSG00000238474 |
. |
50895 |
0.12 |
| chr19_980614_980765 | 0.21 |
WDR18 |
WD repeat domain 18 |
3642 |
0.1 |
| chr5_157810587_157810765 | 0.21 |
ENSG00000222626 |
. |
406712 |
0.01 |
| chr17_42160007_42160158 | 0.21 |
G6PC3 |
glucose 6 phosphatase, catalytic, 3 |
7980 |
0.1 |
| chr7_75592437_75593112 | 0.21 |
POR |
P450 (cytochrome) oxidoreductase |
4387 |
0.19 |
| chr2_71940971_71941122 | 0.21 |
DYSF |
dysferlin |
247214 |
0.02 |
| chr19_36822233_36822591 | 0.21 |
ENSG00000222730 |
. |
1061 |
0.46 |
| chr19_48983392_48983543 | 0.21 |
CYTH2 |
cytohesin 2 |
7858 |
0.1 |
| chr20_37055054_37055242 | 0.21 |
ENSG00000225091 |
. |
937 |
0.29 |
| chr9_138944765_138944996 | 0.21 |
NACC2 |
NACC family member 2, BEN and BTB (POZ) domain containing |
2454 |
0.33 |
| chr18_68090011_68090162 | 0.21 |
ENSG00000264059 |
. |
60627 |
0.13 |
| chr13_111047343_111047494 | 0.20 |
ENSG00000238629 |
. |
19134 |
0.21 |
| chr1_53131017_53131168 | 0.20 |
FAM159A |
family with sequence similarity 159, member A |
32076 |
0.12 |
| chr17_48424304_48424608 | 0.20 |
XYLT2 |
xylosyltransferase II |
960 |
0.41 |
| chr3_101894275_101894426 | 0.20 |
ZPLD1 |
zona pellucida-like domain containing 1 |
76262 |
0.12 |
| chr17_44820432_44820772 | 0.20 |
NSF |
N-ethylmaleimide-sensitive factor |
16680 |
0.21 |
| chr12_52661903_52662054 | 0.20 |
KRT86 |
keratin 86 |
6481 |
0.1 |
| chr2_114081410_114081561 | 0.20 |
PAX8 |
paired box 8 |
44958 |
0.12 |
| chr3_141163153_141163392 | 0.20 |
ZBTB38 |
zinc finger and BTB domain containing 38 |
12707 |
0.22 |
| chr5_53812584_53812820 | 0.20 |
SNX18 |
sorting nexin 18 |
887 |
0.71 |
| chr19_18887031_18887461 | 0.20 |
COMP |
cartilage oligomeric matrix protein |
14868 |
0.14 |
| chr16_66957071_66957222 | 0.20 |
RRAD |
Ras-related associated with diabetes |
1709 |
0.21 |
| chr4_152339822_152339973 | 0.20 |
FAM160A1 |
family with sequence similarity 160, member A1 |
9393 |
0.27 |
| chr6_91085605_91085756 | 0.20 |
ENSG00000266760 |
. |
63219 |
0.12 |
| chr21_37583709_37583860 | 0.20 |
ENSG00000265882 |
. |
2352 |
0.25 |
| chr11_78507410_78507561 | 0.20 |
TENM4 |
teneurin transmembrane protein 4 |
8971 |
0.26 |
| chr18_32958619_32958770 | 0.20 |
ZNF396 |
zinc finger protein 396 |
1393 |
0.48 |
| chr16_30927719_30927870 | 0.19 |
FBXL19 |
F-box and leucine-rich repeat protein 19 |
6582 |
0.08 |
| chr17_1011220_1011906 | 0.19 |
ABR |
active BCR-related |
777 |
0.69 |
| chrX_153673360_153673855 | 0.19 |
FAM50A |
family with sequence similarity 50, member A |
800 |
0.36 |
| chr12_115272734_115272885 | 0.19 |
ENSG00000252459 |
. |
97300 |
0.08 |
| chr6_17968817_17968981 | 0.19 |
KIF13A |
kinesin family member 13A |
18795 |
0.26 |
| chr11_111427694_111427845 | 0.19 |
LAYN |
layilin |
15492 |
0.12 |
| chr3_14494059_14494243 | 0.19 |
SLC6A6 |
solute carrier family 6 (neurotransmitter transporter), member 6 |
19599 |
0.22 |
| chr1_109841299_109841450 | 0.19 |
MYBPHL |
myosin binding protein H-like |
8289 |
0.16 |
| chr1_112275803_112276182 | 0.19 |
FAM212B |
family with sequence similarity 212, member B |
5883 |
0.18 |
| chr15_75335507_75335751 | 0.19 |
PPCDC |
phosphopantothenoylcysteine decarboxylase |
13 |
0.97 |
| chr15_37124741_37124892 | 0.19 |
ENSG00000212511 |
. |
20025 |
0.26 |
| chr11_116861004_116861155 | 0.19 |
ENSG00000264344 |
. |
25166 |
0.14 |
| chr1_23720599_23720750 | 0.19 |
TCEA3 |
transcription elongation factor A (SII), 3 |
6762 |
0.14 |
| chr10_71234648_71234799 | 0.19 |
TSPAN15 |
tetraspanin 15 |
8901 |
0.22 |
| chr19_13231005_13231157 | 0.19 |
NACC1 |
nucleus accumbens associated 1, BEN and BTB (POZ) domain containing |
1930 |
0.18 |
| chr7_1196429_1196848 | 0.18 |
ZFAND2A |
zinc finger, AN1-type domain 2A |
730 |
0.57 |
| chr16_66001590_66001741 | 0.18 |
ENSG00000201999 |
. |
334047 |
0.01 |
| chr9_137118247_137118684 | 0.18 |
ENSG00000221676 |
. |
88779 |
0.07 |
| chr3_120217656_120217807 | 0.18 |
FSTL1 |
follistatin-like 1 |
47631 |
0.16 |
| chr5_138350290_138350441 | 0.18 |
CTB-46B19.2 |
|
2026 |
0.27 |
| chr19_1412580_1412731 | 0.18 |
CTB-25B13.13 |
|
1245 |
0.22 |
| chr5_14880723_14880874 | 0.18 |
ANKH |
ANKH inorganic pyrophosphate transport regulator |
8911 |
0.23 |
| chr2_67791651_67791912 | 0.18 |
ETAA1 |
Ewing tumor-associated antigen 1 |
167330 |
0.04 |
| chr1_214666364_214666654 | 0.18 |
PTPN14 |
protein tyrosine phosphatase, non-receptor type 14 |
28363 |
0.24 |
| chr3_30619633_30619797 | 0.18 |
TGFBR2 |
transforming growth factor, beta receptor II (70/80kDa) |
28279 |
0.25 |
| chr2_28914757_28915042 | 0.18 |
ENSG00000222232 |
. |
8057 |
0.18 |
| chr11_10346579_10346730 | 0.18 |
AMPD3 |
adenosine monophosphate deaminase 3 |
7436 |
0.17 |
| chr19_43582973_43583124 | 0.18 |
PSG2 |
pregnancy specific beta-1-glycoprotein 2 |
3489 |
0.28 |
| chr2_191587157_191587308 | 0.18 |
AC006460.2 |
|
13790 |
0.24 |
| chr11_20057587_20057860 | 0.18 |
NAV2 |
neuron navigator 2 |
13087 |
0.19 |
| chr19_46141846_46142168 | 0.18 |
EML2 |
echinoderm microtubule associated protein like 2 |
124 |
0.73 |
| chr3_45719910_45720061 | 0.18 |
LIMD1-AS1 |
LIMD1 antisense RNA 1 |
10389 |
0.18 |
| chr10_3088991_3089142 | 0.18 |
PFKP |
phosphofructokinase, platelet |
19459 |
0.26 |
| chr1_199712128_199712279 | 0.18 |
ENSG00000263805 |
. |
127795 |
0.06 |
| chr1_16466515_16466919 | 0.18 |
RP11-276H7.2 |
|
14989 |
0.11 |
| chr11_1715317_1715638 | 0.18 |
KRTAP5-6 |
keratin associated protein 5-6 |
2948 |
0.16 |
| chr8_57452711_57452862 | 0.18 |
RP11-17A4.2 |
|
51129 |
0.16 |
| chr19_13957292_13957443 | 0.18 |
ENSG00000207980 |
. |
9894 |
0.08 |
| chr19_18385307_18385507 | 0.18 |
KIAA1683 |
KIAA1683 |
88 |
0.94 |
| chr10_21625403_21625600 | 0.18 |
ENSG00000207264 |
. |
14611 |
0.25 |
| chr18_43373207_43373358 | 0.18 |
SIGLEC15 |
sialic acid binding Ig-like lectin 15 |
32195 |
0.15 |
| chr3_44866927_44867078 | 0.18 |
KIF15 |
kinesin family member 15 |
26323 |
0.11 |
| chr2_20793195_20793346 | 0.17 |
HS1BP3-IT1 |
HS1BP3 intronic transcript 1 (non-protein coding) |
962 |
0.61 |
| chr15_65148885_65149111 | 0.17 |
AC069368.3 |
Uncharacterized protein |
14851 |
0.13 |
| chr1_26678105_26678256 | 0.17 |
AIM1L |
absent in melanoma 1-like |
2441 |
0.17 |
| chr7_116963620_116964059 | 0.17 |
WNT2 |
wingless-type MMTV integration site family member 2 |
496 |
0.84 |
| chr12_15789011_15789254 | 0.17 |
EPS8 |
epidermal growth factor receptor pathway substrate 8 |
26076 |
0.19 |
| chr5_77189202_77189495 | 0.17 |
TBCA |
tubulin folding cofactor A |
24744 |
0.26 |
| chr2_110459905_110460056 | 0.17 |
SOWAHC |
sosondowah ankyrin repeat domain family member C |
88069 |
0.08 |
| chr12_1689271_1689422 | 0.17 |
WNT5B |
wingless-type MMTV integration site family, member 5B |
5523 |
0.23 |
| chr22_38177520_38177671 | 0.17 |
ENSG00000238569 |
. |
3938 |
0.13 |
| chr6_11194834_11194985 | 0.17 |
RP3-510L9.1 |
|
21224 |
0.19 |
| chr9_112832863_112833014 | 0.17 |
AKAP2 |
A kinase (PRKA) anchor protein 2 |
19539 |
0.26 |
| chr3_101882560_101882711 | 0.17 |
ZPLD1 |
zona pellucida-like domain containing 1 |
64547 |
0.14 |
| chr18_46351372_46351523 | 0.17 |
RP11-484L8.1 |
|
9694 |
0.27 |
| chr8_102039132_102039343 | 0.17 |
ENSG00000252736 |
. |
63966 |
0.1 |
| chr7_73294883_73295034 | 0.17 |
WBSCR28 |
Williams-Beuren syndrome chromosome region 28 |
19469 |
0.16 |
| chr6_3325224_3325507 | 0.17 |
RP11-506K6.4 |
|
13469 |
0.22 |
| chr1_172320814_172321075 | 0.17 |
ENSG00000252354 |
. |
3661 |
0.22 |
| chr6_109792757_109793021 | 0.17 |
MICAL1 |
microtubule associated monooxygenase, calponin and LIM domain containing 1 |
5718 |
0.12 |
| chr7_43912051_43912202 | 0.17 |
MRPS24 |
mitochondrial ribosomal protein S24 |
2970 |
0.21 |
| chr11_126199011_126199162 | 0.17 |
RP11-712L6.5 |
Uncharacterized protein |
24873 |
0.1 |
| chr20_34055509_34055660 | 0.17 |
CEP250 |
centrosomal protein 250kDa |
5494 |
0.12 |
| chr11_131758279_131758430 | 0.16 |
AP004372.1 |
|
8648 |
0.25 |
| chr8_119113494_119113873 | 0.16 |
EXT1 |
exostosin glycosyltransferase 1 |
8970 |
0.33 |
| chr17_70687944_70688095 | 0.16 |
ENSG00000222545 |
. |
22591 |
0.21 |
| chr15_101611256_101611507 | 0.16 |
RP11-505E24.2 |
|
14890 |
0.21 |
| chr9_137268626_137268825 | 0.16 |
ENSG00000263897 |
. |
2532 |
0.35 |
| chr19_43526713_43526864 | 0.16 |
PSG11 |
pregnancy specific beta-1-glycoprotein 11 |
3812 |
0.27 |
| chr21_45230405_45230556 | 0.16 |
AP001053.11 |
|
238 |
0.92 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0006562 | proline catabolic process(GO:0006562) |
| 0.1 | 0.2 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.1 | 0.2 | GO:0061072 | iris morphogenesis(GO:0061072) |
| 0.0 | 0.1 | GO:0071436 | sodium ion export(GO:0071436) |
| 0.0 | 0.1 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.1 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.0 | 0.1 | GO:0060662 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.0 | 0.2 | GO:0031274 | pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.2 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.1 | GO:0010901 | regulation of very-low-density lipoprotein particle remodeling(GO:0010901) |
| 0.0 | 0.0 | GO:0048669 | collateral sprouting in absence of injury(GO:0048669) |
| 0.0 | 0.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.1 | GO:0005997 | xylulose metabolic process(GO:0005997) |
| 0.0 | 0.1 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.0 | GO:0009169 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.0 | 0.1 | GO:0097061 | dendritic spine morphogenesis(GO:0060997) regulation of dendritic spine morphogenesis(GO:0061001) dendritic spine organization(GO:0097061) |
| 0.0 | 0.1 | GO:0034201 | response to oleic acid(GO:0034201) |
| 0.0 | 0.1 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.1 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.0 | 0.1 | GO:0002513 | tolerance induction to self antigen(GO:0002513) |
| 0.0 | 0.1 | GO:0090047 | obsolete positive regulation of transcription regulator activity(GO:0090047) |
| 0.0 | 0.1 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 0.1 | GO:0009296 | obsolete flagellum assembly(GO:0009296) |
| 0.0 | 0.1 | GO:0045915 | positive regulation of catecholamine metabolic process(GO:0045915) positive regulation of dopamine metabolic process(GO:0045964) |
| 0.0 | 0.1 | GO:0070670 | response to interleukin-4(GO:0070670) |
| 0.0 | 0.1 | GO:0002540 | arachidonic acid metabolite production involved in inflammatory response(GO:0002538) leukotriene production involved in inflammatory response(GO:0002540) |
| 0.0 | 0.1 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 0.0 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.1 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.1 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.0 | 0.1 | GO:0051503 | adenine nucleotide transport(GO:0051503) |
| 0.0 | 0.0 | GO:0071072 | negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 0.0 | 0.1 | GO:0021801 | cerebral cortex radial glia guided migration(GO:0021801) telencephalon glial cell migration(GO:0022030) |
| 0.0 | 0.0 | GO:0044415 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.0 | 0.0 | GO:0006168 | adenine salvage(GO:0006168) purine nucleobase salvage(GO:0043096) adenine metabolic process(GO:0046083) adenine biosynthetic process(GO:0046084) |
| 0.0 | 0.2 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.0 | 0.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.1 | GO:0032717 | negative regulation of interleukin-8 production(GO:0032717) |
| 0.0 | 0.0 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.1 | GO:0014819 | regulation of skeletal muscle contraction(GO:0014819) |
| 0.0 | 0.1 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.0 | 0.0 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.0 | 0.0 | GO:0060242 | contact inhibition(GO:0060242) |
| 0.0 | 0.0 | GO:0039656 | modulation by virus of host gene expression(GO:0039656) |
| 0.0 | 0.1 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) |
| 0.0 | 0.0 | GO:0071830 | chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) |
| 0.0 | 0.1 | GO:0018101 | protein citrullination(GO:0018101) |
| 0.0 | 0.0 | GO:0060831 | smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:0060831) |
| 0.0 | 0.1 | GO:0032469 | endoplasmic reticulum calcium ion homeostasis(GO:0032469) |
| 0.0 | 0.0 | GO:0007132 | meiotic metaphase I(GO:0007132) |
| 0.0 | 0.0 | GO:0030011 | maintenance of cell polarity(GO:0030011) |
| 0.0 | 0.0 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.1 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 0.0 | GO:0000101 | sulfur amino acid transport(GO:0000101) L-cystine transport(GO:0015811) |
| 0.0 | 0.0 | GO:0032455 | nerve growth factor processing(GO:0032455) |
| 0.0 | 0.1 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.0 | GO:0022605 | oogenesis stage(GO:0022605) |
| 0.0 | 0.0 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.0 | GO:0034145 | positive regulation of toll-like receptor 4 signaling pathway(GO:0034145) |
| 0.0 | 0.3 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.0 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.0 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 0.1 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.0 | 0.1 | GO:0008655 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine nucleoside salvage(GO:0043097) |
| 0.0 | 0.0 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.1 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.0 | GO:0072131 | kidney mesenchyme morphogenesis(GO:0072131) metanephric mesenchyme morphogenesis(GO:0072133) |
| 0.0 | 0.0 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.1 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.0 | 0.0 | GO:0010587 | miRNA metabolic process(GO:0010586) miRNA catabolic process(GO:0010587) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.1 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.1 | GO:0031312 | extrinsic component of organelle membrane(GO:0031312) |
| 0.0 | 0.1 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.1 | GO:0098651 | basement membrane collagen trimer(GO:0098651) |
| 0.0 | 0.0 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.1 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 0.0 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.0 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.0 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.1 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.0 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.0 | 0.1 | GO:0036454 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.0 | 0.0 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.0 | GO:0032449 | CBM complex(GO:0032449) |
| 0.0 | 0.1 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.1 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.0 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.0 | 0.0 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.1 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.1 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.1 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.1 | GO:0042627 | chylomicron(GO:0042627) |
| 0.0 | 0.0 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.2 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.0 | 0.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.1 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.1 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.1 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.1 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.1 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.2 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.0 | 0.1 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.1 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.0 | 0.1 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.0 | 0.4 | GO:1901682 | sulfur compound transmembrane transporter activity(GO:1901682) |
| 0.0 | 0.1 | GO:0004515 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.0 | 0.1 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.1 | GO:0051636 | obsolete Gram-negative bacterial cell surface binding(GO:0051636) |
| 0.0 | 0.1 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.0 | 0.1 | GO:0042285 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.1 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.1 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.1 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.0 | 0.1 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.1 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.0 | GO:0019958 | C-X-C chemokine binding(GO:0019958) interleukin-8 binding(GO:0019959) |
| 0.0 | 0.0 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.1 | GO:0003711 | obsolete transcription elongation regulator activity(GO:0003711) |
| 0.0 | 0.0 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.0 | 0.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.1 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.1 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.1 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.0 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.1 | GO:0016717 | oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water(GO:0016717) |
| 0.0 | 0.0 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.1 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.0 | GO:0050542 | icosanoid binding(GO:0050542) fatty acid derivative binding(GO:1901567) |
| 0.0 | 0.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.1 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.0 | 0.2 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.0 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.0 | 0.0 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.0 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.1 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.1 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.0 | 0.1 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.1 | GO:0000832 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.0 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.0 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.2 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.3 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.2 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.2 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.2 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.1 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |