Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
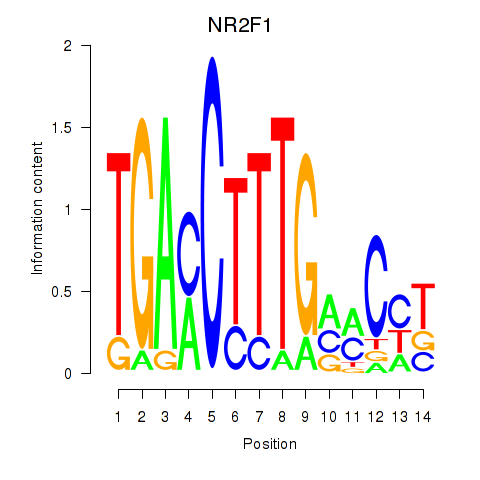
Results for NR2F1
Z-value: 0.80
Transcription factors associated with NR2F1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR2F1
|
ENSG00000175745.7 | nuclear receptor subfamily 2 group F member 1 |
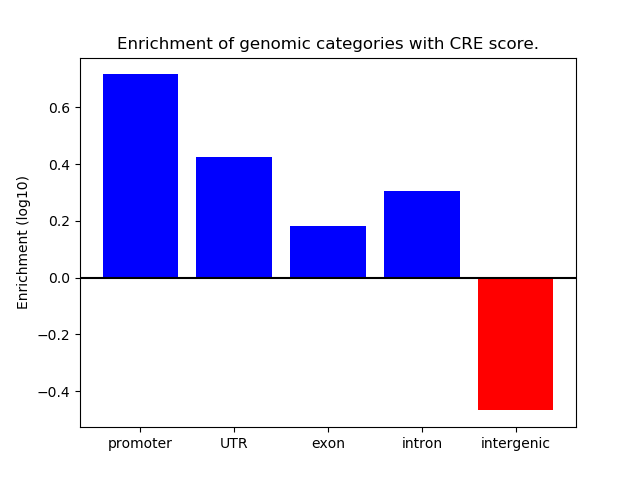
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr5_92919241_92919787 | NR2F1 | 471 | 0.788139 | 0.46 | 2.1e-01 | Click! |
| chr5_92918713_92918890 | NR2F1 | 242 | 0.914673 | 0.06 | 8.7e-01 | Click! |
| chr5_92918371_92918624 | NR2F1 | 546 | 0.718874 | -0.01 | 9.8e-01 | Click! |
Activity of the NR2F1 motif across conditions
Conditions sorted by the z-value of the NR2F1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
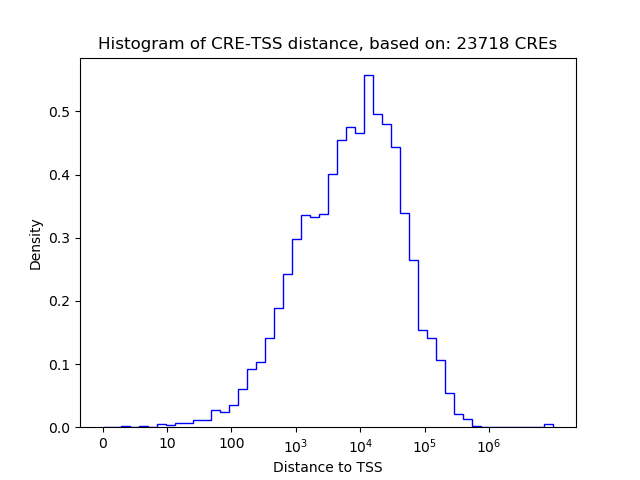
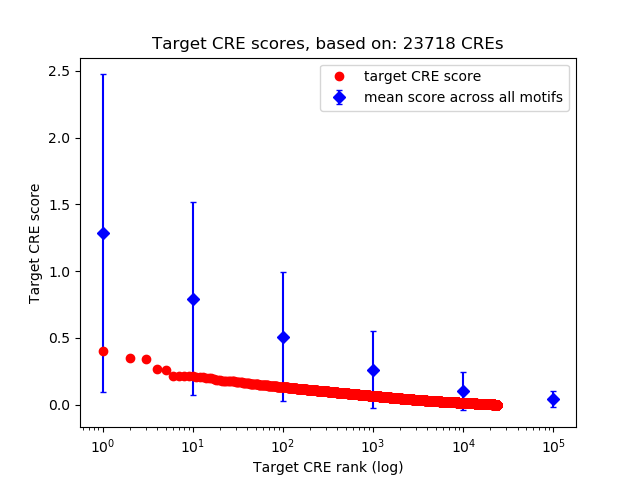
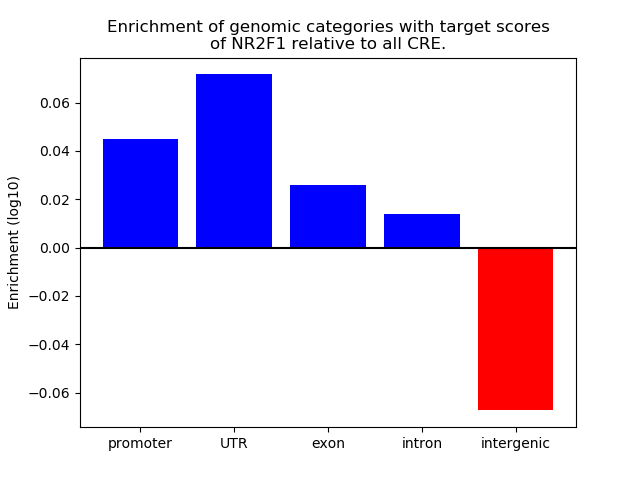
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr19_49099565_49099716 | 0.40 |
SPACA4 |
sperm acrosome associated 4 |
10360 |
0.08 |
| chr14_98635190_98635420 | 0.35 |
ENSG00000222066 |
. |
162782 |
0.04 |
| chr11_3035028_3035343 | 0.34 |
CARS-AS1 |
CARS antisense RNA 1 |
15439 |
0.11 |
| chr6_112348251_112348537 | 0.26 |
WISP3 |
WNT1 inducible signaling pathway protein 3 |
26881 |
0.19 |
| chr19_39369781_39369998 | 0.26 |
RINL |
Ras and Rab interactor-like |
970 |
0.3 |
| chr1_26873022_26873700 | 0.22 |
RPS6KA1 |
ribosomal protein S6 kinase, 90kDa, polypeptide 1 |
1018 |
0.45 |
| chr22_42689321_42689472 | 0.22 |
TCF20 |
transcription factor 20 (AR1) |
50226 |
0.13 |
| chr3_131220905_131221118 | 0.22 |
MRPL3 |
mitochondrial ribosomal protein L3 |
770 |
0.63 |
| chr6_137674077_137674372 | 0.21 |
IFNGR1 |
interferon gamma receptor 1 |
133638 |
0.05 |
| chr1_227949174_227949396 | 0.21 |
SNAP47 |
synaptosomal-associated protein, 47kDa |
13521 |
0.16 |
| chr10_112123592_112123865 | 0.21 |
SMNDC1 |
survival motor neuron domain containing 1 |
59019 |
0.12 |
| chr19_3171495_3171646 | 0.21 |
S1PR4 |
sphingosine-1-phosphate receptor 4 |
7166 |
0.12 |
| chr6_154539920_154540071 | 0.21 |
IPCEF1 |
interaction protein for cytohesin exchange factors 1 |
27995 |
0.26 |
| chr5_74405479_74405669 | 0.20 |
GCNT4 |
glucosaminyl (N-acetyl) transferase 4, core 2 |
78850 |
0.1 |
| chr21_33028994_33029191 | 0.20 |
SOD1 |
superoxide dismutase 1, soluble |
2843 |
0.24 |
| chr16_82686086_82686245 | 0.20 |
CDH13 |
cadherin 13 |
25467 |
0.25 |
| chr16_68786591_68786784 | 0.20 |
ENSG00000200558 |
. |
10304 |
0.16 |
| chr3_111329937_111330088 | 0.19 |
ZBED2 |
zinc finger, BED-type containing 2 |
15722 |
0.2 |
| chr12_76371998_76372996 | 0.19 |
ENSG00000243420 |
. |
20587 |
0.2 |
| chr14_77588217_77588634 | 0.18 |
TMEM63C |
transmembrane protein 63C |
5514 |
0.16 |
| chr15_75069001_75069262 | 0.18 |
CSK |
c-src tyrosine kinase |
5267 |
0.13 |
| chr2_131582514_131582674 | 0.18 |
AC133785.1 |
|
11973 |
0.16 |
| chr19_14141891_14142068 | 0.18 |
CTB-55O6.4 |
|
390 |
0.54 |
| chr3_132377398_132377593 | 0.18 |
UBA5 |
ubiquitin-like modifier activating enzyme 5 |
815 |
0.49 |
| chr13_40779977_40780128 | 0.18 |
ENSG00000207458 |
. |
20912 |
0.28 |
| chr8_129406385_129406536 | 0.17 |
ENSG00000201782 |
. |
173710 |
0.03 |
| chr9_79018760_79018993 | 0.17 |
RFK |
riboflavin kinase |
9455 |
0.24 |
| chr5_35902239_35902390 | 0.17 |
IL7R |
interleukin 7 receptor |
26798 |
0.15 |
| chr1_27675350_27676002 | 0.17 |
SYTL1 |
synaptotagmin-like 1 |
4312 |
0.13 |
| chr3_66427121_66427272 | 0.17 |
ENSG00000241587 |
. |
52917 |
0.15 |
| chr3_111263954_111264147 | 0.17 |
CD96 |
CD96 molecule |
3053 |
0.33 |
| chr11_65324541_65325002 | 0.17 |
LTBP3 |
latent transforming growth factor beta binding protein 3 |
465 |
0.62 |
| chr1_181060923_181061127 | 0.17 |
IER5 |
immediate early response 5 |
3387 |
0.29 |
| chr9_92445809_92446274 | 0.17 |
GADD45G |
growth arrest and DNA-damage-inducible, gamma |
226088 |
0.02 |
| chr2_85050169_85050320 | 0.17 |
TRABD2A |
TraB domain containing 2A |
57962 |
0.11 |
| chrX_48771847_48772324 | 0.17 |
PIM2 |
pim-2 oncogene |
927 |
0.37 |
| chr14_99699518_99699950 | 0.17 |
AL109767.1 |
|
29551 |
0.18 |
| chr19_30328650_30328931 | 0.16 |
CCNE1 |
cyclin E1 |
20680 |
0.25 |
| chr5_142608359_142608828 | 0.16 |
ARHGAP26 |
Rho GTPase activating protein 26 |
21828 |
0.23 |
| chr11_48017026_48017177 | 0.16 |
PTPRJ |
protein tyrosine phosphatase, receptor type, J |
14822 |
0.19 |
| chr22_45065462_45065745 | 0.16 |
PRR5 |
proline rich 5 (renal) |
1010 |
0.59 |
| chr15_60875764_60876131 | 0.16 |
RORA |
RAR-related orphan receptor A |
8793 |
0.23 |
| chr10_121052972_121053123 | 0.16 |
RP11-79M19.2 |
|
42647 |
0.13 |
| chr10_8445974_8446289 | 0.16 |
ENSG00000212505 |
. |
252663 |
0.02 |
| chr20_57732386_57732675 | 0.16 |
ZNF831 |
zinc finger protein 831 |
33545 |
0.19 |
| chr11_35174879_35175121 | 0.16 |
CD44 |
CD44 molecule (Indian blood group) |
14147 |
0.16 |
| chrX_128901533_128901839 | 0.16 |
SASH3 |
SAM and SH3 domain containing 3 |
12274 |
0.19 |
| chr2_238621913_238622064 | 0.16 |
LRRFIP1 |
leucine rich repeat (in FLII) interacting protein 1 |
21006 |
0.21 |
| chr18_20419374_20419602 | 0.16 |
RBBP8 |
retinoblastoma binding protein 8 |
41264 |
0.12 |
| chr8_131299195_131299491 | 0.15 |
ASAP1 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1 |
40055 |
0.21 |
| chr1_111176210_111176587 | 0.15 |
KCNA2 |
potassium voltage-gated channel, shaker-related subfamily, member 2 |
2302 |
0.29 |
| chr19_9968157_9968686 | 0.15 |
OLFM2 |
olfactomedin 2 |
423 |
0.71 |
| chr21_43394304_43394514 | 0.15 |
C2CD2 |
C2 calcium-dependent domain containing 2 |
20410 |
0.17 |
| chr4_114680349_114680500 | 0.15 |
CAMK2D |
calcium/calmodulin-dependent protein kinase II delta |
1800 |
0.52 |
| chr5_75594306_75594457 | 0.15 |
RP11-466P24.6 |
|
12906 |
0.28 |
| chr10_126315256_126315705 | 0.15 |
FAM53B-AS1 |
FAM53B antisense RNA 1 |
76714 |
0.09 |
| chr20_31557861_31558084 | 0.15 |
EFCAB8 |
EF-hand calcium binding domain 8 |
10321 |
0.16 |
| chr2_112941553_112941742 | 0.15 |
FBLN7 |
fibulin 7 |
2187 |
0.37 |
| chr17_35081326_35081477 | 0.15 |
MRM1 |
mitochondrial rRNA methyltransferase 1 homolog (S. cerevisiae) |
123010 |
0.04 |
| chr10_47644509_47644783 | 0.15 |
ANTXRLP1 |
anthrax toxin receptor-like pseudogene 1 |
145 |
0.96 |
| chr11_118892048_118892220 | 0.15 |
ENSG00000266398 |
. |
2480 |
0.09 |
| chr22_31676559_31676796 | 0.15 |
PIK3IP1 |
phosphoinositide-3-kinase interacting protein 1 |
11704 |
0.11 |
| chr17_62184395_62184565 | 0.15 |
ERN1 |
endoplasmic reticulum to nucleus signaling 1 |
23005 |
0.15 |
| chr5_55983263_55983414 | 0.15 |
AC022431.2 |
Homo sapiens uncharacterized LOC101928448 (LOC101928448), mRNA. |
81279 |
0.09 |
| chr12_67919530_67919842 | 0.15 |
DYRK2 |
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 2 |
122432 |
0.06 |
| chr2_235337973_235338124 | 0.15 |
ARL4C |
ADP-ribosylation factor-like 4C |
67196 |
0.14 |
| chr3_67733685_67733836 | 0.14 |
SUCLG2 |
succinate-CoA ligase, GDP-forming, beta subunit |
28722 |
0.26 |
| chr9_134536993_134537144 | 0.14 |
RAPGEF1 |
Rap guanine nucleotide exchange factor (GEF) 1 |
35733 |
0.17 |
| chr19_9967610_9967789 | 0.14 |
OLFM2 |
olfactomedin 2 |
1145 |
0.31 |
| chr1_156088512_156088707 | 0.14 |
LMNA |
lamin A/C |
4096 |
0.13 |
| chr11_68108757_68109054 | 0.14 |
LRP5 |
low density lipoprotein receptor-related protein 5 |
28828 |
0.18 |
| chr2_69766242_69766541 | 0.14 |
ENSG00000207016 |
. |
19085 |
0.19 |
| chr5_86570459_86570610 | 0.14 |
RASA1 |
RAS p21 protein activator (GTPase activating protein) 1 |
5773 |
0.18 |
| chr17_29797345_29797496 | 0.14 |
ENSG00000239595 |
. |
4859 |
0.17 |
| chr15_39933456_39933607 | 0.14 |
FSIP1 |
fibrous sheath interacting protein 1 |
40404 |
0.15 |
| chr19_17111504_17111655 | 0.14 |
CTD-2528A14.1 |
|
8420 |
0.14 |
| chr8_143554044_143554195 | 0.14 |
BAI1 |
brain-specific angiogenesis inhibitor 1 |
8742 |
0.22 |
| chr1_206911756_206912053 | 0.14 |
ENSG00000199349 |
. |
9533 |
0.16 |
| chr10_134351874_134352586 | 0.14 |
INPP5A |
inositol polyphosphate-5-phosphatase, 40kDa |
587 |
0.78 |
| chr13_80594406_80594557 | 0.14 |
SPRY2 |
sprouty homolog 2 (Drosophila) |
319313 |
0.01 |
| chrX_70326040_70326438 | 0.14 |
CXorf65 |
chromosome X open reading frame 65 |
216 |
0.87 |
| chr6_33595892_33596043 | 0.14 |
ITPR3 |
inositol 1,4,5-trisphosphate receptor, type 3 |
6806 |
0.15 |
| chr3_33030408_33030590 | 0.14 |
CCR4 |
chemokine (C-C motif) receptor 4 |
37433 |
0.17 |
| chr5_95874776_95874927 | 0.14 |
CAST |
calpastatin |
9326 |
0.22 |
| chr20_13202929_13203194 | 0.14 |
ISM1 |
isthmin 1, angiogenesis inhibitor |
643 |
0.74 |
| chr14_50411900_50412331 | 0.14 |
ENSG00000251929 |
. |
43447 |
0.1 |
| chr2_219760922_219761155 | 0.14 |
WNT10A |
wingless-type MMTV integration site family, member 10A |
14155 |
0.11 |
| chr14_102297856_102298007 | 0.13 |
CTD-2017C7.1 |
|
7937 |
0.16 |
| chr11_45951042_45951193 | 0.13 |
GYLTL1B |
glycosyltransferase-like 1B |
1530 |
0.28 |
| chr2_196362777_196363039 | 0.13 |
ENSG00000202206 |
. |
15852 |
0.28 |
| chr1_59278295_59278446 | 0.13 |
JUN |
jun proto-oncogene |
28585 |
0.2 |
| chr11_72493247_72493789 | 0.13 |
STARD10 |
StAR-related lipid transfer (START) domain containing 10 |
56 |
0.94 |
| chr10_6555656_6555821 | 0.13 |
PRKCQ |
protein kinase C, theta |
66463 |
0.14 |
| chr1_112145850_112146165 | 0.13 |
RAP1A |
RAP1A, member of RAS oncogene family |
16392 |
0.16 |
| chr14_64883279_64883430 | 0.13 |
CTD-2555O16.4 |
|
25602 |
0.12 |
| chr3_195259224_195259482 | 0.13 |
ENSG00000252620 |
. |
9334 |
0.16 |
| chr20_13211744_13212262 | 0.13 |
ISM1-AS1 |
ISM1 antisense RNA 1 |
8318 |
0.2 |
| chr1_206976194_206976444 | 0.13 |
IL19 |
interleukin 19 |
4104 |
0.2 |
| chr15_42229872_42230035 | 0.13 |
CTD-2382E5.4 |
|
16338 |
0.11 |
| chr1_158899632_158899783 | 0.13 |
PYHIN1 |
pyrin and HIN domain family, member 1 |
879 |
0.64 |
| chr11_73686038_73686668 | 0.13 |
UCP2 |
uncoupling protein 2 (mitochondrial, proton carrier) |
1698 |
0.28 |
| chr15_63898445_63898817 | 0.13 |
USP3-AS1 |
USP3 antisense RNA 1 |
4843 |
0.21 |
| chr2_120988966_120989173 | 0.13 |
TMEM185B |
transmembrane protein 185B |
8085 |
0.17 |
| chrX_39111302_39111453 | 0.13 |
ENSG00000207122 |
. |
170839 |
0.04 |
| chr3_187883613_187883764 | 0.13 |
LPP-AS2 |
LPP antisense RNA 2 |
11843 |
0.19 |
| chr14_97874660_97874811 | 0.13 |
ENSG00000240730 |
. |
121775 |
0.07 |
| chr11_72866229_72866512 | 0.13 |
FCHSD2 |
FCH and double SH3 domains 2 |
13064 |
0.2 |
| chr14_35336861_35337012 | 0.13 |
RP11-73E17.2 |
|
6613 |
0.16 |
| chr3_122292430_122292603 | 0.13 |
PARP15 |
poly (ADP-ribose) polymerase family, member 15 |
3933 |
0.19 |
| chr13_99188287_99188438 | 0.13 |
STK24 |
serine/threonine kinase 24 |
14110 |
0.22 |
| chr6_26275131_26275282 | 0.13 |
HIST1H2BI |
histone cluster 1, H2bi |
2062 |
0.13 |
| chr2_233214354_233214654 | 0.13 |
DIS3L2 |
DIS3 mitotic control homolog (S. cerevisiae)-like 2 |
14009 |
0.14 |
| chr7_150418122_150418385 | 0.13 |
GIMAP1 |
GTPase, IMAP family member 1 |
4608 |
0.18 |
| chr7_105537387_105537614 | 0.13 |
ATXN7L1 |
ataxin 7-like 1 |
20450 |
0.25 |
| chr1_39557935_39558086 | 0.13 |
MACF1 |
microtubule-actin crosslinking factor 1 |
8171 |
0.18 |
| chr21_46505200_46505358 | 0.13 |
PRED57 |
HCG401283; PRED57 protein; Uncharacterized protein |
10214 |
0.14 |
| chr13_114549015_114549308 | 0.13 |
GAS6 |
growth arrest-specific 6 |
895 |
0.65 |
| chr1_158902586_158902737 | 0.13 |
PYHIN1 |
pyrin and HIN domain family, member 1 |
1303 |
0.5 |
| chr1_156697065_156697433 | 0.13 |
ISG20L2 |
interferon stimulated exonuclease gene 20kDa-like 2 |
978 |
0.27 |
| chr11_73079045_73079196 | 0.13 |
RELT |
RELT tumor necrosis factor receptor |
8189 |
0.14 |
| chr1_112146942_112147117 | 0.13 |
RAP1A |
RAP1A, member of RAS oncogene family |
15370 |
0.17 |
| chr3_105542619_105542770 | 0.13 |
CBLB |
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
45193 |
0.21 |
| chr10_5567726_5567877 | 0.13 |
RP11-116G8.5 |
|
96 |
0.8 |
| chr15_40744962_40745272 | 0.13 |
RP11-64K12.9 |
|
1721 |
0.2 |
| chr9_140282226_140282466 | 0.13 |
ENSG00000272272 |
. |
33250 |
0.09 |
| chr14_98675744_98675895 | 0.13 |
ENSG00000222066 |
. |
122268 |
0.06 |
| chr4_40249519_40249765 | 0.13 |
RHOH |
ras homolog family member H |
47678 |
0.14 |
| chr1_115611623_115611842 | 0.13 |
TSPAN2 |
tetraspanin 2 |
20303 |
0.22 |
| chr1_112190409_112190571 | 0.13 |
ENSG00000201028 |
. |
2669 |
0.26 |
| chr2_213844900_213845051 | 0.13 |
AC079610.1 |
|
41294 |
0.18 |
| chr1_203625567_203625718 | 0.13 |
ATP2B4 |
ATPase, Ca++ transporting, plasma membrane 4 |
26295 |
0.19 |
| chr15_70593445_70593646 | 0.13 |
ENSG00000200216 |
. |
107970 |
0.07 |
| chr14_98653738_98654035 | 0.12 |
ENSG00000222066 |
. |
144201 |
0.05 |
| chr9_136002577_136002769 | 0.12 |
RALGDS |
ral guanine nucleotide dissociation stimulator |
2115 |
0.24 |
| chr10_63626765_63626916 | 0.12 |
ARID5B |
AT rich interactive domain 5B (MRF1-like) |
34219 |
0.2 |
| chr22_39492254_39492443 | 0.12 |
APOBEC3H |
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3H |
881 |
0.48 |
| chr2_231501479_231501630 | 0.12 |
ENSG00000199791 |
. |
49718 |
0.12 |
| chr6_106529246_106529540 | 0.12 |
PRDM1 |
PR domain containing 1, with ZNF domain |
4802 |
0.27 |
| chr15_38854255_38854406 | 0.12 |
RASGRP1 |
RAS guanyl releasing protein 1 (calcium and DAG-regulated) |
1850 |
0.35 |
| chr10_82233583_82234488 | 0.12 |
TSPAN14 |
tetraspanin 14 |
14977 |
0.2 |
| chr5_134300560_134300711 | 0.12 |
CATSPER3 |
cation channel, sperm associated 3 |
2961 |
0.24 |
| chr20_37502808_37502959 | 0.12 |
ENSG00000240474 |
. |
1470 |
0.43 |
| chr11_11176629_11176780 | 0.12 |
RP11-567I13.1 |
|
197269 |
0.03 |
| chr12_47615301_47615606 | 0.12 |
PCED1B |
PC-esterase domain containing 1B |
1831 |
0.39 |
| chr2_235330726_235331264 | 0.12 |
ARL4C |
ADP-ribosylation factor-like 4C |
74249 |
0.13 |
| chr8_100646172_100646444 | 0.12 |
ENSG00000243254 |
. |
20002 |
0.22 |
| chr10_33401857_33402306 | 0.12 |
ENSG00000263576 |
. |
14517 |
0.23 |
| chr19_18617770_18618877 | 0.12 |
ELL |
elongation factor RNA polymerase II |
14614 |
0.09 |
| chr7_112093917_112094112 | 0.12 |
IFRD1 |
interferon-related developmental regulator 1 |
1542 |
0.44 |
| chr20_4670873_4671024 | 0.12 |
PRNP |
prion protein |
3759 |
0.26 |
| chr7_110669949_110670394 | 0.12 |
LRRN3 |
leucine rich repeat neuronal 3 |
60891 |
0.14 |
| chr2_30457620_30458309 | 0.12 |
LBH |
limb bud and heart development |
2918 |
0.32 |
| chr9_77766797_77767137 | 0.12 |
ENSG00000200041 |
. |
29570 |
0.16 |
| chr13_97867179_97867538 | 0.12 |
MBNL2 |
muscleblind-like splicing regulator 2 |
6330 |
0.31 |
| chr6_130460805_130460956 | 0.12 |
RP11-73O6.3 |
|
1539 |
0.47 |
| chr20_37433246_37433545 | 0.12 |
PPP1R16B |
protein phosphatase 1, regulatory subunit 16B |
953 |
0.57 |
| chr14_22849174_22849325 | 0.12 |
ENSG00000251002 |
. |
52470 |
0.09 |
| chr19_17907446_17907815 | 0.12 |
B3GNT3 |
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 3 |
1664 |
0.26 |
| chr12_6977477_6978263 | 0.12 |
TPI1 |
triosephosphate isomerase 1 |
237 |
0.79 |
| chr7_150671273_150671529 | 0.12 |
KCNH2 |
potassium voltage-gated channel, subfamily H (eag-related), member 2 |
3613 |
0.16 |
| chr3_59438655_59438806 | 0.12 |
C3orf67 |
chromosome 3 open reading frame 67 |
402920 |
0.01 |
| chr8_59170060_59170211 | 0.12 |
UBXN2B |
UBX domain protein 2B |
153688 |
0.04 |
| chr21_45341287_45341484 | 0.12 |
ENSG00000199598 |
. |
1603 |
0.35 |
| chr6_16478449_16478600 | 0.12 |
ENSG00000265642 |
. |
49770 |
0.18 |
| chr16_9192547_9192808 | 0.12 |
RP11-473I1.9 |
|
6028 |
0.16 |
| chr8_128222716_128223004 | 0.12 |
POU5F1B |
POU class 5 homeobox 1B |
203675 |
0.03 |
| chr2_96829307_96829548 | 0.12 |
DUSP2 |
dual specificity phosphatase 2 |
18248 |
0.13 |
| chr16_25124532_25124693 | 0.12 |
LCMT1 |
leucine carboxyl methyltransferase 1 |
1359 |
0.47 |
| chr5_133467358_133467605 | 0.12 |
TCF7 |
transcription factor 7 (T-cell specific, HMG-box) |
8172 |
0.21 |
| chr18_21592600_21592808 | 0.12 |
TTC39C |
tetratricopeptide repeat domain 39C |
1680 |
0.32 |
| chr10_13149653_13149982 | 0.12 |
OPTN |
optineurin |
7592 |
0.2 |
| chr11_5839481_5839676 | 0.12 |
OR52N2 |
olfactory receptor, family 52, subfamily N, member 2 |
1966 |
0.21 |
| chr14_98657122_98657458 | 0.12 |
ENSG00000222066 |
. |
140797 |
0.05 |
| chr2_111622829_111623021 | 0.12 |
ACOXL |
acyl-CoA oxidase-like |
60029 |
0.15 |
| chr19_18334921_18335258 | 0.12 |
PDE4C |
phosphodiesterase 4C, cAMP-specific |
214 |
0.88 |
| chr14_22749602_22749753 | 0.12 |
ENSG00000238634 |
. |
138790 |
0.04 |
| chr8_41814405_41814794 | 0.12 |
KAT6A |
K(lysine) acetyltransferase 6A |
1282 |
0.52 |
| chr21_43894177_43894328 | 0.11 |
SLC37A1 |
solute carrier family 37 (glucose-6-phosphate transporter), member 1 |
21876 |
0.11 |
| chr4_141279191_141279342 | 0.11 |
SCOC |
short coiled-coil protein |
14511 |
0.19 |
| chr6_167534419_167534775 | 0.11 |
CCR6 |
chemokine (C-C motif) receptor 6 |
1660 |
0.37 |
| chr7_12770799_12771102 | 0.11 |
ENSG00000199470 |
. |
30436 |
0.2 |
| chr7_2755094_2755286 | 0.11 |
AMZ1 |
archaelysin family metallopeptidase 1 |
27354 |
0.17 |
| chr2_240673435_240673899 | 0.11 |
AC093802.1 |
Uncharacterized protein |
10887 |
0.29 |
| chr13_21402931_21403082 | 0.11 |
N6AMT2 |
N-6 adenine-specific DNA methyltransferase 2 (putative) |
54918 |
0.12 |
| chr17_60548755_60548906 | 0.11 |
TLK2 |
tousled-like kinase 2 |
527 |
0.8 |
| chr6_16480859_16481010 | 0.11 |
ENSG00000265642 |
. |
52180 |
0.18 |
| chr14_66403868_66404061 | 0.11 |
CTD-2014B16.3 |
Uncharacterized protein |
67277 |
0.13 |
| chr13_52527282_52527433 | 0.11 |
ATP7B |
ATPase, Cu++ transporting, beta polypeptide |
8694 |
0.22 |
| chr20_49543758_49543983 | 0.11 |
ADNP |
activity-dependent neuroprotector homeobox |
1483 |
0.3 |
| chr19_42396817_42397275 | 0.11 |
ARHGEF1 |
Rho guanine nucleotide exchange factor (GEF) 1 |
1319 |
0.31 |
| chr8_97295512_97295663 | 0.11 |
PTDSS1 |
phosphatidylserine synthase 1 |
11833 |
0.16 |
| chr20_30973001_30973152 | 0.11 |
ASXL1 |
additional sex combs like 1 (Drosophila) |
25526 |
0.17 |
| chr5_176451687_176451930 | 0.11 |
ZNF346 |
zinc finger protein 346 |
2072 |
0.25 |
| chr14_53620332_53620723 | 0.11 |
RP11-547D23.1 |
|
455 |
0.42 |
| chr12_107248186_107248337 | 0.11 |
RP11-144F15.1 |
Uncharacterized protein |
79565 |
0.08 |
| chr15_78834636_78834986 | 0.11 |
PSMA4 |
proteasome (prosome, macropain) subunit, alpha type, 4 |
779 |
0.51 |
| chr13_99938059_99938240 | 0.11 |
GPR183 |
G protein-coupled receptor 183 |
21510 |
0.17 |
| chr9_79791700_79791851 | 0.11 |
VPS13A |
vacuolar protein sorting 13 homolog A (S. cerevisiae) |
494 |
0.73 |
| chr20_62600239_62600795 | 0.11 |
ZNF512B |
zinc finger protein 512B |
701 |
0.44 |
| chr17_45637983_45638134 | 0.11 |
ENSG00000206734 |
. |
3279 |
0.22 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0051834 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.1 | 0.2 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.2 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.1 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 0.1 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.0 | 0.1 | GO:0070266 | necroptotic process(GO:0070266) programmed necrotic cell death(GO:0097300) |
| 0.0 | 0.1 | GO:0071569 | protein ufmylation(GO:0071569) |
| 0.0 | 0.1 | GO:0002874 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) |
| 0.0 | 0.1 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) |
| 0.0 | 0.1 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.1 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.0 | 0.1 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.0 | 0.1 | GO:0001911 | negative regulation of leukocyte mediated cytotoxicity(GO:0001911) |
| 0.0 | 0.1 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.1 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.0 | 0.1 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.0 | 0.1 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.0 | 0.0 | GO:0071803 | regulation of podosome assembly(GO:0071801) positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.2 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.2 | GO:0050860 | negative regulation of antigen receptor-mediated signaling pathway(GO:0050858) negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.1 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 0.2 | GO:0045066 | regulatory T cell differentiation(GO:0045066) |
| 0.0 | 0.1 | GO:0052251 | induction by organism of defense response of other organism involved in symbiotic interaction(GO:0052251) |
| 0.0 | 0.1 | GO:0097709 | connective tissue replacement involved in inflammatory response wound healing(GO:0002248) connective tissue replacement(GO:0097709) |
| 0.0 | 0.1 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.0 | 0.1 | GO:0032621 | interleukin-18 production(GO:0032621) regulation of interleukin-18 production(GO:0032661) |
| 0.0 | 0.1 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.0 | 0.1 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.0 | 0.1 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.0 | 0.0 | GO:1904019 | endothelial cell apoptotic process(GO:0072577) epithelial cell apoptotic process(GO:1904019) regulation of epithelial cell apoptotic process(GO:1904035) regulation of endothelial cell apoptotic process(GO:2000351) |
| 0.0 | 0.1 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 0.0 | 0.2 | GO:0048843 | negative regulation of axon extension involved in axon guidance(GO:0048843) |
| 0.0 | 0.1 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.1 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 0.1 | GO:0048302 | regulation of isotype switching to IgG isotypes(GO:0048302) |
| 0.0 | 0.0 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.0 | 0.0 | GO:0032695 | negative regulation of interleukin-12 production(GO:0032695) |
| 0.0 | 0.1 | GO:0002507 | tolerance induction(GO:0002507) |
| 0.0 | 0.1 | GO:0060340 | positive regulation of type I interferon-mediated signaling pathway(GO:0060340) |
| 0.0 | 0.1 | GO:0010957 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.0 | 0.1 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 0.2 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.0 | 0.1 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.1 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.1 | GO:0016553 | base conversion or substitution editing(GO:0016553) |
| 0.0 | 0.1 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) negative regulation of signal transduction by p53 class mediator(GO:1901797) |
| 0.0 | 0.0 | GO:0034122 | negative regulation of toll-like receptor signaling pathway(GO:0034122) |
| 0.0 | 0.1 | GO:0090036 | regulation of protein kinase C signaling(GO:0090036) positive regulation of protein kinase C signaling(GO:0090037) |
| 0.0 | 0.0 | GO:0051571 | positive regulation of histone H3-K4 methylation(GO:0051571) |
| 0.0 | 0.0 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.0 | 0.1 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.1 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.0 | 0.1 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.0 | 0.0 | GO:1901419 | regulation of response to alcohol(GO:1901419) |
| 0.0 | 0.1 | GO:0046185 | aldehyde catabolic process(GO:0046185) |
| 0.0 | 0.0 | GO:0048537 | mucosal-associated lymphoid tissue development(GO:0048537) |
| 0.0 | 0.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.1 | GO:0060836 | lymphatic endothelial cell differentiation(GO:0060836) |
| 0.0 | 0.1 | GO:0042091 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.0 | 0.1 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.0 | 0.1 | GO:0045046 | protein import into peroxisome membrane(GO:0045046) |
| 0.0 | 0.1 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.0 | 0.1 | GO:0060206 | estrous cycle phase(GO:0060206) |
| 0.0 | 0.1 | GO:0070670 | response to interleukin-4(GO:0070670) |
| 0.0 | 0.1 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.0 | 0.2 | GO:0000303 | response to superoxide(GO:0000303) |
| 0.0 | 0.1 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.0 | 0.0 | GO:0002693 | positive regulation of cellular extravasation(GO:0002693) |
| 0.0 | 0.0 | GO:0000042 | protein targeting to Golgi(GO:0000042) establishment of protein localization to Golgi(GO:0072600) |
| 0.0 | 0.0 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.0 | 0.1 | GO:0009103 | lipopolysaccharide biosynthetic process(GO:0009103) |
| 0.0 | 0.0 | GO:0061526 | acetylcholine secretion, neurotransmission(GO:0014055) regulation of acetylcholine secretion, neurotransmission(GO:0014056) acetylcholine secretion(GO:0061526) |
| 0.0 | 0.1 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) |
| 0.0 | 0.0 | GO:0072217 | negative regulation of metanephros development(GO:0072217) |
| 0.0 | 0.1 | GO:0045842 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.0 | 0.1 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.0 | 0.0 | GO:0070602 | centromeric sister chromatid cohesion(GO:0070601) regulation of centromeric sister chromatid cohesion(GO:0070602) |
| 0.0 | 0.0 | GO:0016242 | regulation of macroautophagy(GO:0016241) negative regulation of macroautophagy(GO:0016242) |
| 0.0 | 0.0 | GO:0039656 | modulation by virus of host gene expression(GO:0039656) |
| 0.0 | 0.0 | GO:0046940 | nucleoside monophosphate phosphorylation(GO:0046940) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.1 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.0 | 0.1 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.0 | GO:0002823 | negative regulation of adaptive immune response(GO:0002820) negative regulation of adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains(GO:0002823) |
| 0.0 | 0.2 | GO:0048024 | regulation of mRNA splicing, via spliceosome(GO:0048024) |
| 0.0 | 0.1 | GO:0060291 | long-term synaptic potentiation(GO:0060291) |
| 0.0 | 0.0 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.0 | GO:0046719 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) regulation by virus of viral protein levels in host cell(GO:0046719) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.0 | 0.1 | GO:0043368 | positive T cell selection(GO:0043368) |
| 0.0 | 0.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.0 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) myosin filament assembly(GO:0031034) striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.0 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.0 | GO:0060242 | contact inhibition(GO:0060242) |
| 0.0 | 0.1 | GO:0000768 | syncytium formation by plasma membrane fusion(GO:0000768) myoblast fusion(GO:0007520) |
| 0.0 | 0.0 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.1 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 0.1 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.0 | GO:0070561 | vitamin D receptor signaling pathway(GO:0070561) |
| 0.0 | 0.0 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.0 | 0.0 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.0 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.1 | GO:0060534 | trachea cartilage development(GO:0060534) |
| 0.0 | 0.0 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.1 | GO:0050687 | negative regulation of defense response to virus(GO:0050687) |
| 0.0 | 0.1 | GO:0015780 | nucleotide-sugar transport(GO:0015780) |
| 0.0 | 0.1 | GO:0002551 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 0.0 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.0 | GO:0042816 | pyridoxine metabolic process(GO:0008614) pyridoxine biosynthetic process(GO:0008615) vitamin B6 metabolic process(GO:0042816) vitamin B6 biosynthetic process(GO:0042819) |
| 0.0 | 0.1 | GO:0009109 | coenzyme catabolic process(GO:0009109) |
| 0.0 | 0.1 | GO:0043267 | negative regulation of potassium ion transport(GO:0043267) |
| 0.0 | 0.0 | GO:0015917 | aminophospholipid transport(GO:0015917) |
| 0.0 | 0.0 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.1 | GO:0042023 | regulation of DNA endoreduplication(GO:0032875) negative regulation of DNA endoreduplication(GO:0032876) DNA endoreduplication(GO:0042023) |
| 0.0 | 0.0 | GO:0032878 | regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.0 | 0.0 | GO:0060413 | atrial septum development(GO:0003283) atrial septum morphogenesis(GO:0060413) |
| 0.0 | 0.0 | GO:0021780 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) oligodendrocyte cell fate specification(GO:0021778) glial cell fate specification(GO:0021780) |
| 0.0 | 0.0 | GO:0033033 | negative regulation of myeloid cell apoptotic process(GO:0033033) |
| 0.0 | 0.0 | GO:2000649 | regulation of sodium:proton antiporter activity(GO:0032415) regulation of sodium ion transmembrane transport(GO:1902305) regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.0 | 0.0 | GO:0031507 | heterochromatin assembly(GO:0031507) heterochromatin organization(GO:0070828) |
| 0.0 | 0.1 | GO:0098927 | early endosome to late endosome transport(GO:0045022) vesicle-mediated transport between endosomal compartments(GO:0098927) |
| 0.0 | 0.2 | GO:1903076 | regulation of establishment of protein localization to plasma membrane(GO:0090003) regulation of protein localization to plasma membrane(GO:1903076) regulation of protein localization to cell periphery(GO:1904375) |
| 0.0 | 0.1 | GO:0002483 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 0.0 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 | 0.0 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.0 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.0 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.0 | 0.1 | GO:0060896 | neural plate pattern specification(GO:0060896) |
| 0.0 | 0.0 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.0 | GO:0046502 | uroporphyrinogen III metabolic process(GO:0046502) |
| 0.0 | 0.0 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.2 | GO:0034199 | activation of protein kinase A activity(GO:0034199) |
| 0.0 | 0.1 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.0 | GO:0035268 | protein mannosylation(GO:0035268) protein O-linked mannosylation(GO:0035269) mannosylation(GO:0097502) |
| 0.0 | 0.0 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 0.1 | GO:0006528 | asparagine metabolic process(GO:0006528) |
| 0.0 | 0.1 | GO:0070170 | regulation of tooth mineralization(GO:0070170) |
| 0.0 | 0.0 | GO:0014052 | gamma-aminobutyric acid secretion(GO:0014051) regulation of gamma-aminobutyric acid secretion(GO:0014052) |
| 0.0 | 0.0 | GO:0007100 | mitotic centrosome separation(GO:0007100) |
| 0.0 | 0.0 | GO:0050651 | dermatan sulfate proteoglycan biosynthetic process(GO:0050651) |
| 0.0 | 0.1 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.0 | 0.0 | GO:0060972 | left/right pattern formation(GO:0060972) |
| 0.0 | 0.0 | GO:0014721 | voluntary skeletal muscle contraction(GO:0003010) twitch skeletal muscle contraction(GO:0014721) |
| 0.0 | 0.0 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.0 | 0.1 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.0 | 0.0 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.0 | 0.1 | GO:0005954 | calcium- and calmodulin-dependent protein kinase complex(GO:0005954) |
| 0.0 | 0.1 | GO:0001652 | granular component(GO:0001652) |
| 0.0 | 0.1 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.1 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.1 | GO:0032449 | CBM complex(GO:0032449) |
| 0.0 | 0.1 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.0 | 0.1 | GO:0031313 | extrinsic component of vacuolar membrane(GO:0000306) extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.1 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.2 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.1 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.1 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.0 | 0.1 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.1 | GO:0000791 | euchromatin(GO:0000791) |
| 0.0 | 0.1 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.1 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.1 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.1 | GO:0070688 | MLL5-L complex(GO:0070688) |
| 0.0 | 0.0 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.1 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 0.3 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.2 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.0 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.0 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.1 | GO:0000300 | obsolete peripheral to membrane of membrane fraction(GO:0000300) |
| 0.0 | 0.1 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.0 | GO:0031312 | extrinsic component of organelle membrane(GO:0031312) |
| 0.0 | 0.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.0 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.0 | GO:0044462 | external encapsulating structure part(GO:0044462) |
| 0.0 | 0.1 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.3 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.1 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.2 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.0 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.0 | 0.1 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.1 | GO:0045261 | proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 0.1 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.0 | GO:1903561 | extracellular exosome(GO:0070062) extracellular vesicle(GO:1903561) |
| 0.0 | 0.0 | GO:0031501 | mannosyltransferase complex(GO:0031501) dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 0.0 | GO:0071942 | XPC complex(GO:0071942) |
| 0.0 | 0.0 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 0.0 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.0 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.0 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.1 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.0 | 0.2 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.0 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.1 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.0 | 0.1 | GO:0010340 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 0.0 | 0.1 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) inositol-1,3,4,5-tetrakisphosphate 5-phosphatase activity(GO:0052659) |
| 0.0 | 0.1 | GO:0031177 | phosphopantetheine binding(GO:0031177) |
| 0.0 | 0.2 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.1 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.1 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.4 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.1 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) |
| 0.0 | 0.1 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.0 | 0.1 | GO:0015925 | galactosidase activity(GO:0015925) |
| 0.0 | 0.2 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.2 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.1 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.1 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.1 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.1 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.1 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.3 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.1 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.1 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.1 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.1 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.0 | GO:0050698 | proteoglycan sulfotransferase activity(GO:0050698) |
| 0.0 | 0.0 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.0 | 0.0 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.0 | 0.2 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.0 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.1 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.0 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.1 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.0 | GO:0043175 | RNA polymerase core enzyme binding(GO:0043175) |
| 0.0 | 0.1 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.2 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.2 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.1 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.1 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.0 | 0.1 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.0 | 0.1 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.0 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.1 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.0 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.0 | 0.1 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.1 | GO:0051766 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) inositol trisphosphate kinase activity(GO:0051766) |
| 0.0 | 0.1 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.0 | 0.0 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.1 | GO:0034593 | phosphatidylinositol bisphosphate phosphatase activity(GO:0034593) |
| 0.0 | 0.0 | GO:0005346 | purine ribonucleotide transmembrane transporter activity(GO:0005346) |
| 0.0 | 0.0 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 0.1 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.0 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.1 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.0 | GO:0004308 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) |
| 0.0 | 0.0 | GO:0005072 | transforming growth factor beta receptor, cytoplasmic mediator activity(GO:0005072) |
| 0.0 | 0.3 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.0 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.0 | 0.0 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.0 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) |
| 0.0 | 0.0 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.0 | 0.0 | GO:0005338 | nucleotide-sugar transmembrane transporter activity(GO:0005338) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.1 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.1 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.1 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.0 | GO:0043047 | single-stranded telomeric DNA binding(GO:0043047) sequence-specific single stranded DNA binding(GO:0098847) |
| 0.0 | 0.1 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 0.0 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.0 | 0.0 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.0 | 0.0 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.0 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.0 | 0.0 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.1 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.0 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.0 | 0.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 0.6 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.0 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.1 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.3 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.4 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.4 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.6 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.1 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.1 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.1 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 0.1 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.5 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.1 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 0.2 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.1 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.3 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.3 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.0 | REACTOME CIRCADIAN CLOCK | Genes involved in Circadian Clock |
| 0.0 | 0.4 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.0 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.3 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.0 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.1 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.0 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.2 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.3 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.1 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.2 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.6 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.1 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.2 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.4 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.0 | 0.2 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.2 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.0 | 0.6 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.1 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.0 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.0 | 0.0 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.0 | 0.1 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |